Ataxia-Telangiectasia Mutated Loss of Heterozygosity in Melanoma
Abstract
1. Introduction
2. Results
2.1. Classification and Distribution of Germline ATM Variants
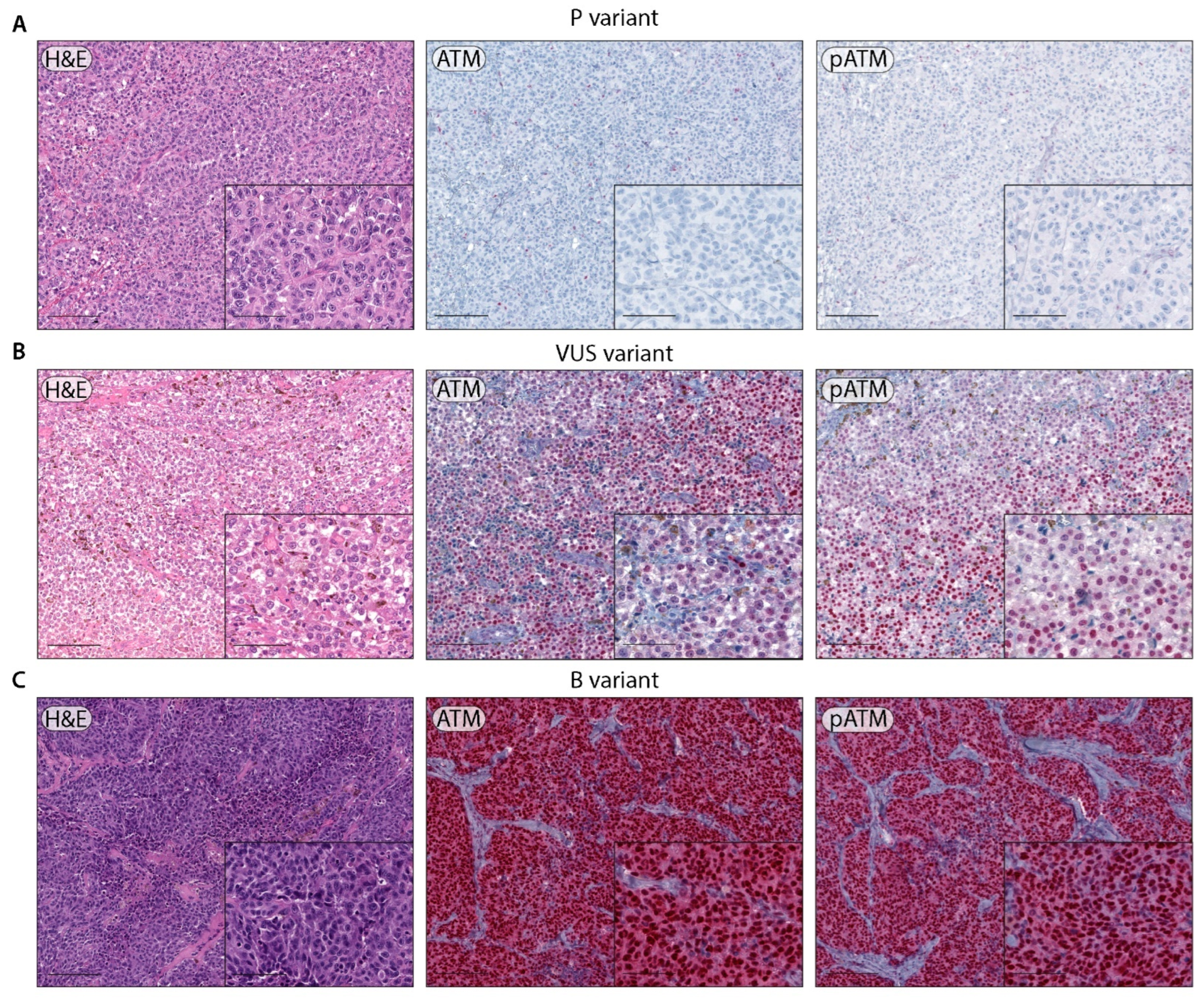
2.2. ATM Immunohistochemistry and LOH Assessment
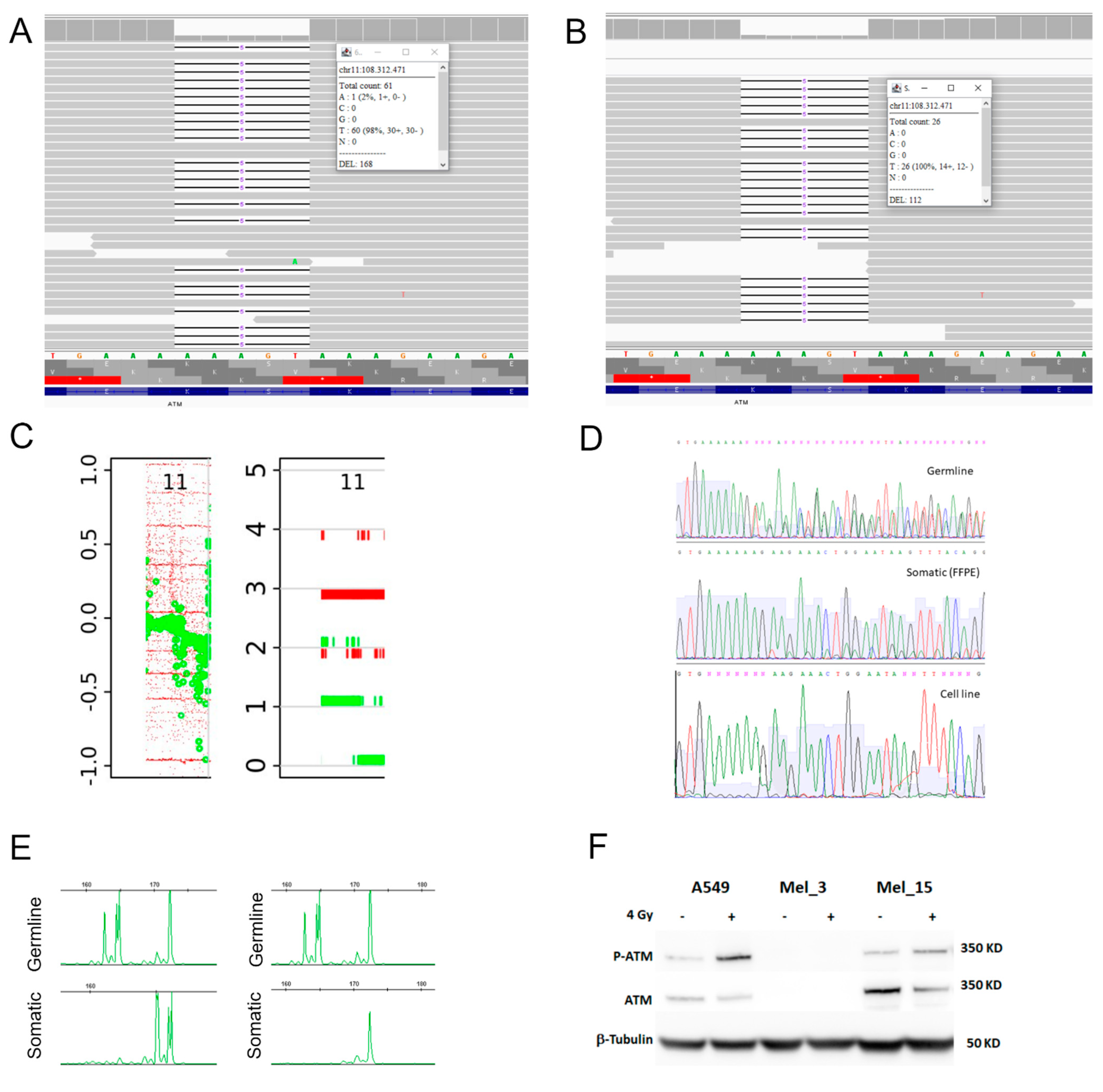
2.3. Western Blotting, DNA, and RNA Sequencing in Melanoma Tissue and Patient-Derived Melanoma Cell Lines
3. Discussion
4. Materials and Methods
4.1. Selection of ATM Variant Carriers and Their Tumors
4.2. Establishment of Melanoma Cell Lines
4.3. Irradiation Conditions
4.4. Immunohistochemistry of Clinical Specimens and Analysis of ATM and P-ATM Staining
4.5. DNA Extraction
4.6. Sanger Sequencing
4.7. Whole-Exome Sequencing (WES) and Copy Number Variation (CNV) Analysis
4.8. RNA Sequencing
4.9. LOH Status by Microsatellite Analysis
4.10. Western Blotting
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shiloh, Y. ATM and Related Protein Kinases: Safeguarding Genome Integrity. Nat. Rev. Cancer 2003, 3, 155–168. [Google Scholar] [CrossRef] [PubMed]
- Putti, S.; Giovinazzo, A.; Merolle, M.; Falchetti, M.L.; Pellegrini, M. ATM Kinase Dead: From Ataxia Telangiectasia Syndrome to Cancer. Cancers 2021, 13, 5498. [Google Scholar] [CrossRef] [PubMed]
- Breast Cancer Association Consortium; Dorling, L.; Carvalho, S.; Allen, J.; González-Neira, A.; Luccarini, C.; Wahlström, C.; Pooley, K.A.; Parsons, M.T.; Fortuno, C.; et al. Breast Cancer Risk Genes—Association Analysis in More than 113,000 Women. N. Engl. J. Med. 2021, 384, 428–439. [Google Scholar] [CrossRef] [PubMed]
- Lesueur, F.; Easton, D.F.; Renault, A.-L.; Tavtigian, S.V.; Bernstein, J.L.; Kote-Jarai, Z.; Eeles, R.A.; Plaseska-Karanfia, D.; Feliubadaló, L.; Spanish ATM working group; et al. First International Workshop of the ATM and Cancer Risk Group (4–5 December 2019). Fam. Cancer 2022, 21, 211–227. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.H.; Iles, M.M.; Harland, M.; Taylor, J.C.; Aitken, J.F.; Andresen, P.A.; Akslen, L.A.; Armstrong, B.K.; Avril, M.-F.; Azizi, E.; et al. Genome-Wide Association Study Identifies Three New Melanoma Susceptibility Loci. Nat. Genet. 2011, 43, 1108–1113. [Google Scholar] [CrossRef] [PubMed]
