The Effects of One-Point Mutation on the New Delhi Metallo Beta-Lactamase-1 Resistance toward Carbapenem Antibiotics and β-Lactamase Inhibitors: An In Silico Systematic Approach
Abstract
1. Introduction
2. Results
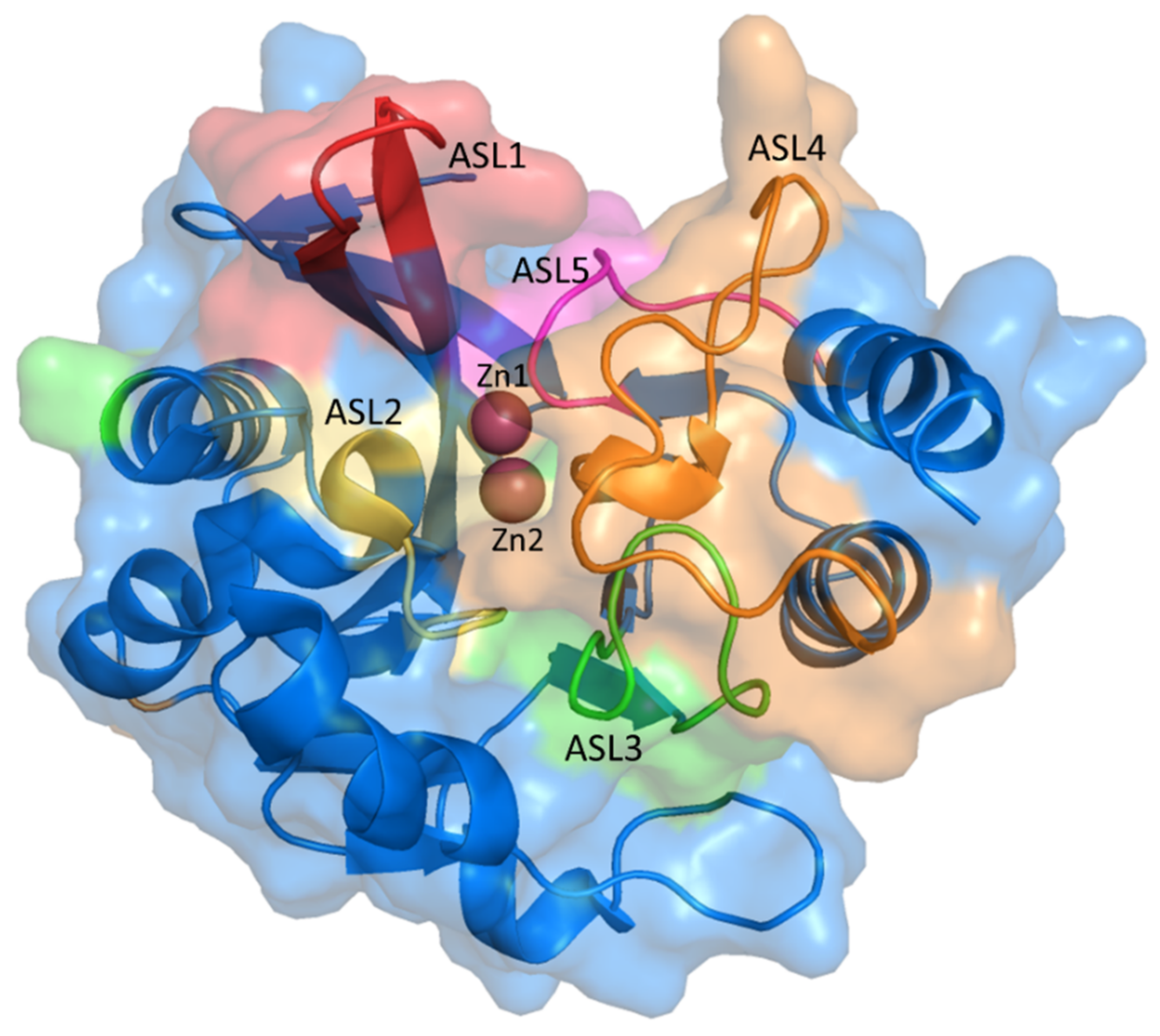
2.1. NDM-1 Wild-Type Protein Structure
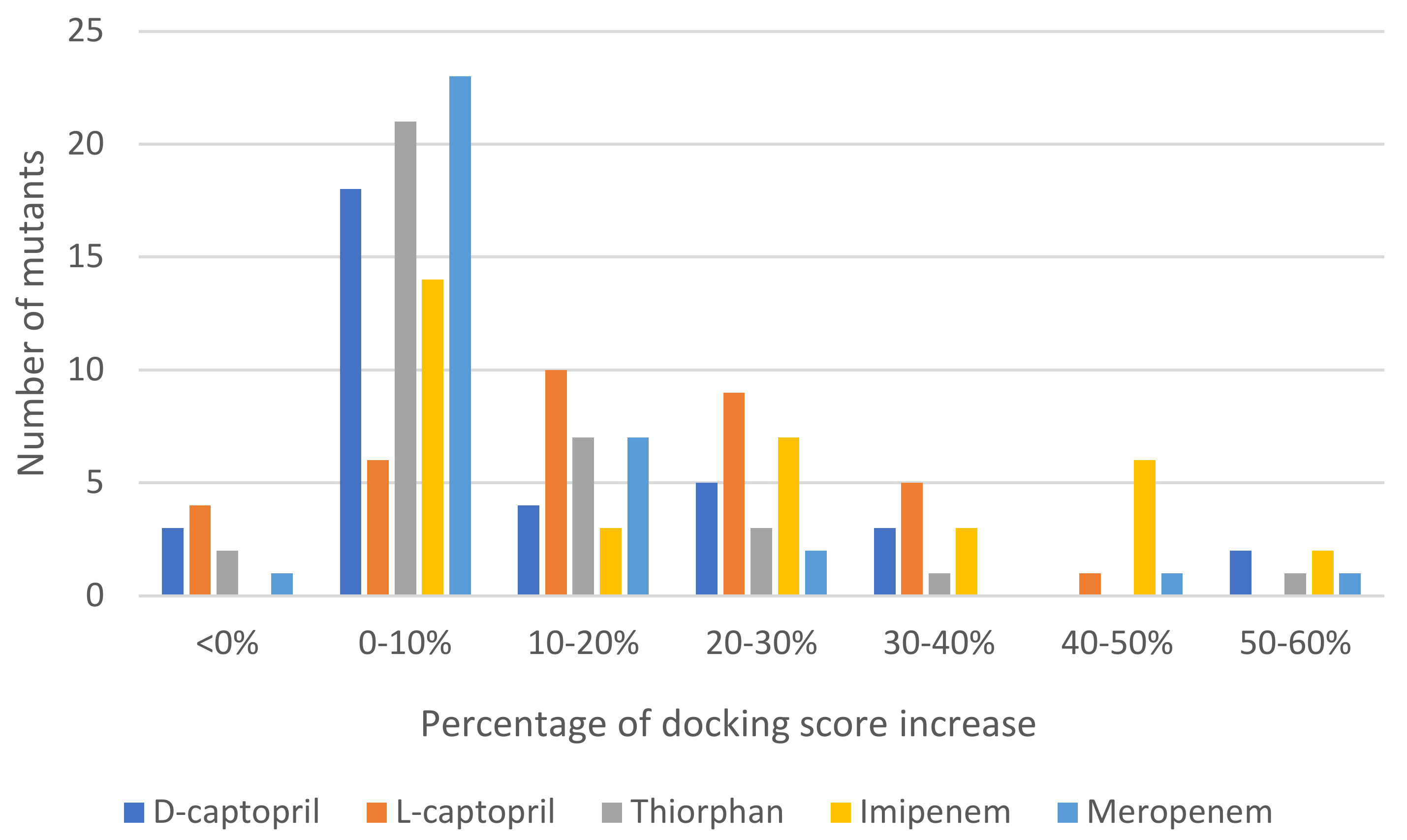
2.2. Binding Affinity of NDM-1 Wild-Type and Mutative Proteins
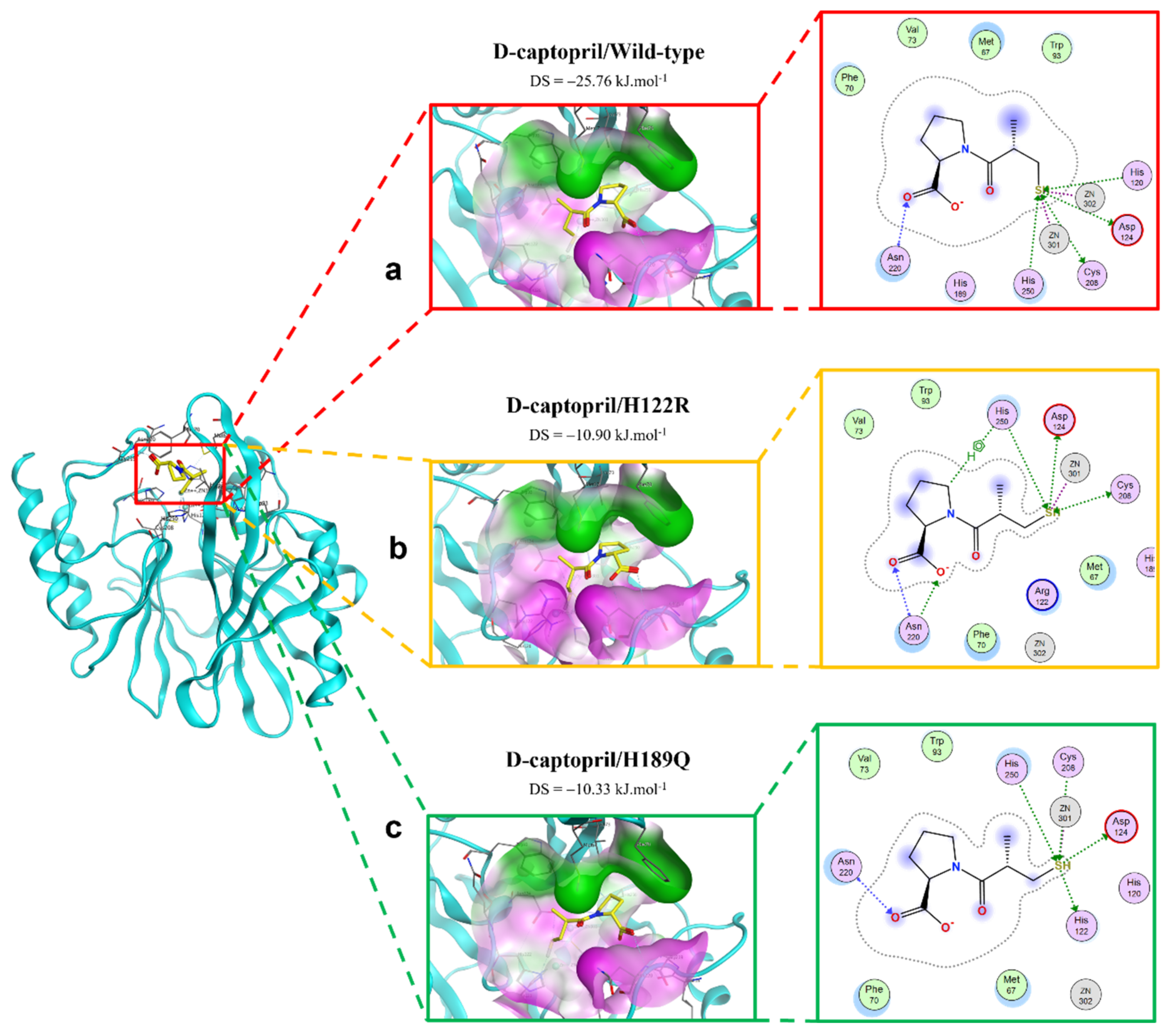
2.2.1. D-Captopril Ligand
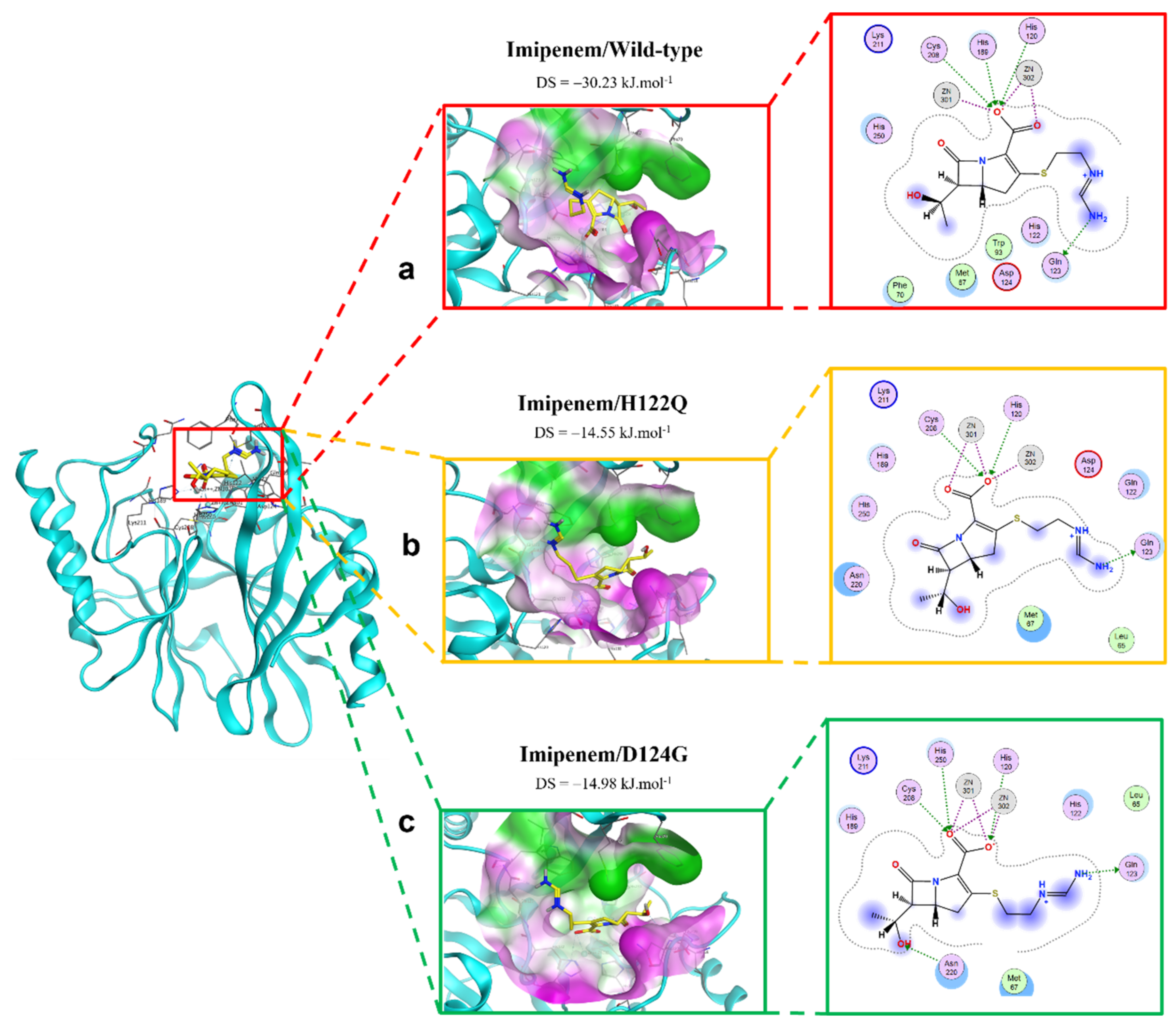
2.2.2. Imipenem Ligand
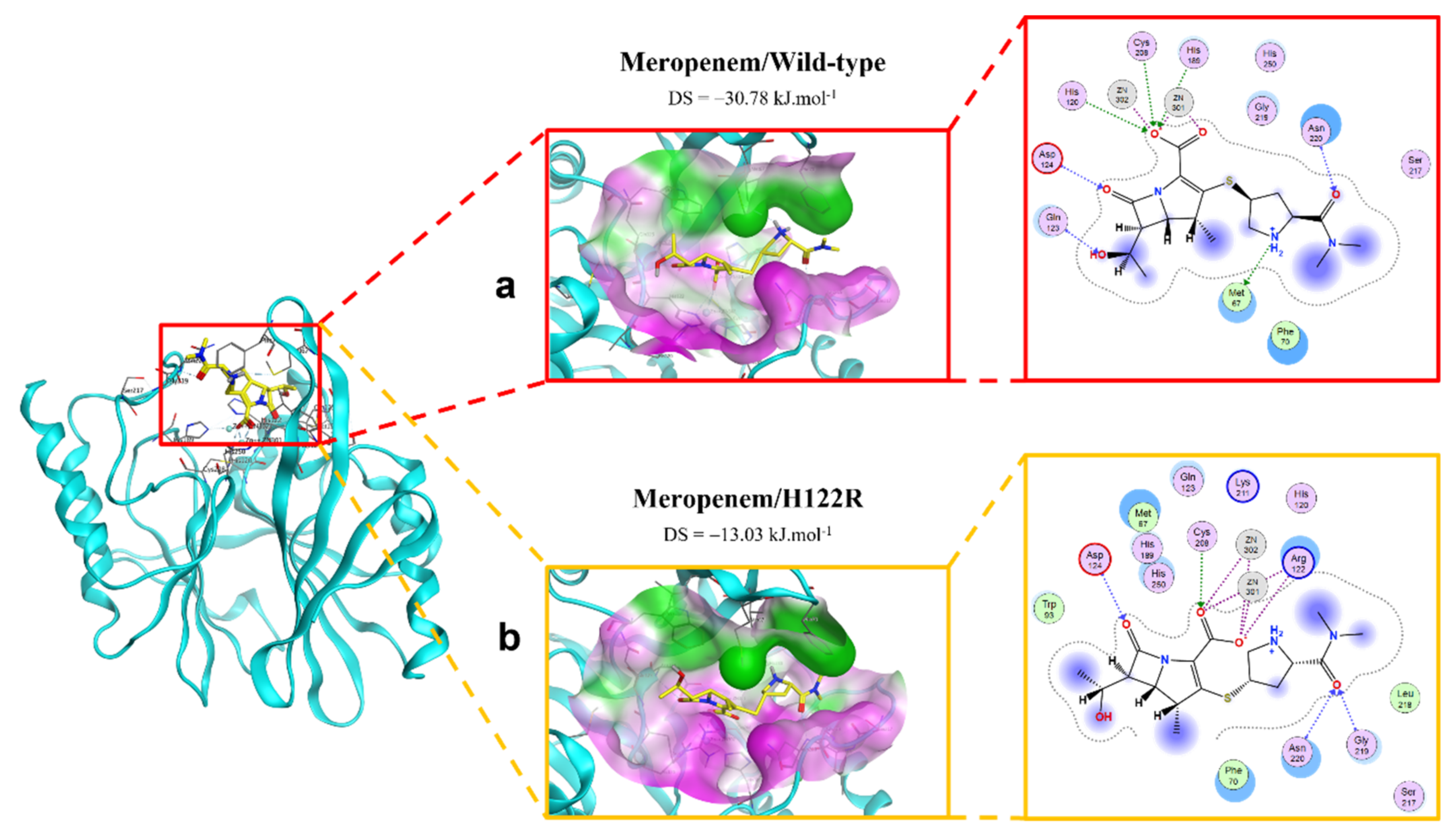
2.2.3. Meropenem Ligand
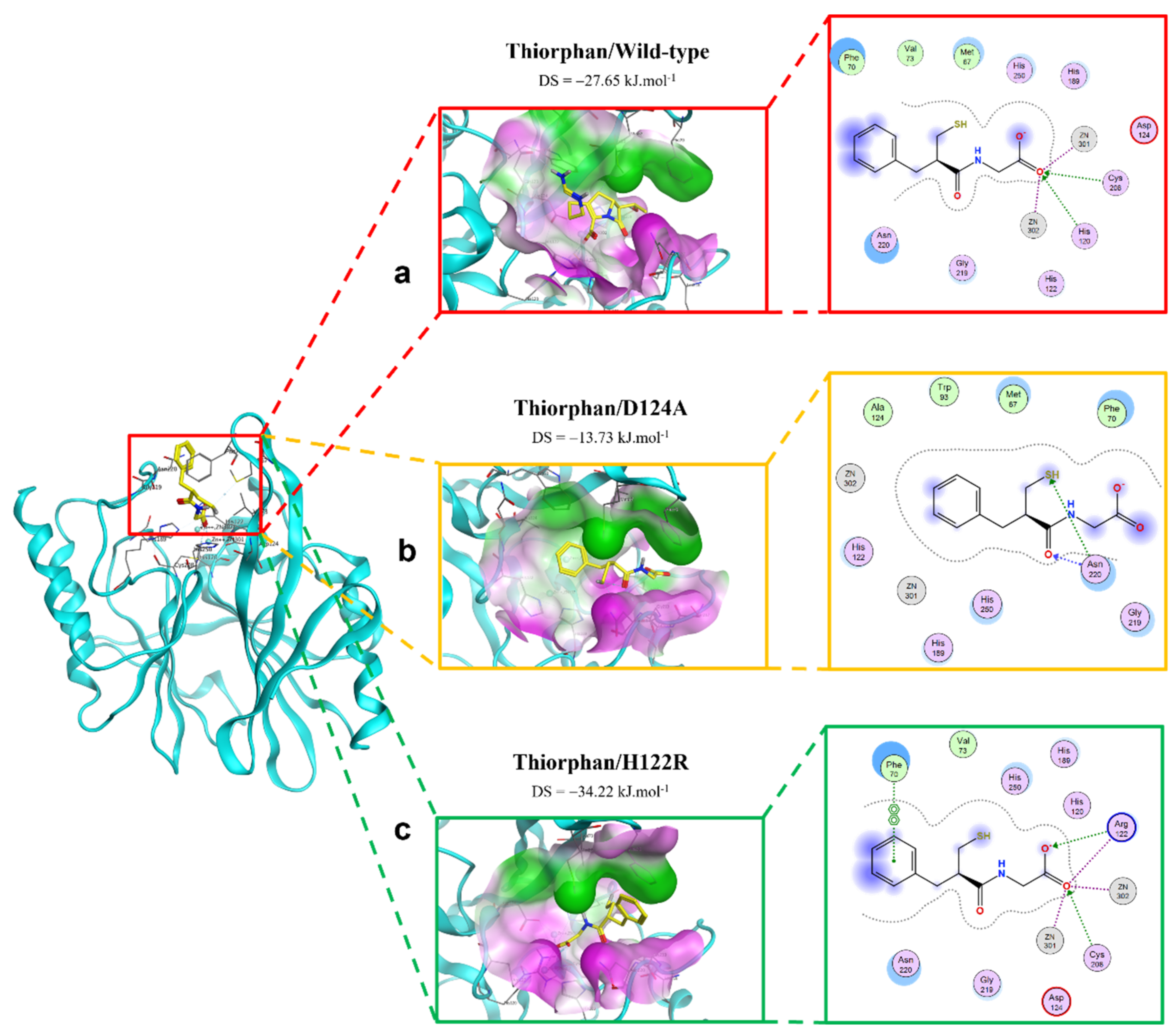
2.2.4. Thiorphan Ligand
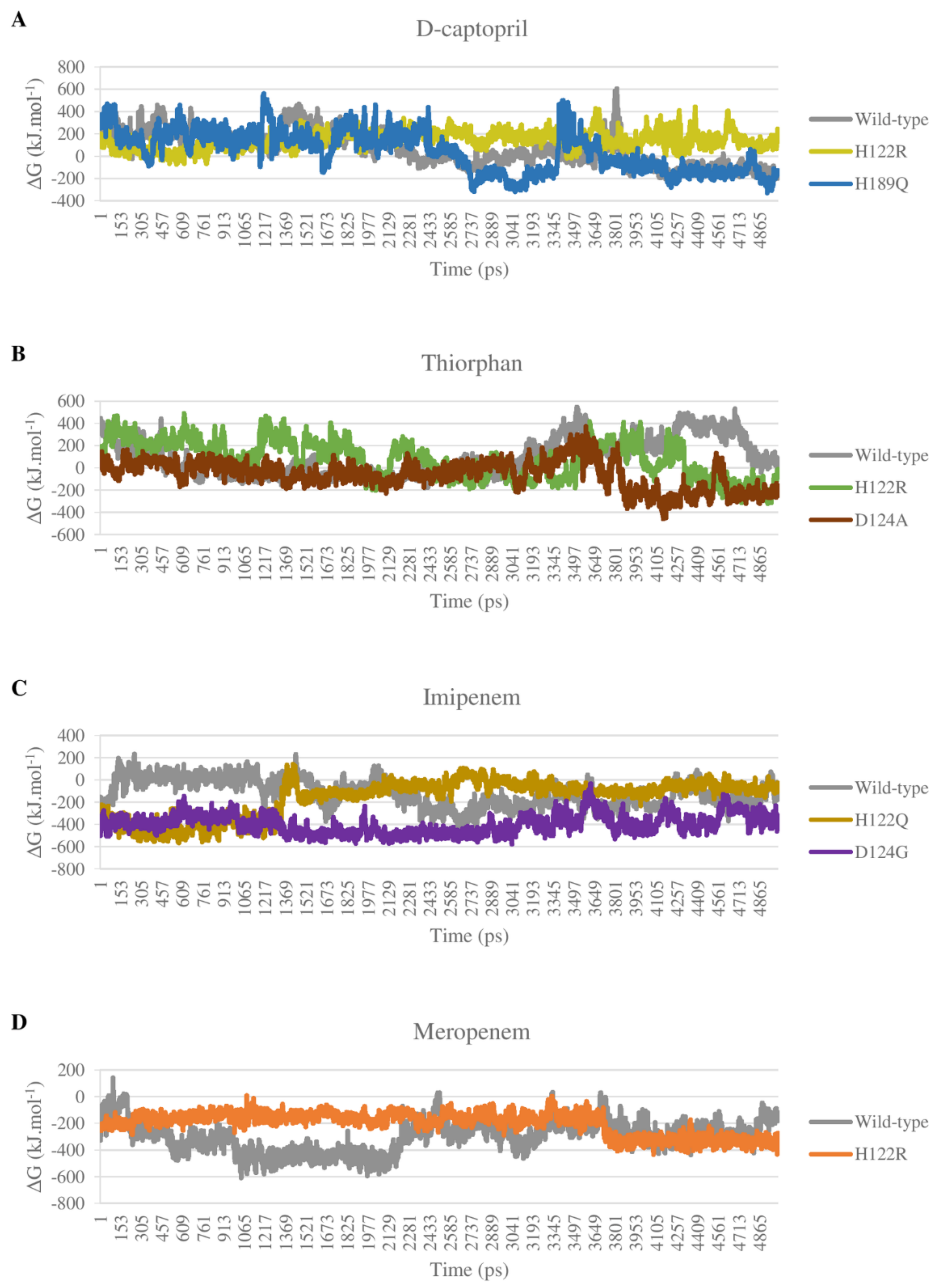
2.3. Molecular Dynamics Simulation
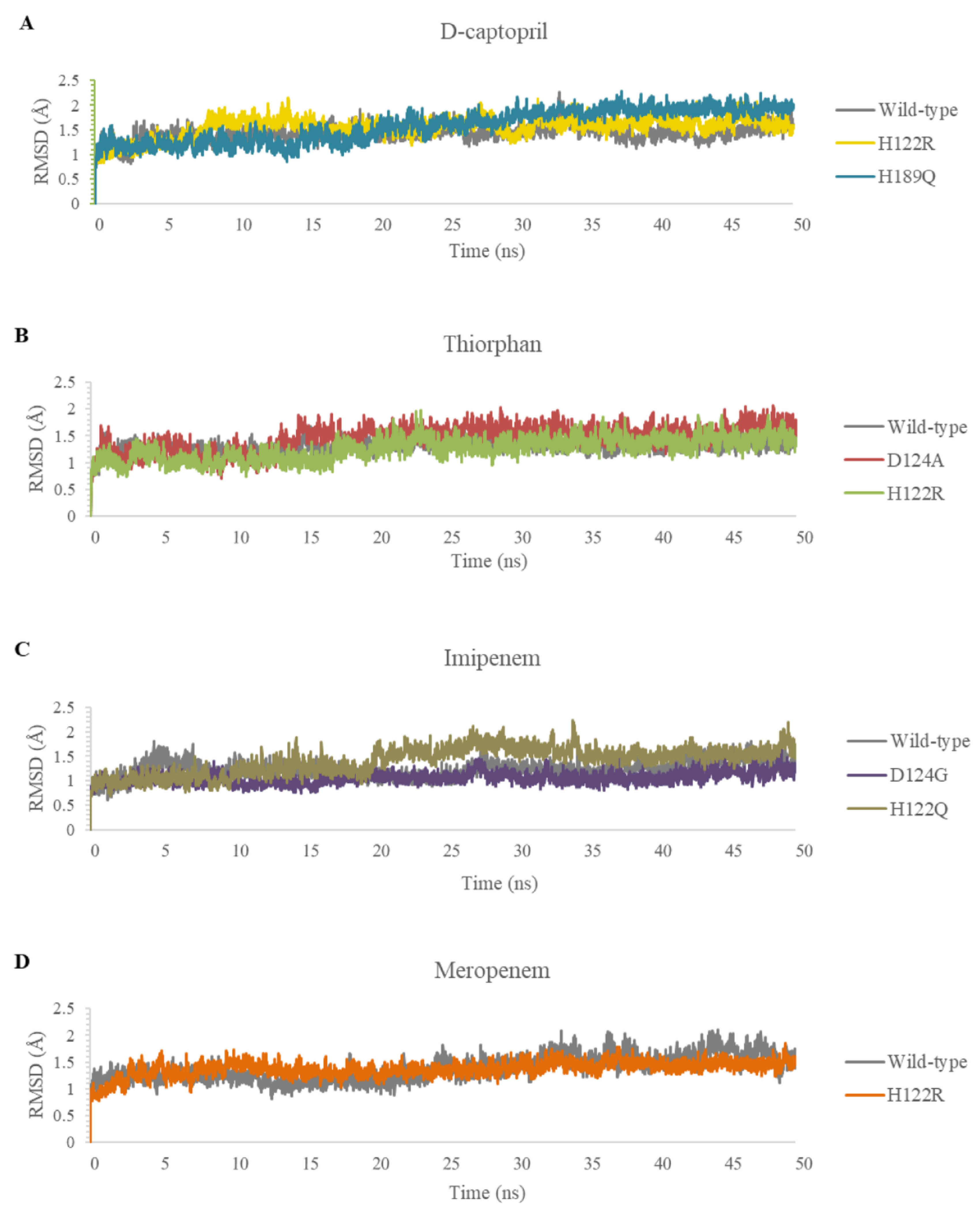
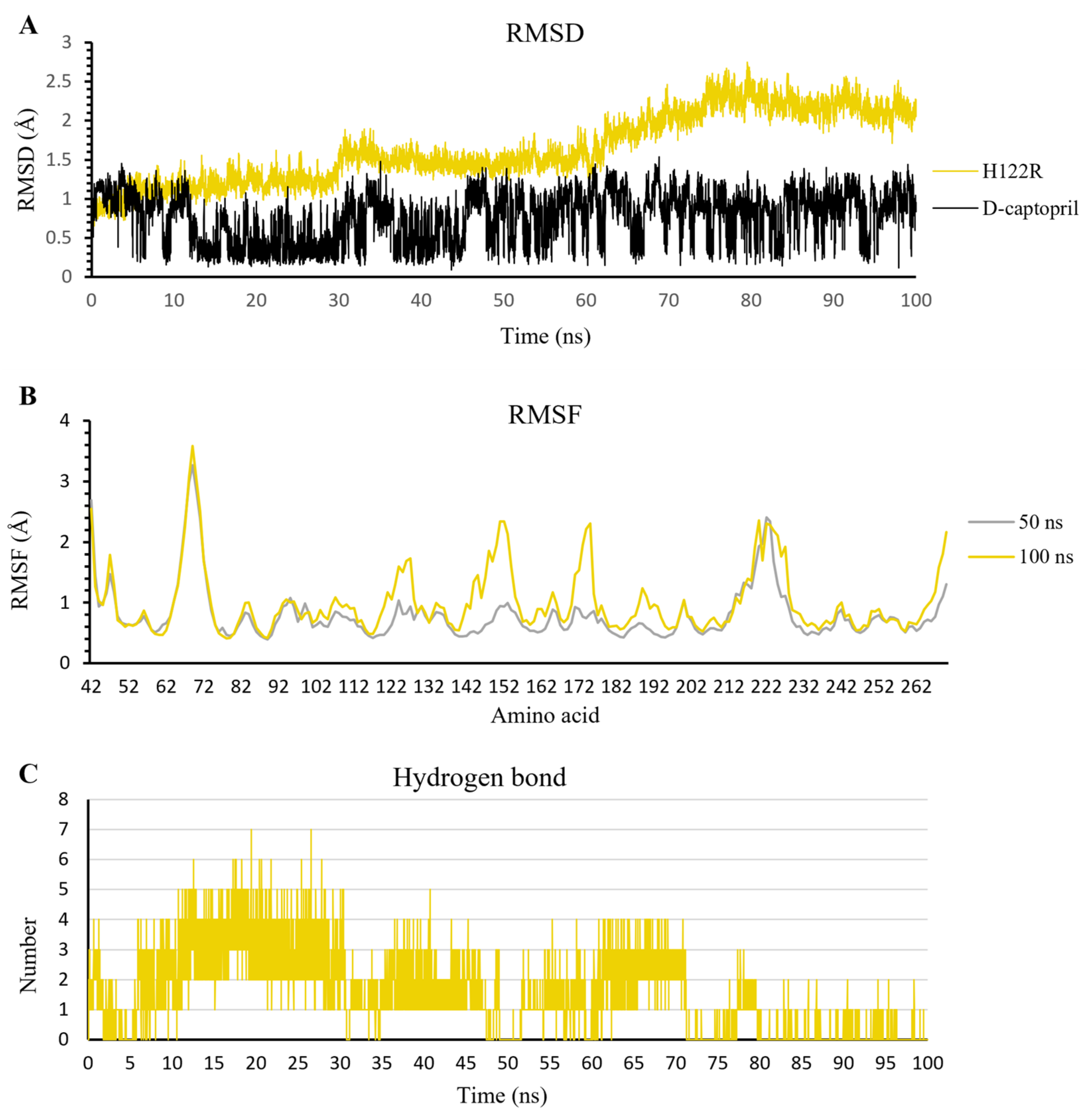
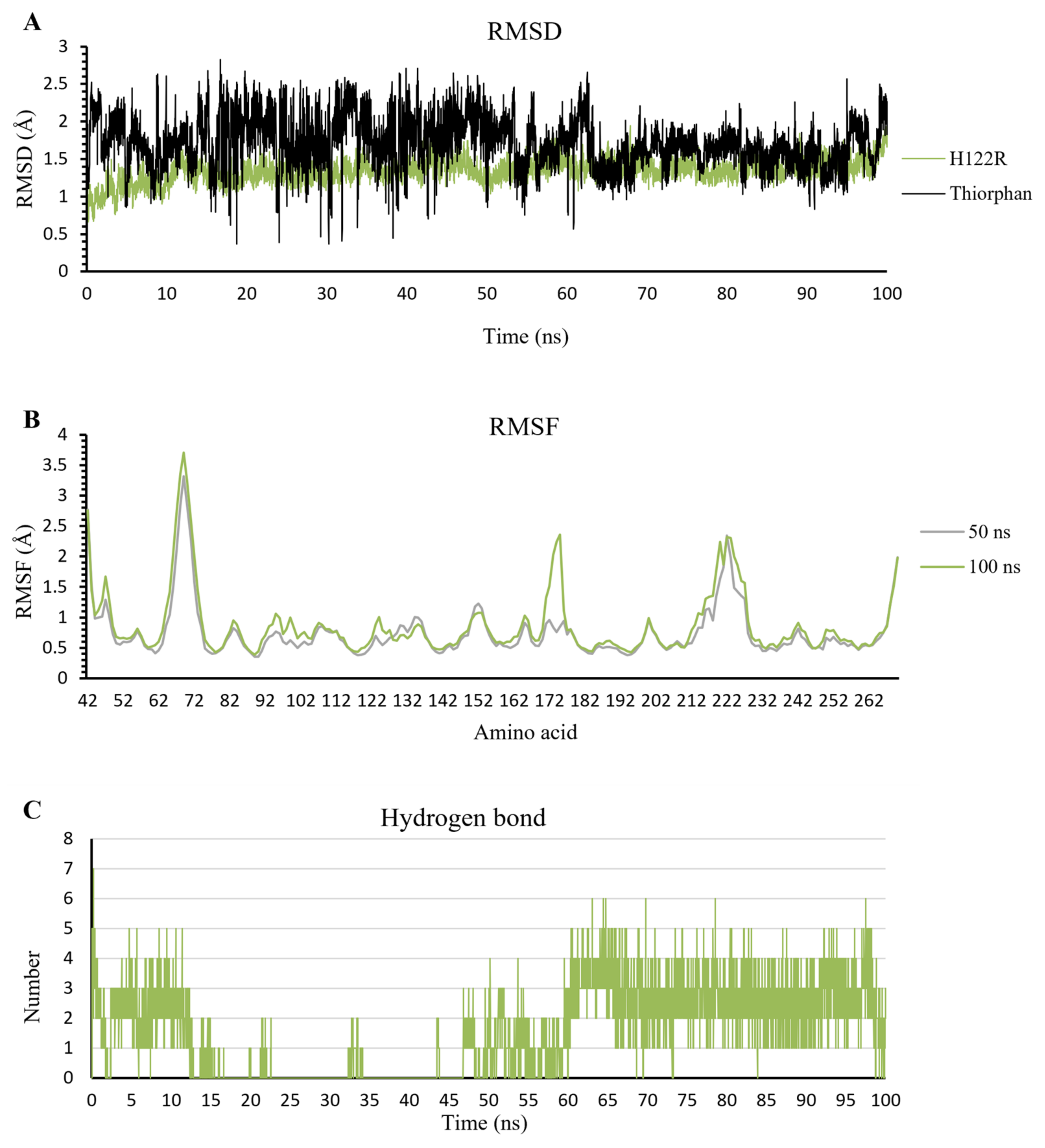
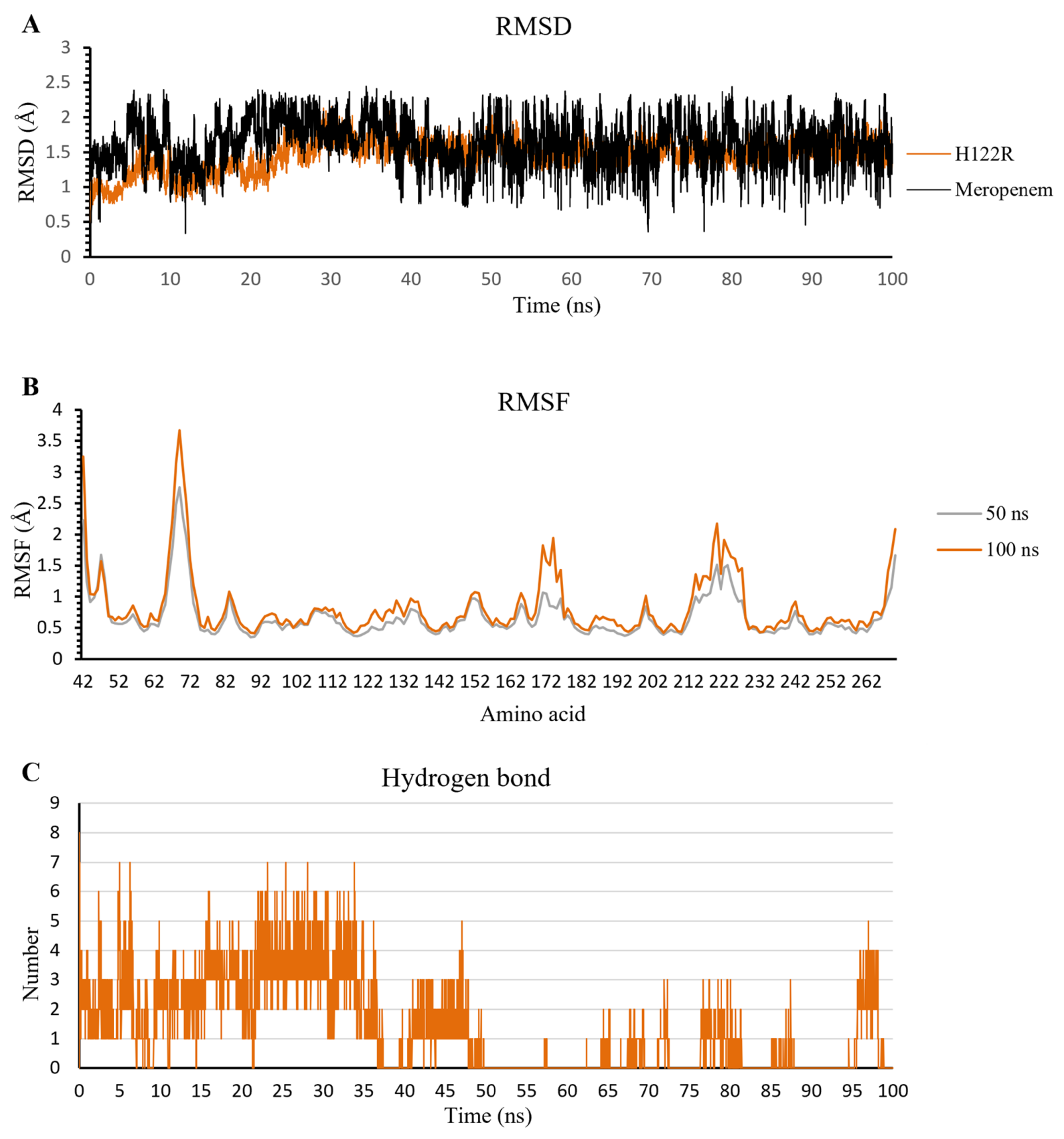
2.3.1. The Stability of Complexes over Simulation Time
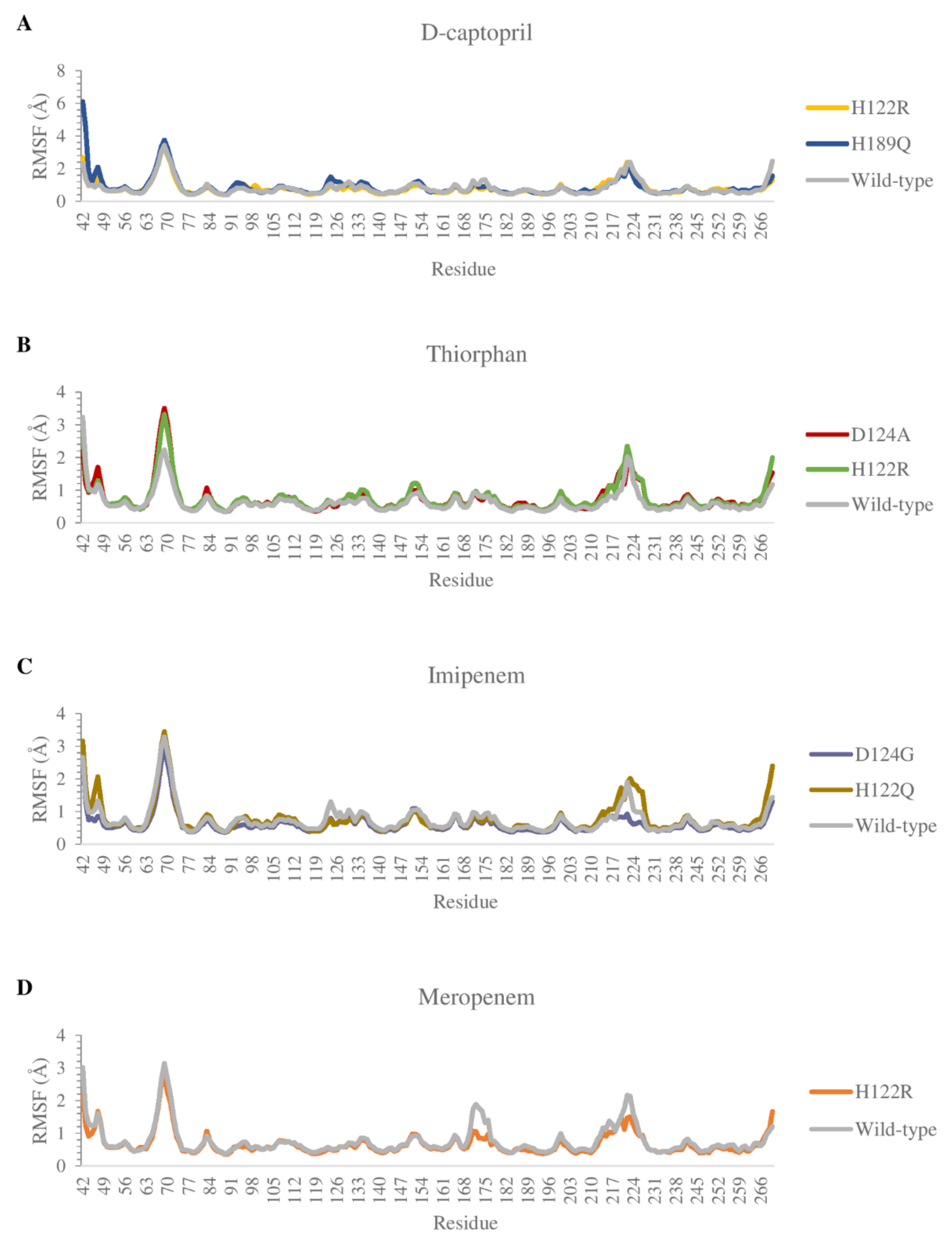
2.3.2. The Flexibility of Residues on the Binding Site
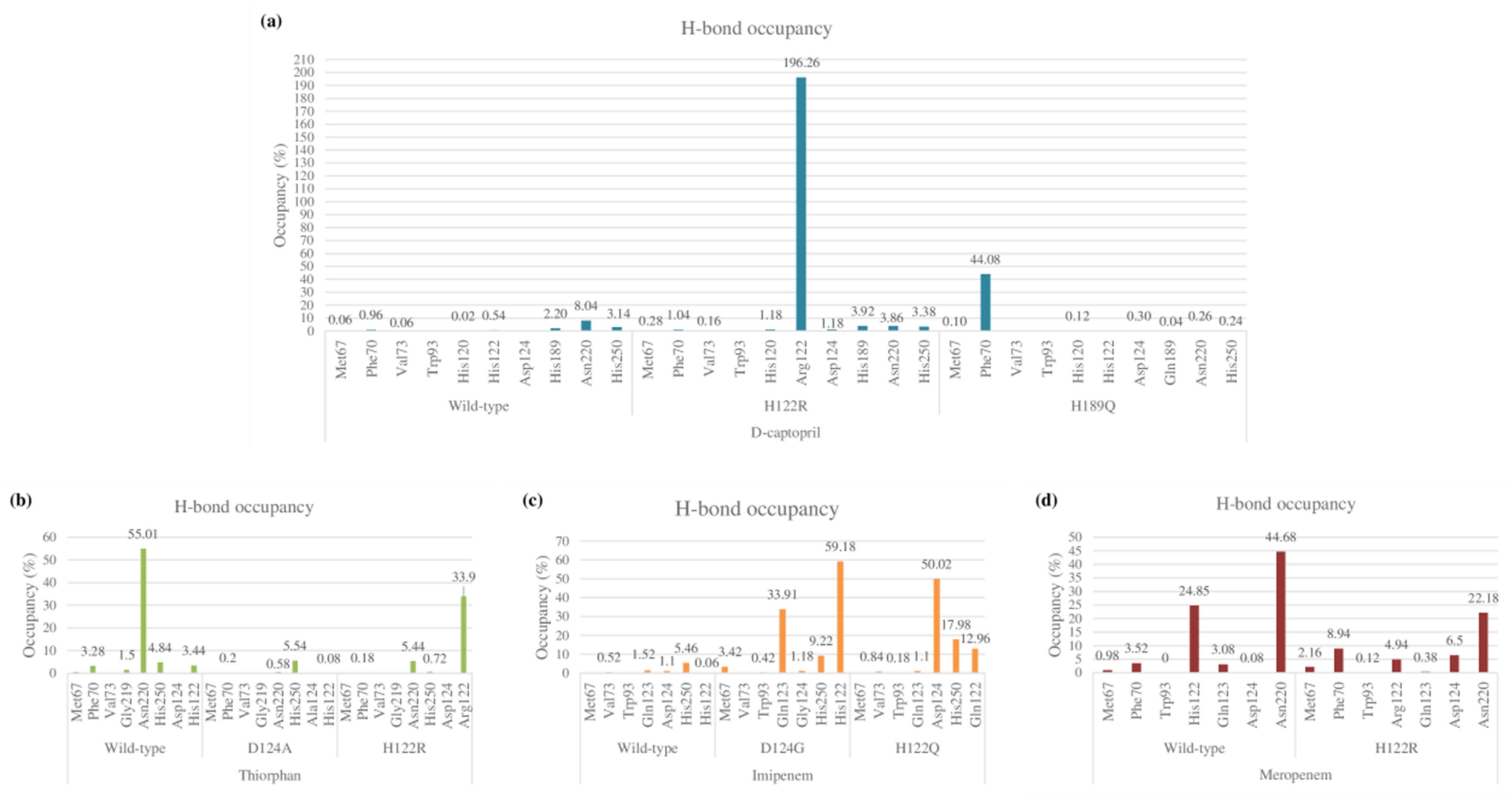
2.3.3. Hydrogen Bonds
D-Captopril
Thiorphan
Imipenem
Meropenem
2.4. Potential H122R Mutant in 100 ns MD Simulation
2.4.1. D-Captopril/H122R Complex
2.4.2. Thiorphan/H122R Complex
2.4.3. Meropenem/H122R Complex
2.5. Binding Free Energy
2.5.1. D-Captopril and Thiorphan
2.5.2. Imipenem and Meropenem
3. Discussion
3.1. NDM-1 Inhibitors
3.2. Antibiotic Ligands
4. Materials and Methods
4.1. Data Collection
4.2. Re-Docking to Find the Most Suitable Structure
4.3. Creating In Silico Mutations
4.4. Molecular Docking
4.5. Molecular Dynamics Simulation
