New Wolbachia pipientis Genotype Increasing Heat Stress Resistance of Drosophila melanogaster Host Is Characterized by a Large Chromosomal Inversion
Abstract
1. Introduction
2. Results and Discussion
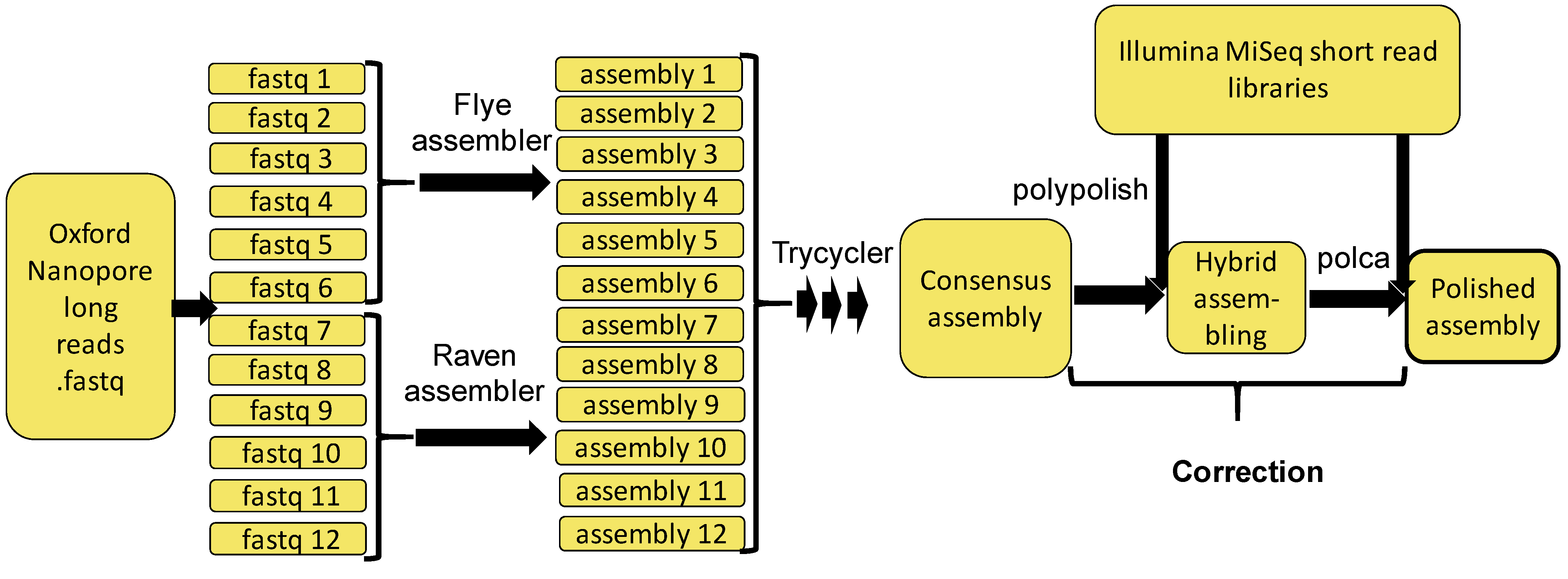
2.1. Hybrid-Assembling Genome of a Novel Wolbachia Strain
2.2. From Assembly to Annotation of Genomes
2.3. Large Chromosomal Inversion as a Probable Cause for a Phenotype of Interest
2.4. Comparative Genomic Analysis of wMel and CS Groups of Wolbachia Pipientis Strains
- 1.
- GroES/WD0308, DnaK/WD0928, Hsp90/WD1277, GroEL/WD0307: genes that show high relative expression in Drosophila embryos, with down-regulation later in the life cycle.
- 2.
- WspB/WD0009, TerC/WD0194, SPFH domain/WD0482, type II secretion/WD0500, HlyD/WD0649, type I secretion/WD0770, VirB3/WD0859, Rhoptry surface protein related/WD1041, WD0191, WD0385, WD0438, WD1213, DksA/WD1094: the up-regulated genes that increase in relative expression starting with the early larval stages and carrying on into adulthood, with decreases at the late larval (12 hr) stage and increases at the white prepupal (2 and 3 d) stages.
- 3.