- Landi, M.T.; Bishop, D.T.; MacGregor, S.; Machiela, M.J.; Stratigos, A.J.; Ghiorzo, P.; Brossard, M.; Calista, D.; Choi, J.; Fargnoli, M.C.; et al. Genome-Wide Association Meta-Analyses Combining Multiple Risk Phenotypes Provide Insights into the Genetic Architecture of Cutaneous Melanoma Susceptibility. Nat. Genet. 2020, 52, 494–504. [Google Scholar] [CrossRef] [PubMed]
- Dalmasso, B.; Pastorino, L.; Nathan, V.; Shah, N.N.; Palmer, J.M.; Howlie, M.; Johansson, P.A.; Freedman, N.D.; Carter, B.D.; Beane-Freeman, L.; et al. Germline ATM Variants Predispose to Melanoma: A Joint Analysis across the GenoMEL and MelaNostrum Consortia. Genet. Med. 2021, 23, 2087–2095. [Google Scholar] [CrossRef]
- Laake, K.; Odegård, A.; Andersen, T.I.; Bukholm, I.K.; Kåresen, R.; Nesland, J.M.; Ottestad, L.; Shiloh, Y.; Børresen-Dale, A.L. Loss of Heterozygosity at 11q23.1 in Breast Carcinomas: Indication for Involvement of a Gene Distal and Close to ATM. Genes Chromosomes Cancer 1997, 18, 175–180. [Google Scholar] [CrossRef]
- Renault, A.-L.; Mebirouk, N.; Fuhrmann, L.; Bataillon, G.; Cavaciuti, E.; Le Gal, D.; Girard, E.; Popova, T.; La Rosa, P.; Beauvallet, J.; et al. Morphology and Genomic Hallmarks of Breast Tumours Developed by ATM Deleterious Variant Carriers. Breast Cancer Res. 2018, 20, 28. [Google Scholar] [CrossRef]
- Bhandaru, M.; Martinka, M.; McElwee, K.J.; Rotte, A. Prognostic Significance of Nuclear Phospho-ATM Expression in Melanoma. PLoS ONE 2015, 10, e0134678. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Pastorino, L.; Andreotti, V.; Dalmasso, B.; Vanni, I.; Ciccarese, G.; Mandalà, M.; Spadola, G.; Pizzichetta, M.A.; Ponti, G.; Tibiletti, M.G.; et al. Insights into Genetic Susceptibility to Melanoma by Gene Panel Testing: Potential Pathogenic Variants in ACD, ATM, BAP1, and POT1. Cancers 2020, 12, 1007. [Google Scholar] [CrossRef] [PubMed]
- Holeckova, K.; Baluchova, K.; Hives, M.; Musak, L.; Kliment, J.; Skerenova, M. Germline Mutations in DNA Repair Genes in Patients With Metastatic Castration-Resistant Prostate Cancer. Vivo 2020, 34, 1773–1778. [Google Scholar] [CrossRef] [PubMed]
- Cavalieri, S.; Funaro, A.; Porcedda, P.; Turinetto, V.; Migone, N.; Gatti, R.A.; Brusco, A. ATM Mutations in Italian Families with Ataxia Telangiectasia Include Two Distinct Large Genomic Deletions. Hum. Mutat. 2006, 27, 1061. [Google Scholar] [CrossRef] [PubMed]