4.6. Binding Free Energy Calculation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khan, S.; Ali, A.; Khan, A.U. Structural and functional insight of New Delhi Metallo β-lactamase-1 variants. Future Med. Chem. 2017, 10, 221–229. [Google Scholar] [CrossRef] [PubMed]
- King, D.T.; Worrall, L.J.; Gruninger, R.; Strynadka, N.C.J. New Delhi Metallo-β-Lactamase: Structural Insights into β-Lactam Recognition and Inhibition. J. Am. Chem. Soc. 2012, 134, 11362–11365. [Google Scholar] [CrossRef] [PubMed]
- Groundwater, P.W.; Xu, S.; Lai, F.; Váradi, L.; Tan, J.; Perry, J.D.; Hibbs, D.E. New Delhi metallo-β-lactamase-1: Structure, inhibitors and detection of producers. Future Med. Chem. 2016, 8, 993–1012. [Google Scholar] [CrossRef] [PubMed]
- Liénard, B.M.; Garau, G.; Horsfall, L.; Karsisiotis, A.I.; Damblon, C.; Lassaux, P.; Papamicael, C.; Roberts, G.; Galleni, M.; Dideberg, O.; et al. Structural basis for the broad-spectrum inhibition of metallo-β-lactamases by thiols. Org. Biomol. Chem. 2008, 6, 2282–2294. [Google Scholar] [CrossRef]
- Li, N.; Xu, Y.; Xia, Q.; Bai, C.; Wang, T.; Wang, L.; He, D.; Xie, N.; Li, L.; Wang, J.; et al. Simplified captopril analogues as NDM-1 inhibitors. Bioorg. Med. Chem. Lett. 2014, 24, 386–389. [Google Scholar] [CrossRef]
- Li, K.; Tan, L.; Zhou, J. HPLC determination of captopril in human plasma and its pharmacokinetic study. Biomed. Chromatogr. 1996, 10, 237–239. [Google Scholar] [CrossRef]
- Eberlin, M.; Mück, T.; Michel, M.C. A comprehensive review of the pharmacodynamics, pharmacokinetics, and clinical effects of the neutral endopeptidase inhibitor racecadotril. Front. Pharmacol. 2012, 3, 93. [Google Scholar] [CrossRef]
