Ethylene Response Factor LlERF110 Mediates Heat Stress Response via Regulation of LlHsfA3A Expression and Interaction with LlHsfA2 in Lilies (Lilium longiflorum)
Abstract
1. Introduction
2. Results
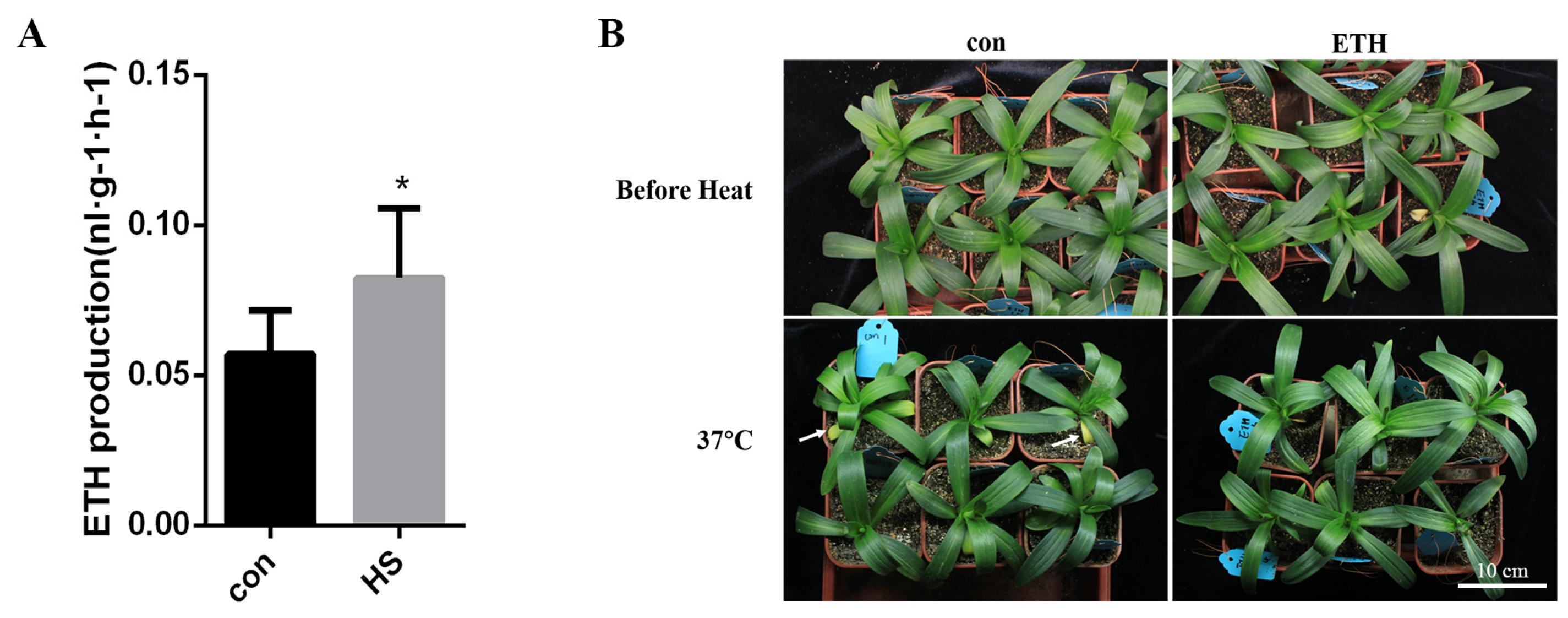
2.1. Heat Stress Induced Ethylene Production
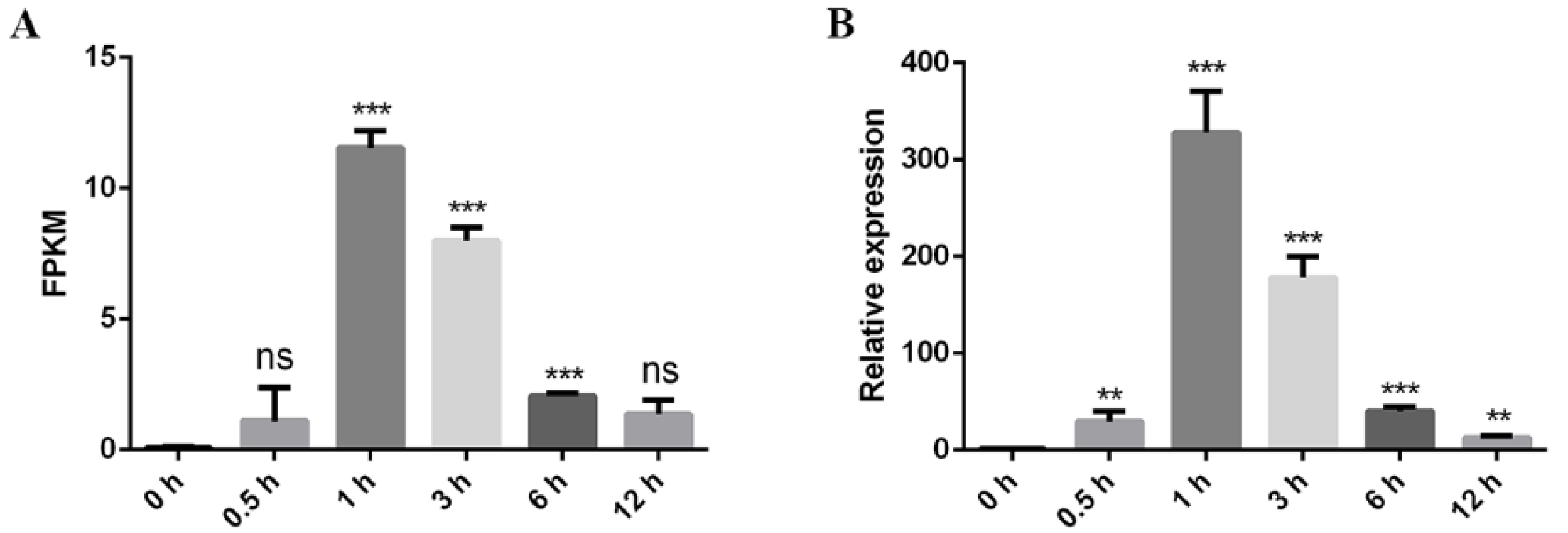
2.2. LlERF110 Was Induced by Heat Stress
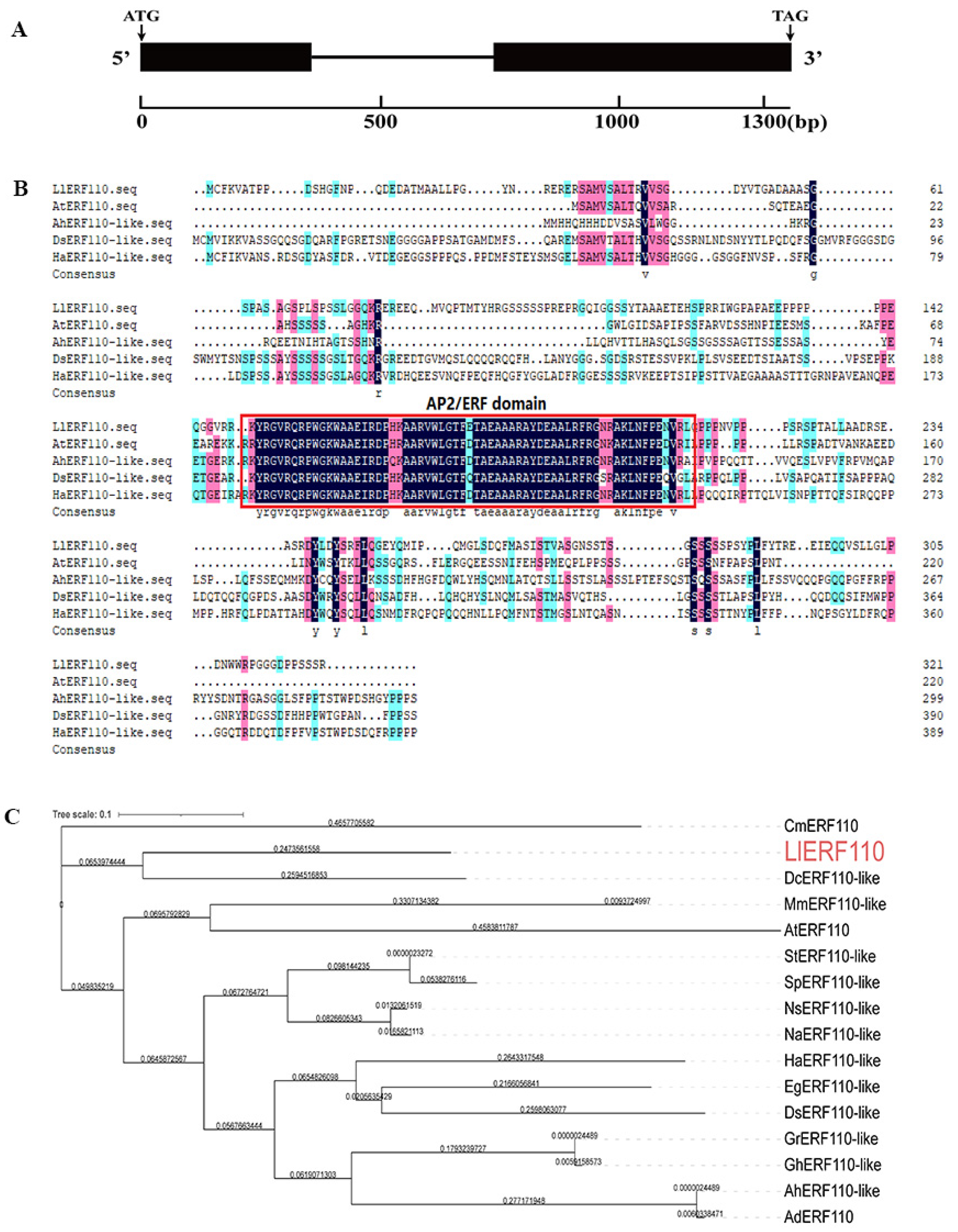
2.3. Molecular Cloning and Sequence Analysis of LlERF110
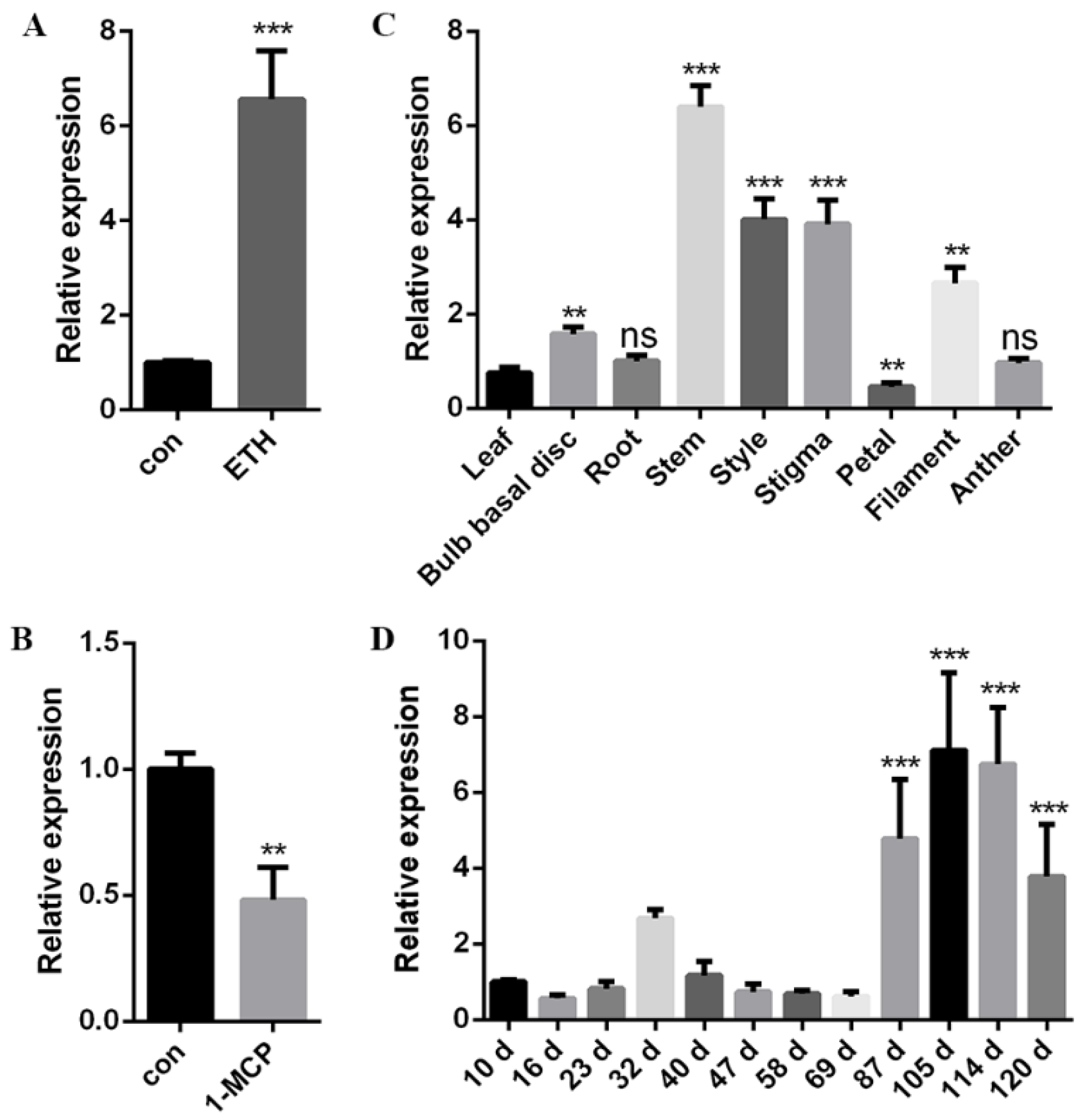
2.4. Expression Analysis of LlERF110
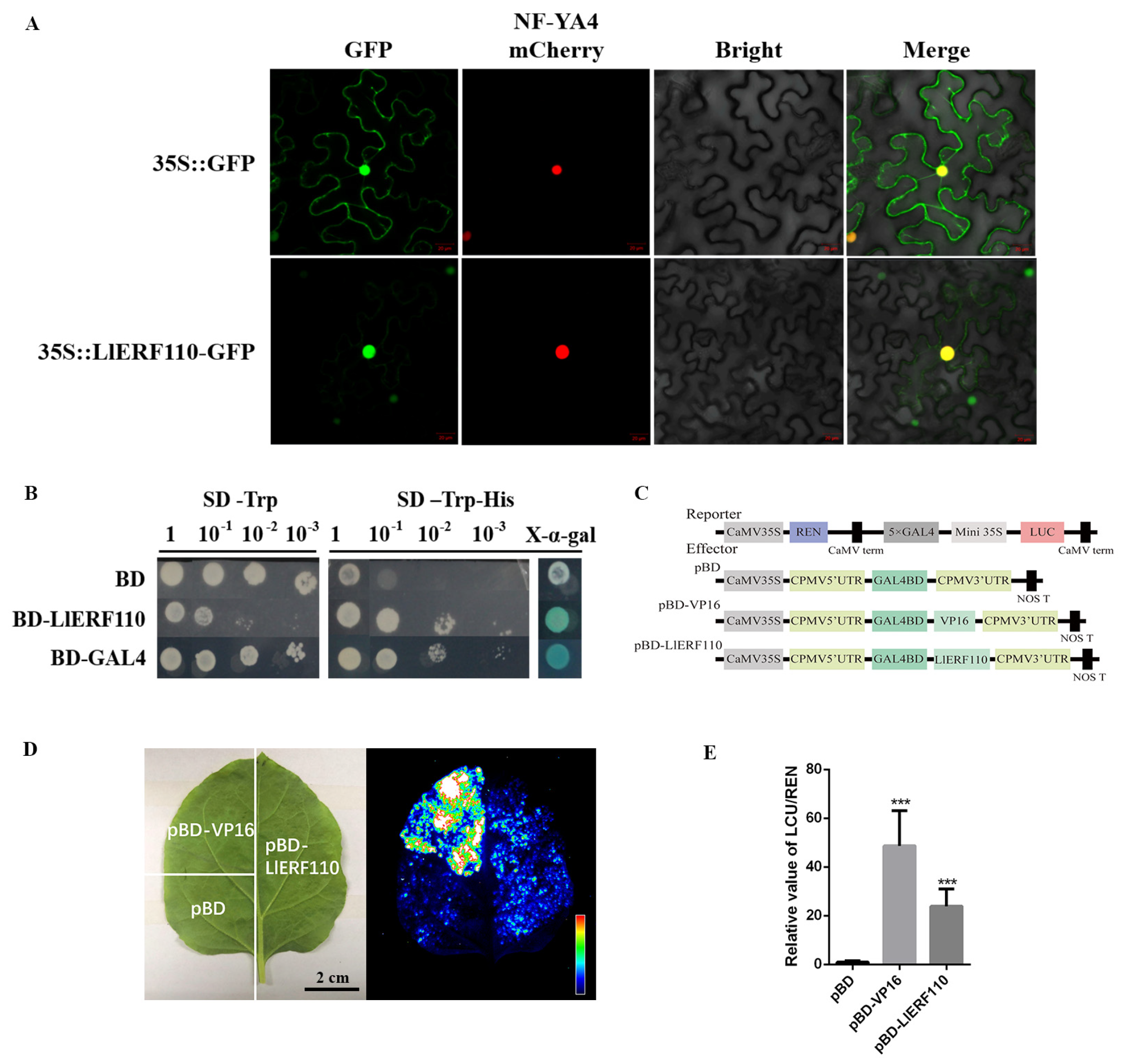
2.5. Subcellular Localization and Transactivation Activity of LlERF110
2.6. Virus-Induced Gene Silencing of LlERF110
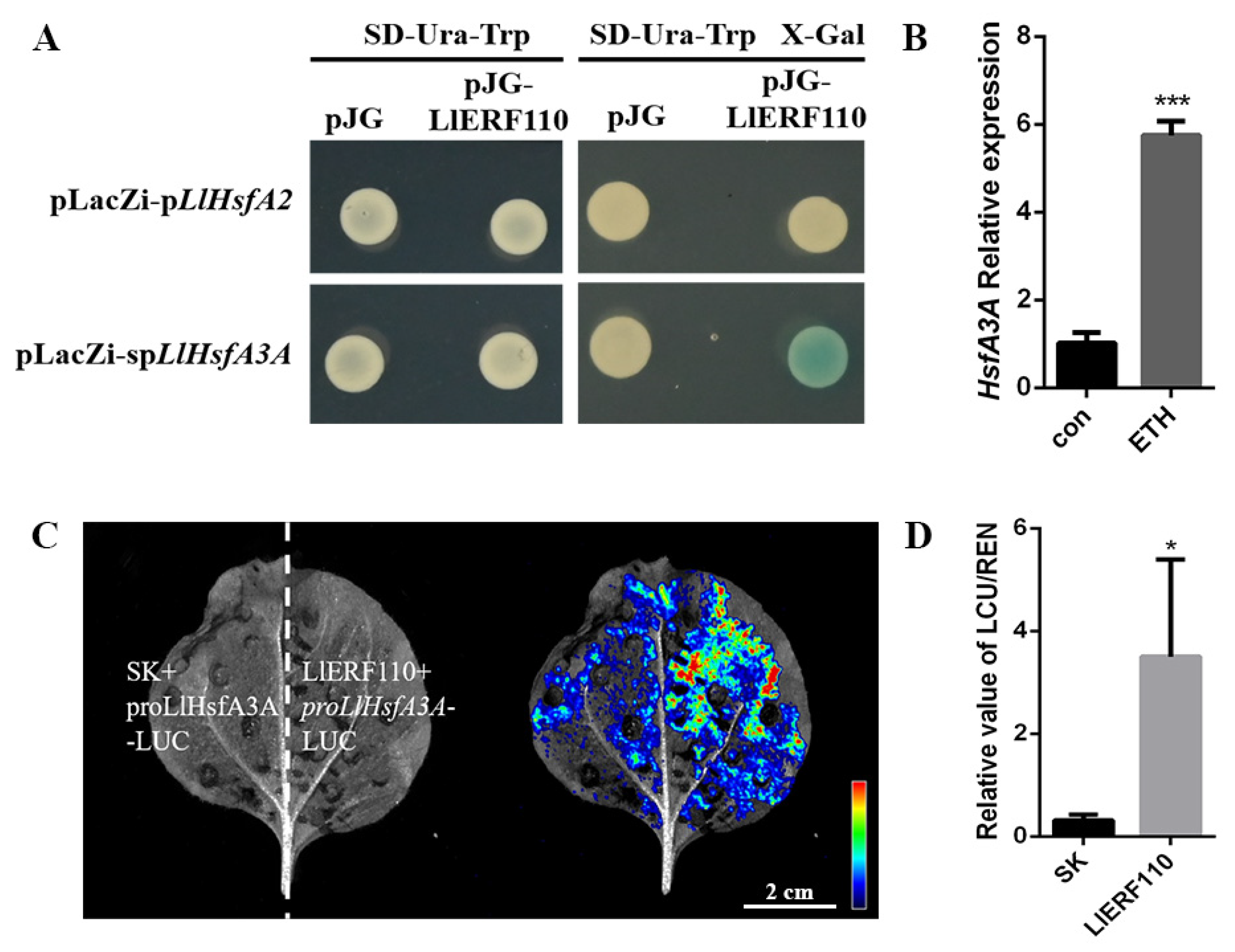
2.7. LlERF110 Binds to the Promoter of LlHsfA3A
2.8. LlERF110 Interacts with LlHsfA2
3. Discussion
3.1. LlERF110 Is a Transcription Factor Involved in Thermotolerance in Lilies
3.2. LlERF110 Is Involved in Ethylene Signaling Pathway
3.3. LlERF110 Regulates the Expression of LlHsfA3A and Interacts with LlHsfA2
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Detection of Ethylene Production, Exogenous Ethylene and 1-MCP Treatment
4.3. Gene Cloning and Sequence Analysis
4.4. Gene Expression Assay
4.5. Subcellular Localization of LlERF110
4.6. Transcriptional Activity Analyses
4.7. Yeast One-Hybrid (Y1H) Assay
4.8. Dual-LUC Reporter Assay
4.9. Yeast Two-Hybrid (Y2H) Assay
4.10. BiFC Assay
4.11. Verification of the Function of LlERF110 in Lily by the BSMV Silencing System
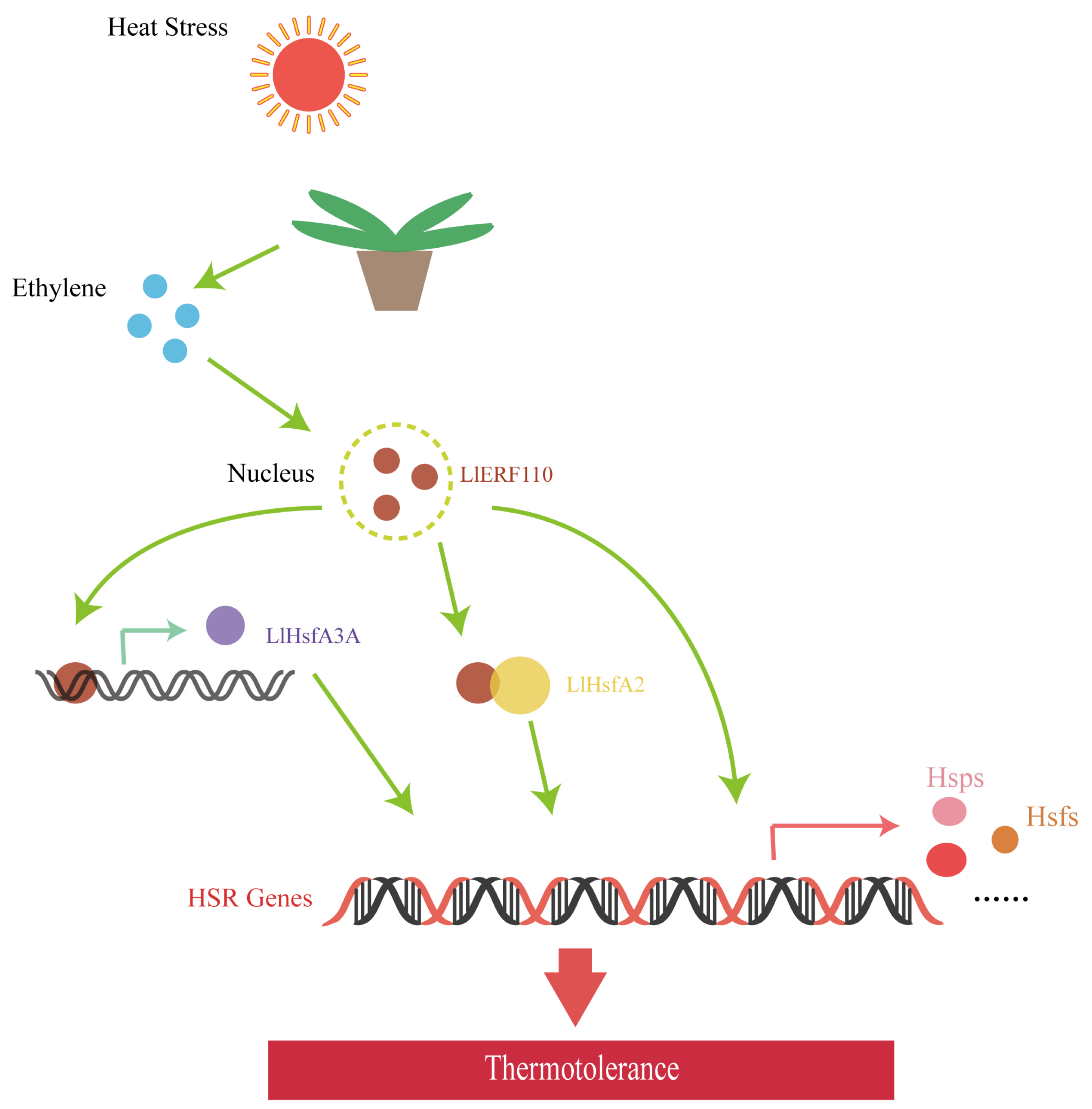
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lobell, D.B.; Schlenker, W.; Costa-Roberts, J. Climate trends and global crop production since 1980. Science 2011, 333, 616–620. [Google Scholar] [CrossRef] [PubMed]
- Schoffl, F.; Prandl, R.; Reindl, A. Regulation of the heat-shock response. Plant Physiol. 1998, 117, 1135–1141. [Google Scholar] [CrossRef] [PubMed]
- Li, B.J.; Gao, K.; Ren, H.M.; Tang, W.Q. Molecular mechanisms governing plant responses to high temperatures. J. Integr. Plant Biol. 2018, 60, 757–779. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.L.; Chen, J.H.; He, N.Y.; Guo, F.Q. Metabolic reprogramming in chloroplasts under heat stress in plants. Int. J. Mol. Sci. 2018, 19, 849. [Google Scholar] [CrossRef] [PubMed]
- Scafaro, A.P.; Fan, Y.Z.; Posch, B.C.; Garcia, A.; Coast, O.; Atkin, O.K. Responses of leaf respiration to heatwaves. Plant Cell Environ. 2021, 44, 2090–2101. [Google Scholar] [CrossRef] [PubMed]
- Perrella, G.; Baurle, I.; van Zanten, M. Epigenetic regulation of thermomorphogenesis and heat stress tolerance. New Phytol. 2022, 234, 1144–1160. [Google Scholar] [CrossRef]