- Carranza, D.; Vega, A.K.; Torres-Rusillo, S.; Montero, E.; Martinez, L.J.; Santamaría, M.; Santos, J.L.; Molina, I.J. Molecular and Functional Characterization of a Cohort of Spanish Patients with Ataxia-Telangiectasia. Neuromolecular. Med. 2017, 19, 161–174. [Google Scholar] [CrossRef]
- Fostira, F.; Kostantopoulou, I.; Apostolou, P.; Papamentzelopoulou, M.S.; Papadimitriou, C.; Faliakou, E.; Christodoulou, C.; Boukovinas, I.; Razis, E.; Tryfonopoulos, D.; et al. One in Three Highly Selected Greek Patients with Breast Cancer Carries a Loss-of-Function Variant in a Cancer Susceptibility Gene. J. Med. Genet. 2020, 57, 53–61. [Google Scholar] [CrossRef]
- Decker, B.; Allen, J.; Luccarini, C.; Pooley, K.A.; Shah, M.; Bolla, M.K.; Wang, Q.; Ahmed, S.; Baynes, C.; Conroy, D.M.; et al. Rare, Protein-Truncating Variants in ATM, CHEK2 and PALB2, but Not XRCC2, Are Associated with Increased Breast Cancer Risks. J. Med. Genet. 2017, 54, 732–741. [Google Scholar] [CrossRef]
- Teraoka, S.N.; Malone, K.E.; Doody, D.R.; Suter, N.M.; Ostrander, E.A.; Daling, J.R.; Concannon, P. Increased Frequency of ATM Mutations in Breast Carcinoma Patients with Early Onset Disease and Positive Family History. Cancer 2001, 92, 479–487. [Google Scholar] [CrossRef]
- Feliubadaló, L.; Moles-Fernández, A.; Santamariña-Pena, M.; Sánchez, A.T.; López-Novo, A.; Porras, L.-M.; Blanco, A.; Capellá, G.; de la Hoya, M.; Molina, I.J.; et al. A Collaborative Effort to Define Classification Criteria for ATM Variants in Hereditary Cancer Patients. Clin. Chem. 2021, 67, 518–533. [Google Scholar] [CrossRef]
- Thorstenson, Y.R.; Roxas, A.; Kroiss, R.; Jenkins, M.A.; Yu, K.M.; Bachrich, T.; Muhr, D.; Wayne, T.L.; Chu, G.; Davis, R.W.; et al. Contributions of ATM Mutations to Familial Breast and Ovarian Cancer. Cancer Res. 2003, 63, 3325–3333. [Google Scholar]
- Grasel, R.S.; Felicio, P.S.; de Paula, A.E.; Campacci, N.; de Oliveira Garcia, F.A.; de Andrade, E.S.; Evangelista, A.F.; Fernandes, G.C.; da Silva Sabato, C.; De Marchi, P.; et al. Using Co-Segregation and Loss of Heterozygosity Analysis to Define the Pathogenicity of Unclassified Variants in Hereditary Breast Cancer Patients. Front. Oncol. 2020, 10, 571330. [Google Scholar] [CrossRef]