- Klingler, F.M.; Wichelhaus, T.A.; Frank, D.; Cuesta-Bernal, J.; El-Delik, J.; Müller, H.F.; Sjuts, H.; Göttig, S.; Koenigs, A.; Pos, K.M.; et al. Approved drugs containing thiols as inhibitors of metallo-β-lactamases: Strategy to combat multidrug-resistant bacteria. J. Med. Chem. 2015, 58, 3626–3630. [Google Scholar] [CrossRef]
- NCBI Pathogen Detection Reference Gene Catalog blaNDM. National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/pathogens/refgene/#blaNDM (accessed on 20 April 2022).
- Bahr, G.; Vitor-Horen, L.; Bethel, C.R.; Bonomo, R.A.; González, L.J.; Vila, A.J. Clinical evolution of New Delhi metallo-β-Lactamase (NDM) optimizes resistance under Zn(II) Deprivation. Antimicrob. Agents Chemother. 2021, 62, e01849-17. [Google Scholar] [CrossRef]
- Stewart, A.C.; Bethel, C.R.; VanPelt, J.; Bergstrom, A.; Cheng, Z.; Miller, C.G.; Williams, C.; Poth, R.; Morris, M.; Lahey, O.; et al. Clinical variants of New Delhi Metallo-β-lactamase are evolving to overcome zinc scarcity. ACS Infect Dis. 2017, 3, 927–940. [Google Scholar] [CrossRef]
- King, D.; Strynadka, N. Crystal structure of New Delhi metallo-β-lactamase reveals molecular basis for antibiotic resistance. Protein Sci. 2011, 20, 1484–1491. [Google Scholar] [CrossRef]
- Ali, A.; Kumar, R.; Iquebal, M.A.; Jaiswal, S.; Kumar, D.; Khan, A.U. The role of conserved residues in the catalytic activity of NDM-1: An approach involving site directed mutagenesis and molecular dynamics. Phys. Chem. Chem. Phys. 2019, 21, 17821–17835. [Google Scholar] [CrossRef]