- Li, N.; Euring, D.J.; Cha, J.Y.; Lin, Z.; Lu, M.Z.; Huang, L.J.; Kim, W.Y. Plant hormone mediated regulation of heat tolerance in response to global climate change. Front. Plant Sci. 2021, 11, 627969. [Google Scholar] [CrossRef]
- Cao, W.H.; Liu, J.; He, X.J.; Mu, R.L.; Zhou, H.L.; Chen, S.Y.; Zhang, J.S. Modulation of ethylene responses affects plant salt-stress responses. Plant Physiol. 2007, 143, 707–719. [Google Scholar] [CrossRef]
- Hattori, Y.; Nagai, K.; Furukawa, S.; Song, X.J.; Kawano, R.; Sakakibara, H.; Wu, J.; Matsumoto, T.; Yoshimura, A.; Kitano, H.; et al. The ethylene response factors snorkel1 and snorkel2 allow rice to adapt to deep water. Nature 2009, 460, 1026–1030. [Google Scholar] [CrossRef]
- Dong, H.; Zhen, Z.; Peng, J.; Chang, L.; Gong, Q.; Wang, N.N. Loss of ACS7 confers abiotic stress tolerance by modulating ABA sensitivity and accumulation in arabidopsis. J. Exp. Bot. 2011, 62, 4875–4887. [Google Scholar] [CrossRef]
- Jiang, C.; Belfield, E.J.; Cao, Y.; Smith, J.A.; Harberd, N.P. An arabidopsis soil-salinity-tolerance mutation confers ethylene-mediated enhancement of sodium potassium homeostasis. Plant Cell 2013, 25, 3535–3552. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.W.; Abel, S.; Keller, J.A.; Shen, N.F.; Theologis, A. The 1-aminocyclopropane-1-carboxylate synthase gene family of Arabidopsis-thaliana. Proc. Natl. Acad. Sci. USA 1992, 89, 11046–11050. [Google Scholar] [CrossRef] [PubMed]
- Arteca, J.M.; Arteca, R.N. A multi-responsive gene encoding 1-aminocyclopropane-1-carboxylate synthase (ACS6) in mature arabidopsis leaves. Plant Mol. Biol. 1999, 39, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Poor, P.; Nawaz, K.; Gupta, R.; Ashfaque, F.; Khan, M.I.R. Ethylene involvement in the regulation of heat stress tolerance in plants. Plant Cell Rep. 2022, 41, 675–698. [Google Scholar] [CrossRef]
- Firon, N.; Pressman, E.; Meir, S.; Khoury, R.; Altahan, L. Ethylene is involved in maintaining tomato (Solanum lycopersicum) pollen quality under heat-stress conditions. Aob Plants 2012, 2012, pls024. [Google Scholar] [CrossRef]
- Jegadeesan, S.; Beery, A.; Altahan, L.; Meir, S.; Pressman, E.; Firon, N. Ethylene production and signaling in tomato (Solanum lycopersicum) pollen grains is responsive to heat stress conditions. Plant Reprod. 2018, 31, 367–383. [Google Scholar] [CrossRef]
- Jegadeesan, S.; Chaturvedi, P.; Ghatak, A.; Pressman, E.; Meir, S.; Faigenboim, A.; Rutley, N.; Beery, A.; Harel, A.; Weckwerth, W.; et al. Proteomics of heat-stress and ethylene-mediated thermotolerance mechanisms in tomato pollen grains. Front. Plant Sci. 2018, 9, 1558. [Google Scholar] [CrossRef]