- Shindo, K.; Yu, J.; Suenaga, M.; Fesharakizadeh, S.; Cho, C.; Macgregor-Das, A.; Siddiqui, A.; Witmer, P.D.; Tamura, K.; Song, T.J.; et al. Deleterious Germline Mutations in Patients With Apparently Sporadic Pancreatic Adenocarcinoma. J. Clin. Oncol. 2017, 35, 3382–3390. [Google Scholar] [CrossRef] [PubMed]
- Tiao, G.; Improgo, M.R.; Kasar, S.; Poh, W.; Kamburov, A.; Landau, D.-A.; Tausch, E.; Taylor-Weiner, A.; Cibulskis, C.; Bahl, S.; et al. Rare Germline Variants in ATM Are Associated with Chronic Lymphocytic Leukemia. Leukemia 2017, 31, 2244–2247. [Google Scholar] [CrossRef] [PubMed]
- Pylkas, K.; Tommiska, J.; Syrjakoski, K.; Kere, J.; Gatei, M.; Waddell, N.; Allinen, M.; Karppinen, S.-M.; Rapakko, K.; Kaariainen, H.; et al. Evaluation of the Role of Finnish Ataxia-Telangiectasia Mutations in Hereditary Predisposition to Breast Cancer. Carcinogenesis 2006, 28, 1040–1045. [Google Scholar] [CrossRef] [PubMed]
- van Os, N.J.H.; Chessa, L.; Weemaes, C.M.R.; van Deuren, M.; Fiévet, A.; van Gaalen, J.; Mahlaoui, N.; Roeleveld, N.; Schrader, C.; Schindler, D.; et al. Genotype-Phenotype Correlations in Ataxia Telangiectasia Patients with ATM c.3576G>A and c.8147T>C Mutations. J. Med. Genet. 2019, 56, 308–316. [Google Scholar] [CrossRef] [PubMed]
- Verhagen, M.M.M.; Last, J.I.; Hogervorst, F.B.L.; Smeets, D.F.C.M.; Roeleveld, N.; Verheijen, F.; Catsman-Berrevoets, C.E.; Wulffraat, N.M.; Cobben, J.M.; Hiel, J.; et al. Presence of ATM Protein and Residual Kinase Activity Correlates with the Phenotype in Ataxia-Telangiectasia: A Genotype-Phenotype Study. Hum. Mutat. 2012, 33, 561–571. [Google Scholar] [CrossRef]
- Li, M.M.; Datto, M.; Duncavage, E.J.; Kulkarni, S.; Lindeman, N.I.; Roy, S.; Tsimberidou, A.M.; Vnencak-Jones, C.L.; Wolff, D.J.; Younes, A.; et al. Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer: A Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J. Mol. Diagn. 2017, 19, 4–23. [Google Scholar] [CrossRef]
- Bruno, W.; Dalmasso, B.; Barile, M.; Andreotti, V.; Elefanti, L.; Colombino, M.; Vanni, I.; Allavena, E.; Barbero, F.; Passoni, E.; et al. Predictors of Germline Status for Hereditary Melanoma: 5 Years of Multi-Gene Panel Testing within the Italian Melanoma Intergroup. ESMO Open 2022, 7, 100525. [Google Scholar] [CrossRef]