- WD0291, WD0292, WD0438: genes that show up-regulation primarily in D. melanogaster adults, with higher expression in adult males relative to adult females, at the same age.
3. Materials and Methods
3.1. Drosophila Lines and Rearing
3.2. Genomic DNA Extraction and Sequencing
3.3. Genome Assembly, Polishing and Annotation Pipeline
3.4. Comparative Genome Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hosokawa, T.; Koga, R.; Kikuchi, Y.; Meng, X.-Y.; Fukatsu, T. Wolbachia as a bacteriocyte-associated nutritional mutualist. Proc. Natl. Acad. Sci. USA 2010, 107, 769–774. [Google Scholar] [CrossRef] [PubMed]
- McFall-Ngai, M.; Hadfield, M.G.; Bosch, T.C.G.; Carey, H.V.; Domazet-Lošo, T.; Douglas, A.E.; Dubilier, N.; Eberl, G.; Fukami, T.; Gilbert, S.F.; et al. Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. USA 2013, 110, 3229–3236. [Google Scholar] [CrossRef] [PubMed]
- Capobianco, F., III; Nandkumar, S.; Parker, J.D. Wolbachia affects survival to different oxidative stressors dependent upon the genetic background in Drosophila melanogaster. Physiol. Entomol. 2018, 43, 239–244. [Google Scholar] [CrossRef]
- Burdina, E.V.; Bykov, R.A.; Menshanov, P.N.; Ilinsky, Y.Y.; Gruntenko, N.E. Unique Wolbachia strain wMelPlus increases heat stress resistance in Drosophila melanogaster. Arch. Insect Biochem. Physiol. 2021, 106, e21776. [Google Scholar] [CrossRef]
- Hertig, M.; Wolbach, S.B. Studies on Rickettsia-like Micro-Organisms in Insects. J. Med. Res. 1924, 44, 329–374.7. [Google Scholar] [PubMed]
- Hertig, M. The Rickettsia, Wolbachia pipientis (gen. et sp.n.) and Associated Inclusions of the Mosquito, Culex pipiens. Parasitology 1936, 28, 453–486. [Google Scholar] [CrossRef]
- Kaur, R.; Shropshire, J.D.; Cross, K.L.; Leigh, B.; Mansueto, A.J.; Stewart, V.; Bordenstein, S.R.; Bordenstein, S.R. Living in the endosymbiotic world of Wolbachia: A centennial review. Cell Host Microbe 2021, 29, 879–893. [Google Scholar] [CrossRef]
- O’Neill, S.L. Wolbachia pipientis: Symbiont or parasite? Parasitol. Today 1995, 11, 168–169. [Google Scholar] [CrossRef]
- Burdina, E.V.; Gruntenko, N.E. Physiological Aspects of Wolbachia pipientis–Drosophila melanogaster Relationship. J. Evol. Biochem. Physiol. 2022, 58, 303–317. [Google Scholar] [CrossRef]
- Hoffmann, A.A.; Turelli, M. Cytoplasmic incompatibility in insects. Influ. Passeng. Inherit. Microorg. Arthropod Reprod. 1997, 42–80. [Google Scholar]
- Min, K.T.; Benzer, S. Wolbachia, normally a symbiont of Drosophila, can be virulent, causing degeneration and early death. Proc. Natl. Acad. Sci. USA 1997, 94, 10792–10796. [Google Scholar] [CrossRef] [PubMed]
- Harcombe, W.; Hoffmann, A.A. Wolbachia effects in Drosophila melanogaster: In search of fitness benefits. J. Invertebr. Pathol. 2004, 87, 45–50. [Google Scholar] [CrossRef]
- Chrostek, E.; Marialva, M.S.P.; Esteves, S.S.; Weinert, L.A.; Martinez, J.; Jiggins, F.M.; Teixeira, L. Wolbachia Variants Induce Differential Protection to Viruses in Drosophila melanogaster: A Phenotypic and Phylogenomic Analysis. PLoS Genet. 2013, 9, e1003896. [Google Scholar] [CrossRef] [PubMed]
- Pimentel, A.C.; Cesar, C.S.; Martins, M.; Cogni, R. The Antiviral Effects of the Symbiont Bacteria Wolbachia in Insects. Front. Immunol. 2021, 11, 626329. [Google Scholar] [CrossRef] [PubMed]