- Browne, R.G.; Li, S.F.; Iacuone, S.; Dolferus, R.; Parish, R.W. Differential responses of anthers of stress tolerant and sensitive wheat cultivars to high temperature stress. Planta 2021, 254, 4. [Google Scholar] [CrossRef]
- Wingler, A.; Purdy, S.; MacLean, J.A.; Pourtau, N. The role of sugars in integrating environmental signals during the regulation of leaf senescence. J. Exp. Bot. 2006, 57, 391–399. [Google Scholar] [CrossRef]
- Wingler, A.; Masclaux-Daubresse, C.; Fischer, A.M. Sugars, senescence, and ageing in plants and heterotrophic organisms. J. Exp. Bot. 2009, 60, 1063–1066. [Google Scholar] [CrossRef]
- Xia, X.J.; Zhou, Y.H.; Shi, K.; Zhou, J.; Foyer, C.H.; Yu, J.Q. Interplay between reactive oxygen species and hormones in the control of plant development and stress tolerance. J. Exp. Bot. 2015, 66, 2839–2856. [Google Scholar] [CrossRef] [PubMed]
- Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Ap2/ERF family transcription factors in plant abiotic stress responses. BBA-Gene Regul. Mech. 2012, 1819, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.C.; Liao, P.M.; Kuo, W.W.; Lin, T.P. The arabidopsis ethylene response factor1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol. 2013, 162, 1566–1582. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; He, R.J.; Xie, Q.L.; Zhao, X.H.; Deng, X.M.; He, J.B.; Song, L.; He, J.; Marchant, A.; Chen, X.Y.; et al. Ethylene response factor 74 (ERF74) plays an essential role in controlling a respiratory burst oxidase homolog d (RBOHd)-dependent mechanism in response to different stresses in arabidopsis. New Phytol. 2017, 213, 1667–1681. [Google Scholar] [CrossRef] [PubMed]
- Scharf, K.D.; Berberich, T.; Ebersberger, I.; Nover, L. The plant heat stress transcription factor (hsf) family: Structure, function and evolution. Biochim. Biophys. Acta 2012, 1819, 104–119. [Google Scholar] [CrossRef]
- Chan-Schaminet, K.Y.; Baniwal, S.K.; Bublak, D.; Nover, L.; Scharf, K.D. Specific interaction between tomato HsfA1 and HsfA2 creates hetero-oligomeric superactivator complexes for synergistic activation of heat stress gene expression. J. Biol. Chem. 2009, 284, 20848–20857. [Google Scholar] [CrossRef]
- Xin, H.; Zhang, H.; Chen, L.; Li, X.; Lian, Q.; Yuan, X.; Hu, X.; Cao, L.; He, X.; Yi, M. Cloning and characterization of HsfA2 from lily (Lilium longiflorum). Plant Cell Rep. 2010, 29, 875–885. [Google Scholar] [CrossRef]