- Ghiorzo, P.; Gargiulo, S.; Pastorino, L.; Nasti, S.; Cusano, R.; Bruno, W.; Gliori, S.; Sertoli, M.R.; Burroni, A.; Savarino, V.; et al. Impact of E27X, a Novel CDKN2A Germ Line Mutation, on P16 and P14ARF Expression in Italian Melanoma Families Displaying Pancreatic Cancer and Neuroblastoma. Hum. Mol. Genet. 2006, 15, 2682–2689. [Google Scholar] [CrossRef][Green Version]
- Bruno, W.; Martinuzzi, C.; Andreotti, V.; Pastorino, L.; Spagnolo, F.; Dalmasso, B.; Cabiddu, F.; Gualco, M.; Ballestrero, A.; Bianchi-Scarrà, G.; et al. Heterogeneity and Frequency of BRAF Mutations in Primary Melanoma: Comparison between Molecular Methods and Immunohistochemistry. Oncotarget 2017, 8, 8069–8082. [Google Scholar] [CrossRef]
- Bruno, W.; Martinuzzi, C.; Dalmasso, B.; Andreotti, V.; Pastorino, L.; Cabiddu, F.; Gualco, M.; Spagnolo, F.; Ballestrero, A.; Queirolo, P.; et al. Combining Molecular and Immunohistochemical Analyses of Key Drivers in Primary Melanomas: Interplay between Germline and Somatic Variations. Oncotarget 2018, 9, 5691–5702. [Google Scholar] [CrossRef][Green Version]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet J. 2011, 17, 10. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional Annotation of Genetic Variants from High-Throughput Sequencing Data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef] [PubMed]
- Talevich, E.; Shain, A.H.; Botton, T.; Bastian, B.C. CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing. PLoS Comput. Biol. 2016, 12, e1004873. [Google Scholar] [CrossRef] [PubMed]
- Van Loo, P.; Nordgard, S.H.; Lingjærde, O.C.; Russnes, H.G.; Rye, I.H.; Sun, W.; Weigman, V.J.; Marynen, P.; Zetterberg, A.; Naume, B.; et al. Allele-Specific Copy Number Analysis of Tumors. Proc. Natl. Acad. Sci. USA 2010, 107, 16910–16915. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]

| Patient ID | Nucleotide Change | Protein Change | ACMG Classification | GnomAd NFE | Personal History of Cancer (Age of Diagnosis) | Other Cancer in Family (Age of Diagnosis) | Co-Segregation |
|---|---|---|---|---|---|---|---|