- de Crespigny, F.E.C.; Pitt, T.D.; Wedell, N. Increased male mating rate in Drosophila is associated with Wolbachia infection. J. Evol. Biol. 2006, 19, 1964–1972. [Google Scholar] [CrossRef]
- Evans, O.; Caragata, E.P.; McMeniman, C.J.; Woolfit, M.; Green, D.C.; Williams, C.R.; Franklin, C.E.; O’Neill, S.L.; McGraw, E.A. Increased locomotor activity and metabolism of Aedes aegypti infected with a life-shortening strain of Wolbachia pipientis. J. Exp. Biol. 2009, 212, 1436–1441. [Google Scholar] [CrossRef]
- Gazla, I.N.; Carracedo, M.C. Effect of intracellular Wolbachia on interspecific crosses between Drosophila melanogaster and Drosophila simulans. Genet. Mol. Res. 2009, 8, 861–869. [Google Scholar] [CrossRef]
- Okayama, K.; Katsuki, M.; Sumida, Y.; Okada, K. Costs and benefits of symbiosis between a bean beetle and Wolbachia. Anim. Behav. 2016, 119, 19–26. [Google Scholar] [CrossRef]
- Fry, A.J.; Rand, D.M. Wolbachia interactions that determine Drosophila melanogaster survival. Evolution 2002, 56, 1976–1981. [Google Scholar] [CrossRef]
- Dean, M.D. A Wolbachia-associated fitness benefit depends on genetic background in Drosophila simulans. Proc. R. Soc. B Biol. Sci. 2006, 273, 1415–1420. [Google Scholar] [CrossRef]
- Riegler, M.; Sidhu, M.; Miller, W.J.; O’Neill, S.L. Evidence for a global Wolbachia replacement in Drosophila melanogaster. Curr. Biol. 2005, 15, 1428–1433. [Google Scholar] [CrossRef] [PubMed]
- Ilinsky, Y. Coevolution of Drosophila melanogaster mtDNA and Wolbachia Genotypes. PLoS ONE 2013, 8, e54373. [Google Scholar] [CrossRef] [PubMed]
- Woolfit, M.; Iturbe-Ormaetxe, I.; Brownlie, J.C.; Walker, T.; Riegler, M.; Seleznev, A.; Popovici, J.; Rancès, E.; Wee, B.A.; Pavlides, J.; et al. Genomic Evolution of the Pathogenic Wolbachia Strain, wMelPop. Genome Biol. Evol. 2013, 5, 2189–2204. [Google Scholar] [CrossRef] [PubMed]
- Hague, M.T.J.; Shropshire, J.D.; Caldwell, C.N.; Statz, J.P.; Stanek, K.A.; Conner, W.R.; Cooper, B.S. Temperature effects on cellular host-microbe interactions explain continent-wide endosymbiont prevalence. Curr. Biol. 2022, 32, 878–888.e8. [Google Scholar] [CrossRef]
- Iturbe-Ormaetxe, I.; Riegler, M.; O’Neill, S.L. New names for old strains? Wolbachia wSim is actually wRi. Genome Biol. 2005, 6, 401. [Google Scholar] [CrossRef][Green Version]
- Chrostek, E.; Teixeira, L. Mutualism Breakdown by Amplification of Wolbachia Genes. PLoS Biol. 2015, 13, e1002065. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef]
- Cozzarelli, N.R. DNA gyrase and the supercoiling of DNA. Science 1980, 207, 953–960. [Google Scholar] [CrossRef]
- De Palmenaer, D.; Siguier, P.; Mahillon, J. IS4 family goes genomic. BMC Evol. Biol. 2008, 8, 18. [Google Scholar] [CrossRef]
- Midonet, C.; Barre, F.-X. Xer Site-Specific Recombination: Promoting Vertical and Horizontal Transmission of Genetic Information. Microbiol. Spectr. 2014, 2, 163–182. [Google Scholar] [CrossRef]
- Daveran-Mingot, M.L.; Campo, N.; Ritzenthaler, P.; Le Bourgeois, P. A natural large chromosomal inversion in Lactococcus lactis is mediated by homologous recombination between two insertion sequences. J. Bacteriol. 1998, 180, 4834–4842. [Google Scholar] [CrossRef] [PubMed]