- Ali, A.; Gupta, D.; Khan, A.U. Role of non-active site residues in maintaining New Delhi metallo-β-lactamase-1(NDM-1) function: An approach of site-directed mutagenesis and docking. FEMS Microbiol. Lett. 2021, 368, fnz003. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Le, M.-T.; Mai, T.T.; Huynh, P.N.H.; Tran, T.-D.; Thai, K.-M.; Nguyen, Q.-T. Structure-based discovery of interleukin-33 inhibitors: A pharmacophore modelling, molecular docking, and molecular dynamics simulation approach. SAR QSAR Environ. Res. 2020, 31, 883–904. [Google Scholar] [CrossRef]
- Tran-Nguyen, V.K.; Le, M.T.; Tran, T.D.; Truong, V.D.; Thai, K.M. Peramivir binding affinity with influenza A neuraminidase and research on its mutations using an induced-fit docking approach. SAR QSAR Environ. Res. 2019, 30, 899–917. [Google Scholar] [CrossRef]
- Le, M.T.; Hoang, V.N.; Nguyen, D.N.; Bui, T.H.; Phan, T.V.; Huynh, P.N.; Tran, T.D.; Thai, K.M. Structure-based discovery of ABCG2 inhibitors: A homology protein-based pharmacophore modeling and molecular docking approach. Molecules 2021, 26, 3115. [Google Scholar] [CrossRef]
- Thai, K.-M.; Le, D.-P.; Tran, N.-V.-K.; Nguyen, T.-T.-H.; Tran, T.-D.; Le, M.-T. Computational assay of Zanamivir binding affinity with original and mutant influenza neuraminidase 9 using molecular docking. J. Theor. Biol. 2015, 385, 31–39. [Google Scholar] [CrossRef]
- Hersh, R.T. Atlas of protein sequence and structure, 1966. Syst. Biol 1967, 16, 262–263. [Google Scholar] [CrossRef]
- Dayhoff, M.O. National Biomedical Research Foundation. In Atlas of Protein Sequence and Structure; National Biomedical Research Foundation: Washington, DC, USA, 1979. [Google Scholar]