| Mel_1 | c.3576G>A | p.Ser1135_Lys1192del | P (PM2,PP5) | 3.52 × 10−5 | CM (40) breast (46) | CM I (80) | YES |
| Mel_2 | c.4451delT | p.Met1484fs | P (PVS1,PM2,PP5) | ND | 3 CM (45), pancreas (50) | CM III (33), pancreas I (79) | YES |
| Mel_3 | c.5979_5983del | p.Ser1993ArgfsTer23 | P (PVS1,PM2,PP5) | 1.76 × 10−5 | CM (47) | ||
| Mel_4 | c.1073A>T | p.Asn358Ile | VUS (PM2) | ND | CM (64), endometrial (61) | CM I (81) | ND |
| Mel_5 | c.1916A>G | p.Asp639Gly | VUS (PM2) | ND | 4 CM (46) | ||
| Mel_6 | c.2386A>C | p.Asn796His | VUS (PM2) | 8.80 × 10−6 | CM (67) | ||
| Mel_7 | c.3704C>T | p.Pro1235Leu | VUS (PM2,PP3) | ND | CM (59), CRCC (56) | CM I (44), glioblastoma I (67) | ND |
| Mel_8 | c.4306C>T | p.His1436Tyr | VUS (PM2) | 4.41 × 10−5 | 2 CM (21) | ||
| Mel_9 | c.4709T>C | p.Val1570Ala | VUS (PM2) | 7.30 × 10−4 | 6 CM (47) | CM II (na) | ND |
| Mel_10 | c.8617G>A | p.Val2873Ile | VUS (PM2) | 1.77 × 10−5 | 2 CM (40), prostate (52) | CM I (na), CM II (76) | ND |
| Mel_11 | c.8734A>G | p.Arg2912Gly | VUS (PM2,PP3) | 3.87 × 10−4 | 2 CM (29) | ||
| Mel_12 | c.8734A>G | p.Arg2912Gly | VUS (PM2,PP3) | 3.87 × 10−4 | 6 CM (42), breast (51), lung (69) | CM I (36) | YES |
| Mel_13 | c.1810C>T | p.Pro604Ser | B (BS2,BP6) | 1.55 × 10−3 | 2 CM (42) | CM I (69), breast I (65), CM II (52) | ND |
| Mel_14 | WT | CM (71) | CM III (43) | ||||
| CTRL_1 | c.6067G>A | p.Gly2023Arg | LB (BP6) | 2.30 × 10−3 |
| Patient ID | Histology | Stage Group | TNM Staging | Protein Change | ACMG Classification | ATM Expression by IHC (Ab32420) | p-ATM Expression by IHC (Ab81292) | LOH by Microsatellite Analysis/SS |
|---|---|---|---|---|---|---|---|---|
| Mel_1 | Primary melanoma | IA | pT1a N0M0 | p.Ser1135_Lys1192del | P | negative | negative | |
| Breast Ductal Carcinoma | 0 | pTis N0 MX G2 | negative | negative | YES | |||
| Mel_2 | Primary melanoma | 0 | pTisN0M0 | p.Met1484fs | P | 99% | 70% | |
| Mel_3 | Melanoma—cutaneous metastasis | IV | - | p.Ser1993ArgfsTer23 | P | 1% | 1% | YES |
| Mel_4 | Primary melanoma | 0 | pTisN0M0 | p.Asn358Ile | VUS | 20% | 15% | |
| Mel_5 | Primary melanoma | 0 | pTisN0M0 | p.Asp639Gly | VUS | 85% | 80% | |
| Mel_6 | Melanoma—subperitoneal metastasis | IV | - | p.Asn796His | VUS | 50% | 50% | YES |