- Gong, B.; Yi, J.; Wu, J.; Sui, J.; Khan, M.A.; Wu, Z.; Zhong, X.; Seng, S.; He, J.; Yi, M. LlHsfA1, a novel heat stress transcription factor in lily (Lilium longiflorum), can interact with llhsfa2 and enhance the thermotolerance of transgenic Arabidopsis thaliana. Plant Cell Rep. 2014, 33, 1519–1533. [Google Scholar] [CrossRef]
- Xin, H.B.; Zhang, H.; Zhong, X.H.; Lian, Q.L.; Dong, A.X.; Cao, L.; Yi, M.F.; Cong, R.C. Over-expression of LlHsfA2b, a lily heat shock transcription factor lacking trans-activation activity in yeast, can enhance tolerance to heat and oxidative stress in transgenic arabidopsis seedlings. Plant Cell Tiss. Org. 2017, 130, 617–629. [Google Scholar] [CrossRef]
- Schramm, F.; Larkindale, J.; Kiehlmann, E.; Ganguli, A.; Englich, G.; Vierling, E.; von Koskull-Doring, P. A cascade of transcription factor DREB2a and heat stress transcription factor HsfA3 regulates the heat stress response of arabidopsis. Plant J. 2008, 53, 264–274. [Google Scholar] [CrossRef]
- Wu, Z.; Liang, J.; Wang, C.; Zhao, X.; Zhong, X.; Cao, X.; Li, G.; He, J.; Yi, M. Overexpression of lily HsfA3s in arabidopsis confers increased thermotolerance and salt sensitivity via alterations in proline catabolism. J. Exp. Bot. 2018, 69, 2005–2021. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.S.; Li, Z.Y.; Chen, Y.; Chen, M.; Li, L.C.; Ma, Y.Z. Heat shock protein 90 in plants: Molecular mechanisms and roles in stress responses. Int. J. Mol. Sci. 2012, 13, 15706–15723. [Google Scholar] [CrossRef] [PubMed]
- Ul Haq, S.; Khan, A.; Ali, M.; Khattak, A.M.; Gai, W.X.; Zhang, H.X.; Wei, A.M.; Gong, Z.H. Heat shock proteins: Dynamic biomolecules to counter plant biotic and abiotic stresses. Int. J. Mol. Sci. 2019, 20, 5321. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.Z.; Wang, Y.; Xu, F.X.; Song, C.X.; Yang, X.; Zhang, Z.; Yi, M.F.; Ma, N.; Zhou, X.F.; He, J.N. Small hsps play an important role in crosstalk between hsf-hsp and ROS pathways in heat stress response through transcriptomic analysis in lilies (Lilium longiflorum). BMC Plant Biol. 2022, 22, 202. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Shi, Y.; Yang, S. Molecular regulation of plant responses to environmental temperatures. Mol. Plant 2020, 13, 544–564. [Google Scholar] [CrossRef]
- Gong, Z.; Xiong, L.; Shi, H.; Yang, S.; Herrera-Estrella, L.R.; Xu, G.; Chao, D.-Y.; Li, J.; Wang, P.-Y.; Qin, F.; et al. Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 2020, 63, 635–674. [Google Scholar] [CrossRef]
- Grassotti, A.; Gimelli, F. Bulb and Cut Flower Production in the Genus Lilium: Current Status and the Future; International Society for Horticultural Science (ISHS): Leuven, Belgium, 2011; pp. 21–35. [Google Scholar]