- Badia, J.; Ibanez, E.; Sabate, M.; Baldoma, L.; Aguilar, J. A rare 920-kilobase chromosomal inversion mediated by IS1 transposition causes constitutive expression of the yiaK-S operon for carbohydrate utilization in Escherichia coli. J. Biol. Chem. 1998, 273, 8376–8381. [Google Scholar] [CrossRef] [PubMed]
- Umeda, M.; Ohtsubo, E. Mapping of insertion element IS5 in the Escherichia coli K-12 chromosome. Chromosomal rearrangements mediated by IS5. J. Mol. Biol. 1990, 213, 229–237. [Google Scholar] [CrossRef]
- Rocha, E.P. Order and disorder in bacterial genomes. Curr. Opin. Microbiol. 2004, 7, 519–527. [Google Scholar] [CrossRef] [PubMed]
- Le, V.V.H.; León-Quezada, R.I.; Biggs, P.J.; Rakonjac, J. A large chromosomal inversion affects antimicrobial sensitivity of Escherichia coli to sodium deoxycholate. Microbiology 2022, 168, 001232. [Google Scholar] [CrossRef] [PubMed]
- Achaz, G.; Coissac, E.; Netter, P.; Rocha, E.P.C. Associations Between Inverted Repeats and the Structural Evolution of Bacterial Genomes. Genetics 2003, 164, 1279–1289. [Google Scholar] [CrossRef]
- Duarte, E.H.; Carvalho, A.; López-Madrigal, S.; Costa, J.; Teixeira, L. Forward genetics in Wolbachia: Regulation of Wolbachia proliferation by the amplification and deletion of an addictive genomic island. PLoS Genet. 2021, 17, e1009612. [Google Scholar] [CrossRef]
- Lindsey, A.R.I. Sensing, signaling, and secretion: A review and analysis of systems for regulating host interaction in Wolbachia. Genes 2020, 11, 813. [Google Scholar] [CrossRef]
- Mayoral, J.G.; Hussain, M.; Joubert, D.A.; Iturbe-Ormaetxe, I.; O’Neill, S.L.; Asgari, S. Wolbachia small noncoding RNAs and their role in cross-kingdom communications. Proc. Natl. Acad. Sci. USA 2014, 111, 18721–18726. [Google Scholar] [CrossRef]
- Ferree, P.M.; Frydman, H.M.; Li, J.M.; Cao, J.; Wieschaus, E.; Sullivan, W. Wolbachia Utilizes Host Microtubules and Dynein for Anterior Localization in the Drosophila Oocyte. PLoS Pathog. 2005, 1, e14. [Google Scholar] [CrossRef]
- Gutzwiller, F.; Carmo, C.R.; Miller, D.E.; Rice, D.W.; Newton, I.L.G.; Hawley, R.S.; Teixeira, L.; Bergman, C.M. Dynamics of Wolbachia pipientis Gene Expression across the Drosophila melanogaster Life Cycle. G3 Genes Genomes Genet. 2015, 5, 2843–2856. [Google Scholar] [CrossRef]
- French, S. Consequences of Replication Fork Movement through Transcription Units in Vivo. Science 1992, 258, 1362–1365. [Google Scholar] [CrossRef]
- Merrikh, C.N.; Merrikh, H. Gene inversion potentiates bacterial evolvability and virulence. Nat. Commun. 2018, 9, 4662. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N. Interruptions in gene expression drive highly expressed operons to the leading strand of DNA replication. Nucleic Acids Res. 2005, 33, 3224–3234. [Google Scholar] [CrossRef] [PubMed]
- Rocha, E.P. Is there a role for replication fork asymmetry in the distribution of genes in bacterial genomes? Trends Microbiol. 2002, 10, 393–395. [Google Scholar] [CrossRef]
- Lopez, P.; Philippe, H. Composition strand asymmetries in prokaryotic genomes: Mutational bias and biased gene orientation. Comptes Rendus L’académie Sci. Ser. III-Sci. Vie 2001, 324, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Said, I.; Byrne, A.; Serrano, V.; Cardeno, C.; Vollmers, C.; Corbett-Detig, R. Linked genetic variation and not genome structure causes widespread differential expression associated with chromosomal inversions. Proc. Natl. Acad. Sci. USA 2018, 115, 5492–5497. [Google Scholar] [CrossRef]
