Molecular Mechanisms Underlying Metabolic Resistance to Cyflumetofen and Bifenthrin in Tetranychus urticae Koch on Cowpea
Abstract
1. Introduction
2. Results
2.1. Selection of the Cyflumetofen- and Bifenthrin-Resistant Strains
2.2. The Enzyme Activity in the Cyflumetofen- and Bifenthrin-Resistant Strains
2.3. Transcriptome Sequencing, Data Processing and Differential Gene Expression Analysis
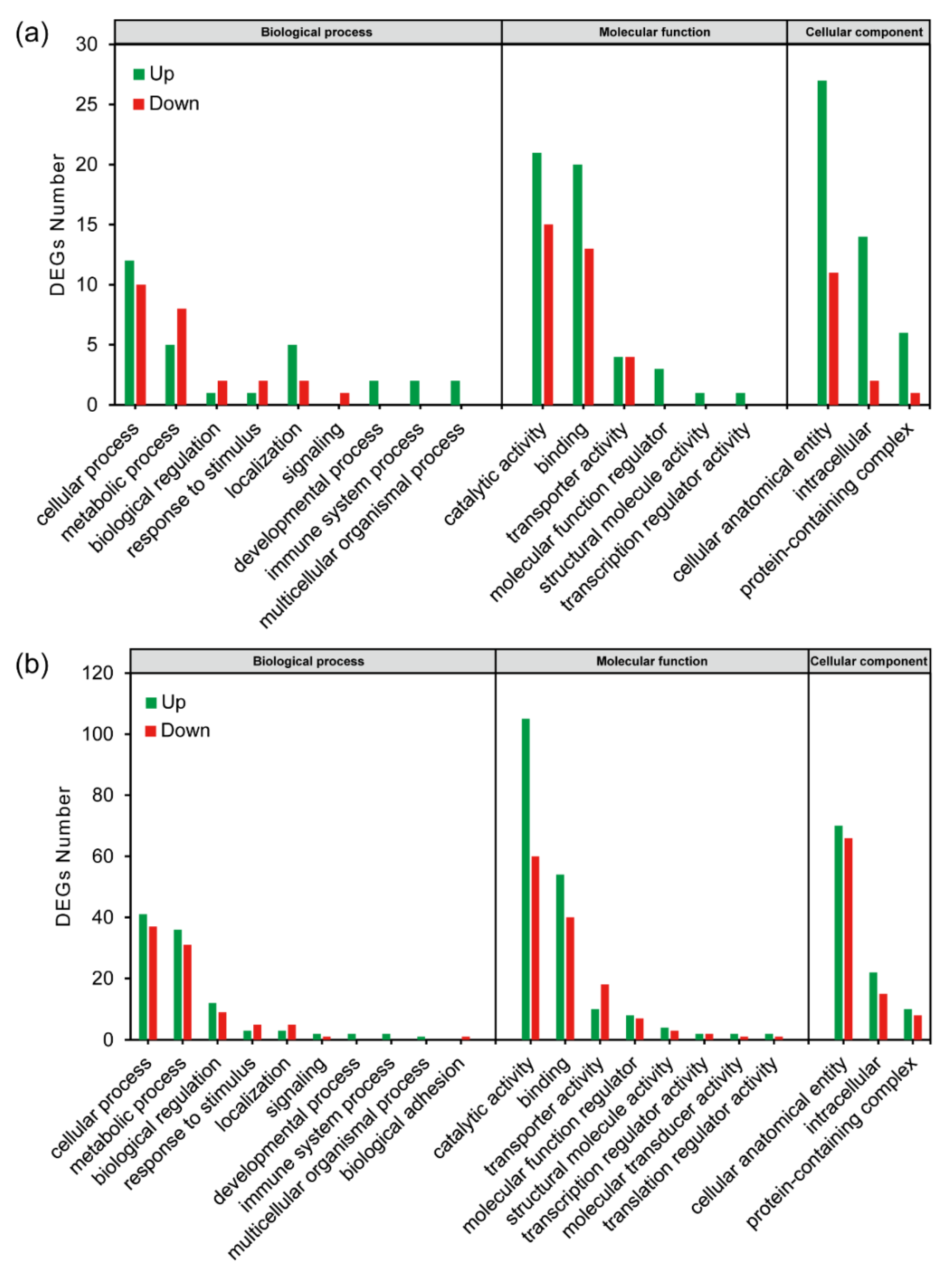
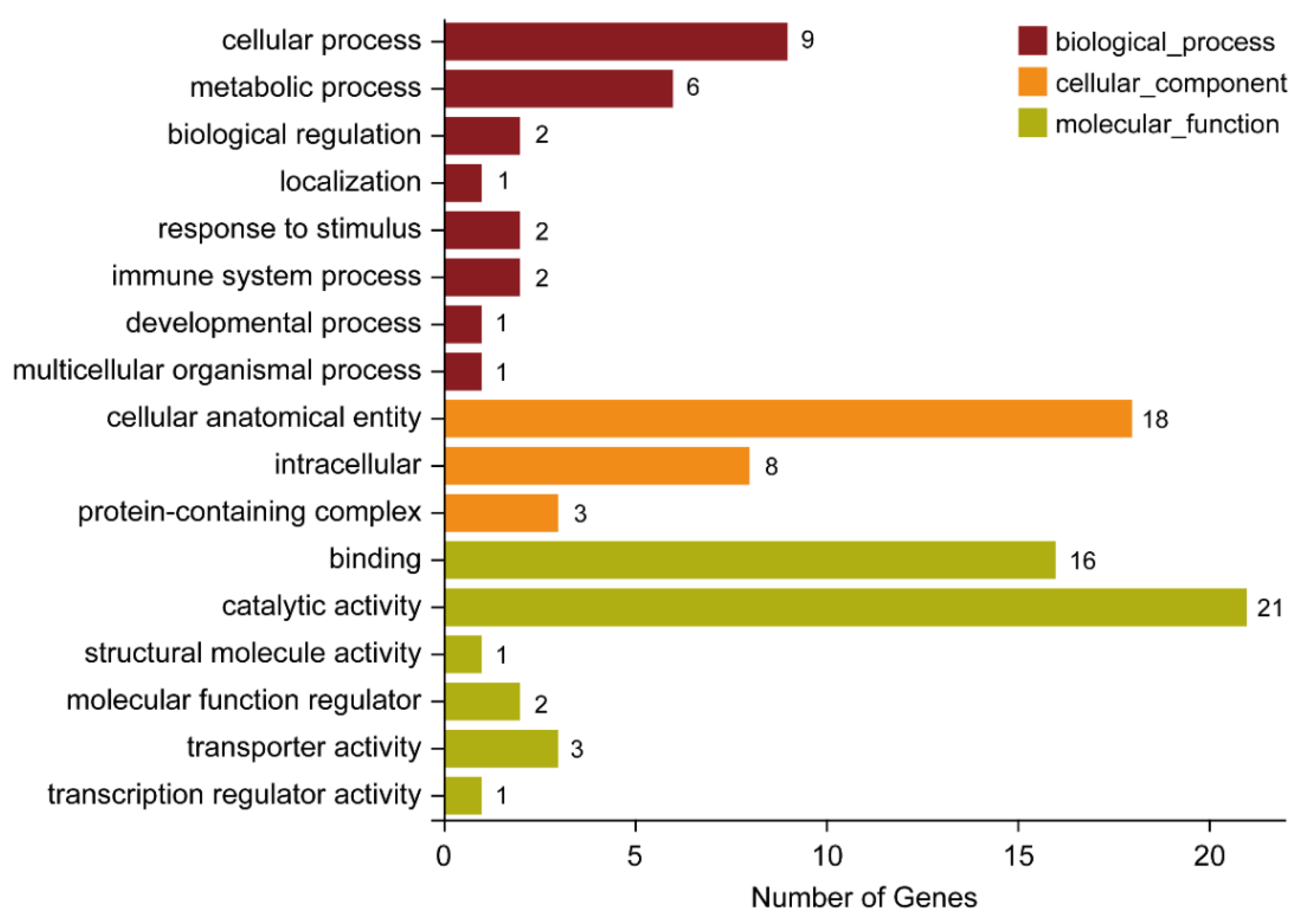
2.4. Functional Annotation
2.4.1. Functional Annotation of the Lab_SS vs R_cfm Group