- Zhang, M.; Jiang, L.; Zhang, D.; Jia, G. De novo transcriptome characterization of lilium ‘sorbonne’ and key enzymes related to the flavonoid biosynthesis. Mol. Genet. Genom. 2015, 290, 399–412. [Google Scholar] [CrossRef]
- Xu, L.F.; Yang, P.P.; Yuan, S.X.; Feng, Y.Y.; Xu, H.; Cao, Y.W.; Ming, J. Transcriptome analysis identifies key candidate genes mediating purple ovary coloration in asiatic hybrid lilies. Int. J. Mol. Sci. 2016, 17, 1881. [Google Scholar] [CrossRef]
- Wang, C.; Zhou, Y.; Yang, X.; Zhang, B.; Xu, F.; Wang, Y.; Song, C.; Yi, M.; Ma, N.; Zhou, X.; et al. The heat stress transcription factor LlHsfA4 enhanced basic thermotolerance through regulating ros metabolism in lilies (Lilium longiflorum). Int. J. Mol. Sci. 2022, 23, 572. [Google Scholar] [CrossRef]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-wide analysis of the ERF gene family in arabidopsis and rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef]
- Liu, J.X.; Howell, S.H. BZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in arabidopsis. Plant Cell 2010, 22, 782–796. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.Y.; Zhao, X.B.; Burger, M.; Wang, Y.R.; Chory, J. Two interacting ethylene response factors regulate heat stress response. Plant Cell 2021, 33, 338–357. [Google Scholar] [CrossRef] [PubMed]
- Steed, C.L.; Harrison, M.A. Regulation of ethylene biosynthesis after short-term heat-treatment in etiolated pea stems. Physiol. Plant. 1993, 87, 103–107. [Google Scholar] [CrossRef]
- Larkindale, J.; Huang, B. Thermotolerance and antioxidant systems in agrostis stolonifera: Involvement of salicylic acid, abscisic acid, calcium, hydrogen peroxide, and ethylene. J. Plant Physiol. 2004, 161, 405–413. [Google Scholar] [CrossRef]
- Wan, L.Y.; Wu, Y.S.; Huang, J.Q.; Dai, X.F.; Lei, Y.; Yan, L.Y.; Jiang, H.F.; Zhang, J.C.; Varshney, R.K.; Liao, B.S. Identification of ERF genes in peanuts and functional analysis of ahERF008 and ahERF019 in abiotic stress response. Funct. Integr. Genom. 2014, 14, 467–477. [Google Scholar] [CrossRef]
- Klay, I.; Gouia, S.; Lu, M.; Mila, I.; Khoudi, H.; Bernadac, A.; Bouzayen, M.; Pirrello, J. Ethylene response factors (ERF) are differentially regulated by different abiotic stress types in tomato plants. Plant Sci. 2018, 274, 137–145. [Google Scholar] [CrossRef]
- Trapero-Mozos, A.; Ducreux, L.J.M.; Bita, C.E.; Morris, W.; Wiese, C.; Morris, J.A.; Paterson, C.; Hedley, P.E.; Hancock, R.D.; Taylor, M. A reversible light-and genotype-dependent acquired thermotolerance response protects the potato plant from damage due to excessive temperature. Planta 2018, 247, 1393. [Google Scholar] [CrossRef]