- Zoete, V.; Cuendet, M.A.; Grosdidier, A.; Michielin, O. SwissParam: A fast force field generation tool for small organic molecules. J. Comput. Chem. 2011, 32, 2359–2368. [Google Scholar] [CrossRef]
- Reva, B.A.; Finkelstein, A.V.; Skolnick, J. What is the probability of a chance prediction of a protein structure with an rmsd of 6 å? Fold Des. 1998, 3, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Kumar, C.V.; Swetha, R.G.; Anbarasu, A.; Ramaiah, S. Computational analysis reveals the association of threonine 118 Methionine mutation in PMP22 resulting in CMT-1A. Adv. Bioinform. 2014, 2014, 502618. [Google Scholar] [CrossRef] [PubMed]
- Sundar, S.; Thangamani, L.; Manivel, G.; Kumar, P.; Piramanayagam, S. Molecular docking, molecular dynamics and MM/PBSA studies of FDA approved drugs for protein kinase a of Mycobacterium tuberculosis; application insights of drug repurposing. Inf. Med. Unlocked 2019, 16, 100210. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Szalewicz, K. Hydrogen Bond. In Encyclopedia of Physical Science and Technology, 3rd ed.; Meyers, R.A., Ed.; Academic Press: Cambridge, MA, USA, 2003; pp. 505–538. [Google Scholar]
- Kumari, R.; Kumar, R.; Lynn, A. g_mmpbsa—A GROMACS Tool for High-Throughput MM-PBSA Calculations. J. Chem. Inf. Model 2014, 54, 1951–1962. [Google Scholar] [CrossRef]
- Wang, C.; Greene, D.; Xiao, L.; Qi, R.; Luo, R. Recent developments and applications of the MMPBSA method. Front. Mol. Biosci. 2018, 4, 87. [Google Scholar] [CrossRef]