- Lato, D.F.; Zeng, Q.; Golding, G.B. Genomic inversions in Escherichia coli alter gene expression and are associated with nucleoid protein binding sites. Genome 2022, 65, 287–299. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Neoh, H.M.; Iwamoto, A.; Hiramatsu, K. Coordinated phenotype switching with large-scale chromosome flip-flop inversion observed in bacteria. Proc. Natl. Acad. Sci. USA 2012, 109, 5–9. [Google Scholar] [CrossRef]
- Wellenreuther, M.; Bernatchez, L. Eco-Evolutionary Genomics of Chromosomal Inversions. Trends Ecol. Evol. 2018, 33, 427–440. [Google Scholar] [CrossRef]
- Hoffman, A.; Sgro, C.; Weeks, A. Chromosomal inversion polymorphisms and adaptation. Trends Ecol. Evol. 2004, 19, 482–488. [Google Scholar] [CrossRef] [PubMed]
- Zivanovic, Y.; Lopez, P.; Philippe, H.; Forterre, P. Pyrococcus genome comparison evidences chromosome shuffling-driven evolution. Nucleic Acids Res. 2002, 30, 1902–1910. [Google Scholar] [CrossRef] [PubMed]
- Eisen, J.A.; Heidelberg, J.F.; White, O.; Salzberg, S.L. Evidence for symmetric chromosomal inversions around the replication origin in bacteria. Genome Biol. 2000, 1, research0011.1. [Google Scholar] [CrossRef] [PubMed]
- Van de Putte, P.; Goosen, N. DNA inversions in phages and bacteria. Trends Genet. 1992, 8, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Andreenkova, O.V.; Shishkina, O.D.; Klimenko, A.I.; Korenskaia, A.E.; Bobrovskikh, M.A.; Shatskaya, N.V.; Vasiliev, G.V.; Gruntenko, N.E. Easy and Effective Method for Extracting and Purifying Wolbachia Genomic DNA. Int. J. Mol. Sci. 2022, 23, 15315. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Judd, L.M.; Cerdeira, L.T.; Hawkey, J.; Méric, G.; Vezina, B.; Wyres, K.L.; Holt, K.E. Trycycler: Consensus long-read assemblies for bacterial genomes. Genome Biol. 2021, 22, 266. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Vaser, R.; Šikić, M. Time- and memory-efficient genome assembly with Raven. Nat. Comput. Sci. 2021, 1, 332–336. [Google Scholar] [CrossRef]
- Wick, R.R.; Holt, K.E. Polypolish: Short-read polishing of long-read bacterial genome assemblies. PLoS Comput. Biol. 2022, 18, e1009802. [Google Scholar] [CrossRef]
- Zimin, A.V.; Salzberg, S.L. The genome polishing tool POLCA makes fast and accurate corrections in genome assemblies. PLoS Comput. Biol. 2020, 16, e1007981. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Manni, M.; Berkeley, M.R.; Seppey, M.; Zdobnov, E.M. BUSCO: Assessing Genomic Data Quality and Beyond. Curr. Protoc. 2021, 1, e323. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Taboada, B.; Estrada, K.; Ciria, R.; Merino, E. Operon-mapper: A web server for precise operon identification in bacterial and archaeal genomes. Bioinformatics 2018, 34, 4118–4120. [Google Scholar] [CrossRef]
- Hallgren, J.; Tsirigos, K.D.; Damgaard Pedersen, M.; Juan, J.; Armenteros, A.; Marcatili, P.; Nielsen, H.; Krogh, A.; Winther, O. DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. bioRxiv 2022. [Google Scholar]