- Pan, C.Z.; Zhang, H.; Ma, Q.M.; Fan, F.J.; Fu, R.S.; Ahammed, G.J.; Yu, J.Q.; Shi, K. Role of ethylene biosynthesis and signaling in elevated CO2-induced heat stress response in tomato. Planta 2019, 250, 563–572. [Google Scholar] [CrossRef]
- Larkindale, J.; Knight, M.R. Protection against heat stress-induced oxidative damage in arabidopsis involves calcium, abscisic acid, ethylene, and salicylic acid. Plant Physiol. 2002, 128, 682–695. [Google Scholar] [CrossRef]
- Kieber, J.J.; Rothenberg, M.; Roman, G.; Feldmann, K.A.; Ecker, J.R. CTR1, a negative regulator of the ethylene response pathway in arabidopsis, encodes a member of the raf family of protein-kinases. Cell 1993, 72, 427–441. [Google Scholar] [CrossRef]
- Alonso, J.M.; Hirayama, T.; Roman, G.; Nourizadeh, S.; Ecker, J.R. EIN2, a bifunctional transducer of ethylene and stress responses in arabidopsis. Science 1999, 284, 2148–2152. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.Y.; Chen, Y.F.; Randlett, M.D.; Zhao, X.C.; Findell, J.L.; Kieber, J.J.; Schaller, G.E. Localization of the Raf-like kinase CTR1 to the endoplasmic reticulum of arabidopsis through participation in ethylene receptor signaling complexes. J. Biol. Chem. 2003, 278, 34725–34732. [Google Scholar] [CrossRef] [PubMed]
- Ju, C.L.; Yoon, G.M.; Shemansky, J.M.; Lin, D.Y.; Ying, Z.I.; Chang, J.H.; Garrett, W.M.; Kessenbrock, M.; Groth, G.; Tucker, M.L.; et al. CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the er membrane to the nucleus in arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 19486–19491. [Google Scholar] [CrossRef]
- Li, W.Y.; Ma, M.D.; Feng, Y.; Li, H.J.; Wang, Y.C.; Ma, Y.T.; Li, M.Z.; An, F.Y.; Guo, H.W. EIN2-directed translational regulation of ethylene signaling in arabidopsis. Cell 2015, 163, 670–683. [Google Scholar] [CrossRef] [PubMed]
- Merchante, C.; Brumos, J.; Yun, J.; Hu, Q.W.; Spencer, K.R.; Enriquez, P.; Binder, B.M.; Heber, S.; Stepanova, A.N.; Alonso, J.M. Gene-specific translation regulation mediated by the hormone-signaling molecule EIN2. Cell 2015, 163, 684–697. [Google Scholar] [CrossRef] [PubMed]
- Leclercq, J.; Adams-Phillips, L.C.; Zegzouti, H.; Jones, B.; Latche, A.; Giovannoni, J.J.; Pech, J.C.; Bouzayen, M. Lectr1, a tomato CTR1-like gene, demonstrates ethylene signaling ability in arabidopsis and novel expression patterns in tomato. Plant Physiol. 2002, 130, 1132–1142. [Google Scholar] [CrossRef]