- Shrivastava, S.; Shrivastava, P.; Ramasamy, J. World health organization releases global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. J. Med. Soc. 2018, 32, 76–77. [Google Scholar] [CrossRef]
| Amino Acid | High Frequent Mutation of NDM-1 | Number | ||||
|---|---|---|---|---|---|---|
| Met67 | M67L | M67V | M67I | M67K | 4 | |
| Phe70 | F70L | F70Y | 2 | |||
| Val73 | V73A | V73I | 2 | |||
| Trp93 | W93R | W93L | W93S | 3 | ||
| His120 | H120R | H120N | H120Q | 3 | ||
| His122 | H122R | H122N | H122Q | 3 | ||
| Asp124 | D124A | D124N | D124E | D124G | 4 | |
| His189 | H189R | H189N | H189Q | 3 | ||
| Cys208 | C208S | 1 | ||||
| Gly219 | G219A | G219S | 2 | |||
| Asn220 | N220G | N220H | N220K | N220S | N220T | 5 |
| His250 | H250R | H250N | H250Q | 3 | ||
| Total | 35 | |||||
| Ligand | Structure | Docking Score (kJ·mol−1) |
|---|---|---|
| D-captopril | 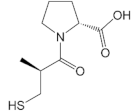 | −25.76 |
| L-captopril | 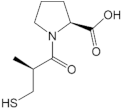 | −20.75 |
| Thiorphan | 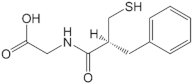 | −27.65 |
| Imipenem | 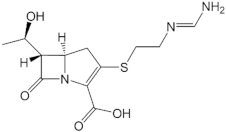 | −30.23 |
| Meropenem |  | −30.78 |
| Ligand | Mutant | |||||
|---|---|---|---|---|---|---|
| D-captopril | H189Q | H122R | H250N | N220G | H250R | |
| Percentage increase in docking score (%) | 59.89 | 57.67 | 39.48 | 32.76 | 32.04 | |
| L-captopril | N220G | H250N | H189Q | N220H | N220S | D124E |
| Percentage increase in docking score (%) | 45.42 | 39.01 | 37.68 | 36.90 | 36.2 | 30.29 |
| Thiorphan | D124A | H189Q | H122R | |||
| Percentage increase in docking score (%) | 50.35 | 30.69 | −23.78 | |||
| Imipenem | H122Q | H122R | D124N | D124E | D124A | H122N |