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar] [CrossRef]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Afgan, E.; Baker, D.; Batut, B.; Van Den Beek, M.; Bouvier, D.; Ech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Sun, L.V.; Vamathevan, J.; Riegler, M.; Deboy, R.; Brownlie, J.C.; McGraw, E.A.; Martin, W.; Esser, C.; Ahmadinejad, N.; et al. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: A streamlined genome overrun by mobile genetic elements. PLoS Biol. 2004, 2, 327–341. [Google Scholar] [CrossRef] [PubMed]
- Genin, S.; Denny, T.P. Pathogenomics of the ralstonia solanacearum species complex. Annu. Rev. Phytopathol. 2012, 50, 67–89. [Google Scholar] [CrossRef] [PubMed]
- Hague, M.T.J.; Caldwell, C.N.; Cooper, B.S. Pervasive effects of wolbachia on host temperature preference. mBio 2020, 11, e01768-20. [Google Scholar] [CrossRef]
- Hague, M.T.J.; Woods, H.A.; Cooper, B.S. Pervasive effects of Wolbachia on host activity. Biol. Lett. 2021, 17, 20210052. [Google Scholar] [CrossRef]
- Darling, A.E.; Mau, B.; Perna, N.T. Progressivemauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 2010, 5, e11147. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef]


| Genome Statistics | wMelPlus_TryCycler | wMelPlus_Polypolish | wMelPlus_Final | wMelCS_TryCycler | wMelCS_Polypolish | wMelCS_Final | wMelCS_ b (Reference) |
|---|---|---|---|---|---|---|---|
| Total length | 1,266,704 | 1,267,811 | 1,267,850 | 1,266,679 | 1,267,816 | 1,267,849 | 1,267,843 |
| GC content (%) | 35.26 | 35.23 | 35.23 | 35.26 | 35.23 | 35.23 | 35.23 |
| Duplication ratio | 0.999 | 1 | 1 | 0.999 | 1 | 1 | N/A |
| misassemblies | 2 | 2 | 2 | 0 | 0 | 0 | 0 |
| mismatches per 100 kbp | 0.95 | 0.47 | 0.95 | 0.63 | 0.39 | 0.39 | N/A |
| indels per 100 kbp | 89.92 | 4.1 | 1.03 | 92.2 | 3.71 | 1.1 | 0 |
| Complete and single-copy BUSCOs | 232 | 361 | 361 | 234 | 361 | 361 | 361 |
| Complete and duplicated BUSCOs | 0 | 1 | 1 | 0 | 1 | 1 | 1 |
| Fragmented BUSCOs | 80 | 0 | 0 | 82 | 0 | 0 | 0 |
| Missing BUSCOs | 52 | 2 | 2 | 48 | 2 | 2 | 2 |
| Number of Genes in the Orthogroup wMelCS112 | Number of Genes in the Orthogroup wMelPlus | Locus_tags in the wMelCS112 Genome | Product | Length (Aminoacid Residues) |
|---|---|---|---|---|
| 1 | 0 | WMELCS112_00485 | Hypothetical | 39 |
| 3 | 2 | WMELCS112_00749, WMELCS112_00771, WMELCS112_00896 | Hypothetical | 58 |
| Position | Impact | Gene | Gene Locus_tag (wMelCS112/wMelPlus) | Product | Mutation | Substitution Type |
|---|---|---|---|---|---|---|
| 104,567 | Synonymous variant | gyrB | WMELCS112_00114/ WMELPLUS_00114 | DNA gyrase subunit B | Ala465Ala | |
| 108,911 | Stop gained | WMELCS112_00118/ WMELPLUS_00118 | IS4 family transposase | Glu443 * | Nonsense change | |
| 256,666 | Missense variant | WMELCS112_00283/ WMELPLUS_00283 | Hypothetical protein | Asn36Asp | Neutral AA is changed to acidic AA | |
| 396,812 | Missense variant | WMELCS112_00444 /WMELPLUS_00458 | Hypothetical protein | Glu616Lys | Acidic AA is changed to basic AA | |
| 624,101 | Missense variant | WMELCS112_0065/ WMELPLUS_00654 | Hypothetical protein | Ala619Val | Same class/polarity/charge substitution | |
| 726,291 | Stop loss and splice-region variant | xerD_1 | WMELCS112_00784/ WMELPLUS_00783 | Tyrozine recombinase XerD | Ter310Glu | Gene extension |
| 1,212,940 | Synonymous variant | WMELCS112_01313/ WMELPLUS_01311 | Gly491Gly |
| Strain | Leading Strand (+) | Lagging Strand (−) | Total Number of CDS | GSBp (%) |