| Mel_7 | Primary melanoma | IA | pT1a N0M0 | p.Pro1235Leu | VUS | 100% | ||
| Mel_8 | Dysplastic nevus | - | p.His1436Tyr | VUS | 80% | 75% | ||
| Mel_9 | Primary melanoma | IA | pT1a N0M0 | p.Val1570Ala | VUS | 99% | ||
| Mel_10 | Primary melanoma | IA | pT1a N0M0 | p.Val2873Ile | VUS | 100% | 70% | |
| Mel_11 | Dysplastic nevus | p.Arg2912Gly | VUS | 95% | 95% | |||
| Primary melanoma | 0 | pTisN0M0 | VUS | 99% | 90% | |||
| Mel_12 | Primary melanoma | IA | pTxN0M0) | p.Arg2912Gly | VUS | 95% | 75% | NO |
| Mel_13 | Primary melanoma | IA | pT1a N0M0 | p.Pro604Ser | B | 99% | 95% | |
| Primary melanoma | IA | pT1a N0M0 | 95% | 90% | ||||
| Mel_14 | Primary melanoma | IB | pT2aN0M0 | - | - | 95% | 60% | NO |
| Melanoma cutaneous metastasis | IV | - | 100% | 100% | ||||
| CTRL_1 | Junctional nevus | - | p.Gly2023Arg | LB | 99% | 80% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pastorino, L.; Dalmasso, B.; Allavena, E.; Vanni, I.; Ugolini, F.; Baroni, G.; Croce, M.; Guadagno, A.; Cabiddu, F.; Andreotti, V.; et al. Ataxia-Telangiectasia Mutated Loss of Heterozygosity in Melanoma. Int. J. Mol. Sci. 2022, 23, 16027. https://doi.org/10.3390/ijms232416027
Pastorino L, Dalmasso B, Allavena E, Vanni I, Ugolini F, Baroni G, Croce M, Guadagno A, Cabiddu F, Andreotti V, et al. Ataxia-Telangiectasia Mutated Loss of Heterozygosity in Melanoma. International Journal of Molecular Sciences. 2022; 23(24):16027. https://doi.org/10.3390/ijms232416027
Chicago/Turabian StylePastorino, Lorenza, Bruna Dalmasso, Eleonora Allavena, Irene Vanni, Filippo Ugolini, Gianna Baroni, Michela Croce, Antonio Guadagno, Francesco Cabiddu, Virginia Andreotti, and et al. 2022. "Ataxia-Telangiectasia Mutated Loss of Heterozygosity in Melanoma" International Journal of Molecular Sciences 23, no. 24: 16027. https://doi.org/10.3390/ijms232416027
APA StylePastorino, L., Dalmasso, B., Allavena, E., Vanni, I., Ugolini, F., Baroni, G., Croce, M., Guadagno, A., Cabiddu, F., Andreotti, V., Bruno, W., Zoppoli, G., Ferrando, L., Tanda, E. T., Spagnolo, F., Menin, C., Gangemi, R., Massi, D., & Ghiorzo, P. (2022). Ataxia-Telangiectasia Mutated Loss of Heterozygosity in Melanoma. International Journal of Molecular Sciences, 23(24), 16027. https://doi.org/10.3390/ijms232416027