| Percentage increase in docking score (%) | 55.98 | 48.81 | 44.36 | 38.91 | 36.07 | 33.87 |
| Meropenem | D124G | H189N | H189Q | H189R | G219S | |
| Percentage increase in docking score (%) | 54.43 | 49.79 | 42.89 | 41.45 | 38.84 | |
| Ligand | Structure | ΔEvdW (kJ·mol−1) | ΔEelec (kJ·mol−1) | ΔGpol (kJ·mol−1) | ΔGnonpol (kJ·mol−1) | ΔGbind (kJ·mol−1) |
|---|---|---|---|---|---|---|
| D-captopril | Wild type | −50.970 ± 22.061 | 80.759 ± 191.470 | 43.346 ± 67.767 | −7.343 ± 2.844 | 65.792 ± 148.849 |
| H122R | −42.950 ± 28.844 | 80.925 ± 92.689 | 114.204 ± 77.055 | −7.774 ± 3.800 | 144.404 ± 75.914 | |
| H189Q | −23.138 ± 27.542 | −11.116 ± 235.609 | 84.664 ± 94.956 | −4.140 ± 3.599 | 46.270 ± 183.341 | |
| Thiorphan | Wild type | −72.670 ± 16.206 | 51.009 ± 263.722 | 121.157 ± 112.454 | −11.658 ± 2.345 | 87.838 ± 167.206 |
| H122R | −32.179 ± 28.586 | 37.285 ± 234.204 | 76.746 ± 101.440 | −5.770 ± 4.223 | 76.083 ± 164.236 | |
| D124A | −54.567 ± 26.739 | −74.501 ± 212.301 | 77.550 ± 96.705 | −7.975 ± 3.283 | −59.492 ± 126.395 | |
| Imipenem | Wild type | −32.915 ± 26.014 | −217.441 ± 214.214 | 137.187 ± 126.093 | −6.442 ± 4.127 | −119.611 ± 122.194 |
| H122Q | −56.520 ± 17.858 | −331.121 ± 166.963 | 235.477 ± 87.403 | −10.508 ± 1.873 | −162.672 ± 165.792 | |
| D124G | −38.956 ± 22.490 | −745.386 ± 143.139 | 397.213 ± 78.758 | −11.414 ± 3.015 | −398.543 ± 86.196 | |
| Meropenem | Wild type | −52.850 ± 30.194 | −399.750 ± 167.545 | 162.424 ± 85.753 | −9.767 ± 4.724 | −299.943 ± 116.132 |