|---|---|---|---|---|
| wMelPop | 684 | 620 | 1304 | 52.45399 |
| wMelPop2 | 684 | 620 | 1304 | 52.45399 |
| wMelPlus | 619 | 646 | 1265 | 48.93281 |
| wMelCS112 | 652 | 615 | 1267 | 51.46014 |
| wMelCS_b | 650 | 618 | 1268 | 51.26183 |
| wMelOctoless | 638 | 611 | 1249 | 51.08086 |
| wMel (GCF_000008025.1) | 633 | 638 | 1271 | 49.8033 |
| wMel (GCF_016584425.1) | 630 | 639 | 1269 | 49.64539 |
| Accession | Isolate/Strain | Assembly Level | Group | Phenotype |
|---|---|---|---|---|
| GCF_000008025.1 | wMel | Complete Genome | wMel | [72] |
| GCF_000475015.1 | wMelPop | Scaffold | wMelCS | It over-replicates, which causes severe life-shortening of its host [73] |
| GCF_014354335.1 | wMelCS | Scaffold | wMelCS | [74] |
| GCF_014354345.1 | wMel | Scaffold | wMel | [74] |
| GCF_016584325.1 | wMelpop | Complete Genome | wMelCS | Causes early death of the host (carries additional copies of Octomom region) [37] |
| GCF_016584355.1 | wMelPop2 | Complete Genome | wMelCS | Causes early death of the host through over-replication (carries additional copies of Octomom region) [37] |
| GCF_016584375.1 | wMelOctoless | Complete Genome | wMelCS | Causes early death of the host through over-replication (the Octoless region is absent) [37] |
| GCF_016584405.1 | wMelCS_b | Complete Genome | wMelCS | [37] |
| GCF_016584425.1 | wMel | Complete Genome | wMel | [37] |
| GCF_017916155.1 | FFD25 | Scaffold | wMelCS | [75] |
| GCF_021347805.1 | wMel_Trop | Scaffold | wMel | Sampled from a tropical climate [24] |
| GCF_021347845.1 | wMel_Temp | Scaffold | wMel | Sampled from a temperate climate [24] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Korenskaia, A.E.; Shishkina, O.D.; Klimenko, A.I.; Andreenkova, O.V.; Bobrovskikh, M.A.; Shatskaya, N.V.; Vasiliev, G.V.; Gruntenko, N.E. New Wolbachia pipientis Genotype Increasing Heat Stress Resistance of Drosophila melanogaster Host Is Characterized by a Large Chromosomal Inversion. Int. J. Mol. Sci. 2022, 23, 16212. https://doi.org/10.3390/ijms232416212
Korenskaia AE, Shishkina OD, Klimenko AI, Andreenkova OV, Bobrovskikh MA, Shatskaya NV, Vasiliev GV, Gruntenko NE. New Wolbachia pipientis Genotype Increasing Heat Stress Resistance of Drosophila melanogaster Host Is Characterized by a Large Chromosomal Inversion. International Journal of Molecular Sciences. 2022; 23(24):16212. https://doi.org/10.3390/ijms232416212
Chicago/Turabian StyleKorenskaia, Aleksandra E., Olga D. Shishkina, Alexandra I. Klimenko, Olga V. Andreenkova, Margarita A. Bobrovskikh, Natalja V. Shatskaya, Gennady V. Vasiliev, and Nataly E. Gruntenko. 2022. "New Wolbachia pipientis Genotype Increasing Heat Stress Resistance of Drosophila melanogaster Host Is Characterized by a Large Chromosomal Inversion" International Journal of Molecular Sciences 23, no. 24: 16212. https://doi.org/10.3390/ijms232416212
APA StyleKorenskaia, A. E., Shishkina, O. D., Klimenko, A. I., Andreenkova, O. V., Bobrovskikh, M. A., Shatskaya, N. V., Vasiliev, G. V., & Gruntenko, N. E. (2022). New Wolbachia pipientis Genotype Increasing Heat Stress Resistance of Drosophila melanogaster Host Is Characterized by a Large Chromosomal Inversion. International Journal of Molecular Sciences, 23(24), 16212. https://doi.org/10.3390/ijms232416212