- Yang, Z.; Tian, L.; Latoszek-Green, M.; Brown, D.; Wu, K. Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Mol. Biol. 2005, 58, 585–596. [Google Scholar] [CrossRef]
- Ma, B.; Zhou, Y.; Chen, H.; He, S.J.; Huang, Y.H.; Zhao, H.; Lu, X.; Zhang, W.K.; Pang, J.H.; Chen, S.Y.; et al. Membrane protein MHZ33 stabilizes OsEIN22 in rice by interacting with its nramp-like domain. Proc. Natl. Acad. Sci. USA 2018, 115, 2520–2525. [Google Scholar] [CrossRef]
- Ashraf, M.F.; Yang, S.; Wu, R.J.; Wang, Y.Z.; Hussain, A.; Noman, A.; Khan, M.I.; Liu, Z.Q.; Qiu, A.L.; Guan, D.Y.; et al. Capsicum annuum HSFb2a positively regulates the response to ralstonia solanacearum infection or high temperature and high humidity forming transcriptional cascade with caWRKY6 and caWRKY40. Plant Cell Physiol. 2018, 59, 2608–2623. [Google Scholar] [CrossRef]
- Shekhawat, K.; Saad, M.M.; Sheikh, A.; Mariappan, K.; Al-Mahmoudi, H.; Abdulhakim, F.; Eida, A.A.; Jalal, R.; Masmoudi, K.; Hirt, H. Root endophyte induced plant thermotolerance by constitutive chromatin modification at heat stress memory gene loci. EMBO Rep. 2021, 22, e51049. [Google Scholar] [CrossRef]
- Potuschak, T.; Lechner, E.; Parmentier, Y.; Yanagisawa, S.; Grava, S.; Koncz, C.; Genschik, P. EIN3-dependent regulation of plant ethylene hormone signaling by two arabidopsis F-box proteins: EBF1 andEBF2. Cell 2003, 115, 679–689. [Google Scholar] [CrossRef] [PubMed]
- Pei, H.X.; Ma, N.; Tian, J.; Luo, J.; Chen, J.W.; Li, J.; Zheng, Y.; Chen, X.; Fei, Z.J.; Gao, J.P. An NAC transcription factor controls ethylene-regulated cell expansion in flower petals. Plant Physiol. 2013, 163, 775–791. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.W.; Zhang, Q.; Wang, Q.G.; Feng, M.; Li, Y.; Meng, Y.L.; Zhang, Y.; Liu, G.Q.; Ma, Z.M.; Wu, H.Z.; et al. RhMKK9, a rose map kinase kinase gene, is involved in rehydration-triggered ethylene production in rose gynoecia. BMC Plant Biol. 2017, 17, 51. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; He, J.N.; Zhao, Y.; Wu, T.; Zhou, X.F.; Ding, Y.L.; Kong, L.Y.; Wang, X.J.; Wang, Y.; Li, J.G.; et al. EAR1 negatively regulates aba signaling by enhancing 2C protein phosphatase activity. Plant Cell 2018, 30, 815–834. [Google Scholar] [CrossRef] [PubMed]
- Sainsbury, F.; Thuenemann, E.C.; Lomonossoff, G.P. Peaq: Versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol. J. 2009, 7, 682–693. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, Z.C.; Feng, M.; Chen, J.W.; Qin, M.Z.; Wang, W.R.; Bao, Y.; Xu, Q.; Ye, Y.; Ma, C.; et al. The circadian-controlled PIF8-BBX28 module regulates petal senescence in rose flowers by governing mitochondrial ros homeostasis at night. Plant Cell 2021, 33, 2716–2735. [Google Scholar] [CrossRef]