| H122R | −95.839 ± 17.361 | −165.440 ± 90.794 | 72.914 ± 40.381 | −12.123 ± 1.984 | −200.489 ± 84.547 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tran, V.-T.; Tran, V.-H.; Nguyen, D.-N.; Do, T.-G.-S.; Vo, T.-P.; Nguyen, T.-T.-N.; Huynh, P.N.H.; Thai, K.-M. The Effects of One-Point Mutation on the New Delhi Metallo Beta-Lactamase-1 Resistance toward Carbapenem Antibiotics and β-Lactamase Inhibitors: An In Silico Systematic Approach. Int. J. Mol. Sci. 2022, 23, 16083. https://doi.org/10.3390/ijms232416083
Tran V-T, Tran V-H, Nguyen D-N, Do T-G-S, Vo T-P, Nguyen T-T-N, Huynh PNH, Thai K-M. The Effects of One-Point Mutation on the New Delhi Metallo Beta-Lactamase-1 Resistance toward Carbapenem Antibiotics and β-Lactamase Inhibitors: An In Silico Systematic Approach. International Journal of Molecular Sciences. 2022; 23(24):16083. https://doi.org/10.3390/ijms232416083
Chicago/Turabian StyleTran, Van-Thanh, Viet-Hung Tran, Dac-Nhan Nguyen, Tran-Giang-Son Do, Thanh-Phuong Vo, Thi-Thao-Nhung Nguyen, Phuong Nguyen Hoai Huynh, and Khac-Minh Thai. 2022. "The Effects of One-Point Mutation on the New Delhi Metallo Beta-Lactamase-1 Resistance toward Carbapenem Antibiotics and β-Lactamase Inhibitors: An In Silico Systematic Approach" International Journal of Molecular Sciences 23, no. 24: 16083. https://doi.org/10.3390/ijms232416083
APA StyleTran, V.-T., Tran, V.-H., Nguyen, D.-N., Do, T.-G.-S., Vo, T.-P., Nguyen, T.-T.-N., Huynh, P. N. H., & Thai, K.-M. (2022). The Effects of One-Point Mutation on the New Delhi Metallo Beta-Lactamase-1 Resistance toward Carbapenem Antibiotics and β-Lactamase Inhibitors: An In Silico Systematic Approach. International Journal of Molecular Sciences, 23(24), 16083. https://doi.org/10.3390/ijms232416083