- Han, Y.C.; Kuang, J.F.; Chen, J.Y.; Liu, X.C.; Xiao, Y.Y.; Fu, C.C.; Wang, J.N.; Wu, K.Q.; Lu, W.J. Banana transcription factor maERF11 recruits histone deacetylase maHDA1 and represses the expression of maACO1 and expansins during fruit ripening. Plant Physiol. 2016, 171, 1070–1084. [Google Scholar] [CrossRef]
- Guo, P.R.; Li, Z.H.; Huang, P.X.; Li, B.S.; Fang, S.; Chu, J.F.; Guo, H.W. A tripartite amplification loop involving the transcription factor WRKY75, salicylic acid, and reactive oxygen species accelerates leaf senescence. Plant Cell 2017, 29, 2854–2870. [Google Scholar] [CrossRef]
- Wei, Q.; Ma, C.; Xu, Y.J.; Wang, T.L.; Chen, Y.Y.; Lu, J.; Zhang, L.L.; Jiang, C.Z.; Hong, B.; Gao, J.P. Control of chrysanthemum flowering through integration with an aging pathway. Nat. Commun. 2017, 8, 829. [Google Scholar] [CrossRef]
- Yuan, C.; Li, C.; Yan, L.J.; Jackson, A.O.; Liu, Z.Y.; Han, C.G.; Yu, J.L.; Li, D.W. A high throughput barley stripe mosaic virus vector for virus induced gene silencing in monocots and dicots. PLoS ONE 2011, 6, e26468. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Zhou, Y.; Wang, R.; Xu, F.; Tong, S.; Song, C.; Shao, Y.; Yi, M.; He, J. Ethylene Response Factor LlERF110 Mediates Heat Stress Response via Regulation of LlHsfA3A Expression and Interaction with LlHsfA2 in Lilies (Lilium longiflorum). Int. J. Mol. Sci. 2022, 23, 16135. https://doi.org/10.3390/ijms232416135
Wang Y, Zhou Y, Wang R, Xu F, Tong S, Song C, Shao Y, Yi M, He J. Ethylene Response Factor LlERF110 Mediates Heat Stress Response via Regulation of LlHsfA3A Expression and Interaction with LlHsfA2 in Lilies (Lilium longiflorum). International Journal of Molecular Sciences. 2022; 23(24):16135. https://doi.org/10.3390/ijms232416135
Chicago/Turabian StyleWang, Yue, Yunzhuan Zhou, Rui Wang, Fuxiang Xu, Shi Tong, Cunxu Song, Yanan Shao, Mingfang Yi, and Junna He. 2022. "Ethylene Response Factor LlERF110 Mediates Heat Stress Response via Regulation of LlHsfA3A Expression and Interaction with LlHsfA2 in Lilies (Lilium longiflorum)" International Journal of Molecular Sciences 23, no. 24: 16135. https://doi.org/10.3390/ijms232416135
APA StyleWang, Y., Zhou, Y., Wang, R., Xu, F., Tong, S., Song, C., Shao, Y., Yi, M., & He, J. (2022). Ethylene Response Factor LlERF110 Mediates Heat Stress Response via Regulation of LlHsfA3A Expression and Interaction with LlHsfA2 in Lilies (Lilium longiflorum). International Journal of Molecular Sciences, 23(24), 16135. https://doi.org/10.3390/ijms232416135