2.4.2. Functional Annotation of the Lab_SS vs R_bft Group
2.4.3. Functional Annotation of the Common DEGs between the Lab_SS vs R_cfm Group and the Lab_SS vs R_bft Group
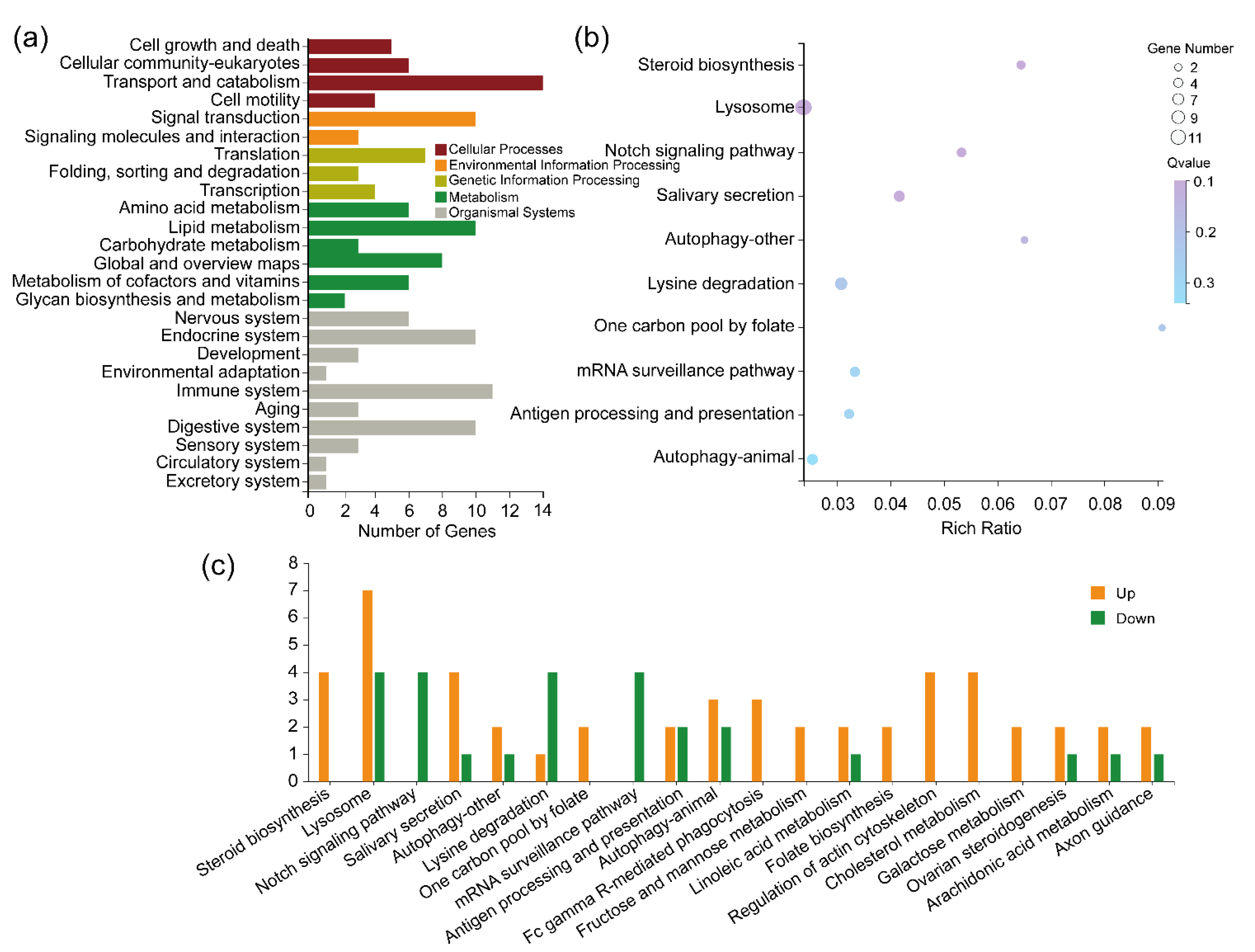
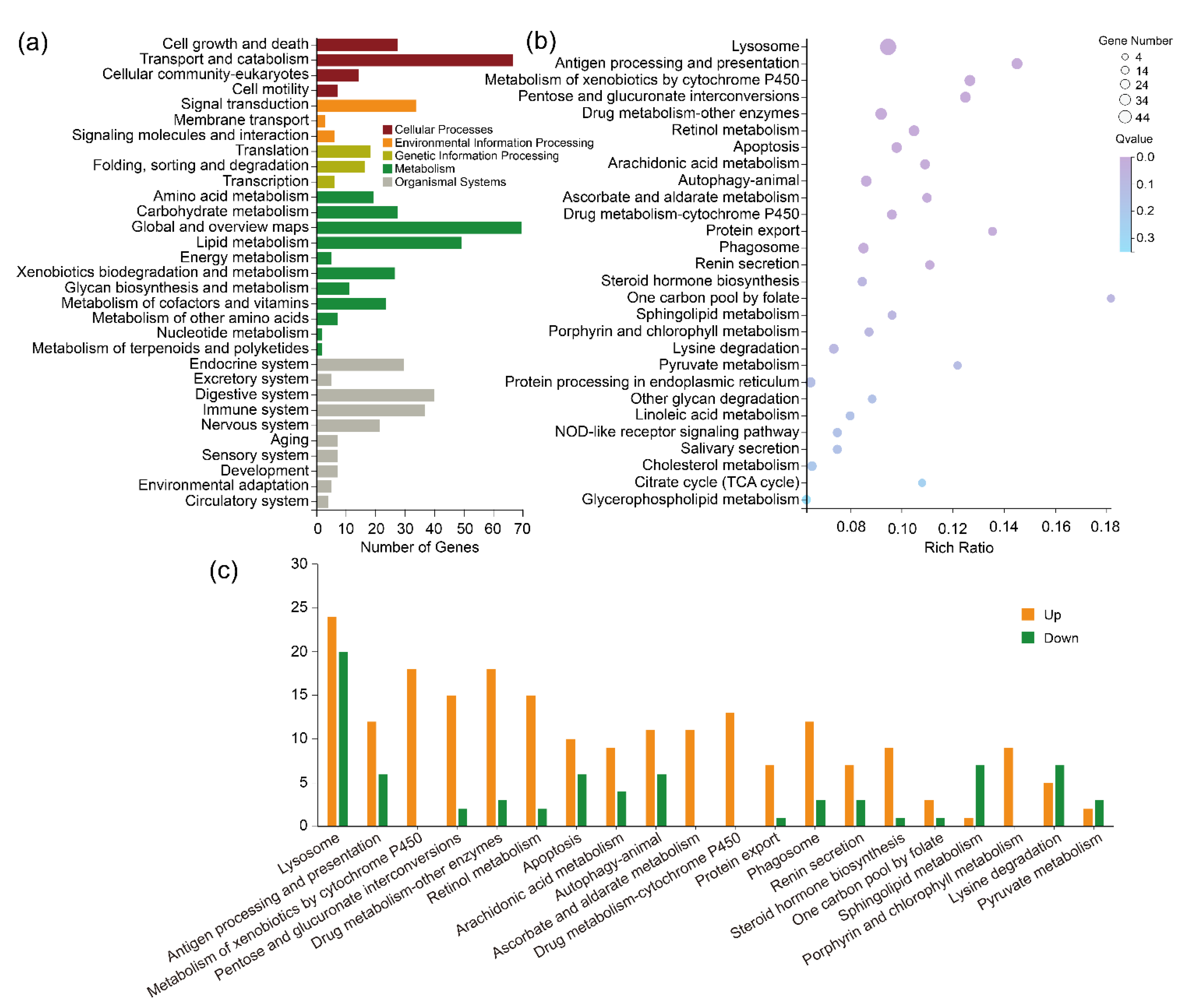
2.5. Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Analysis
2.5.1. KEGG Pathway Analysis of the Lab_SS vs R_cfm Group
2.5.2. KEGG Pathway Analysis of the Lab_SS vs R_bft Group
2.5.3. KEGG Pathway Analysis of the Common DEGs between the Lab_SS vs R_cfm Group and the Lab_SS vs R_bft Group
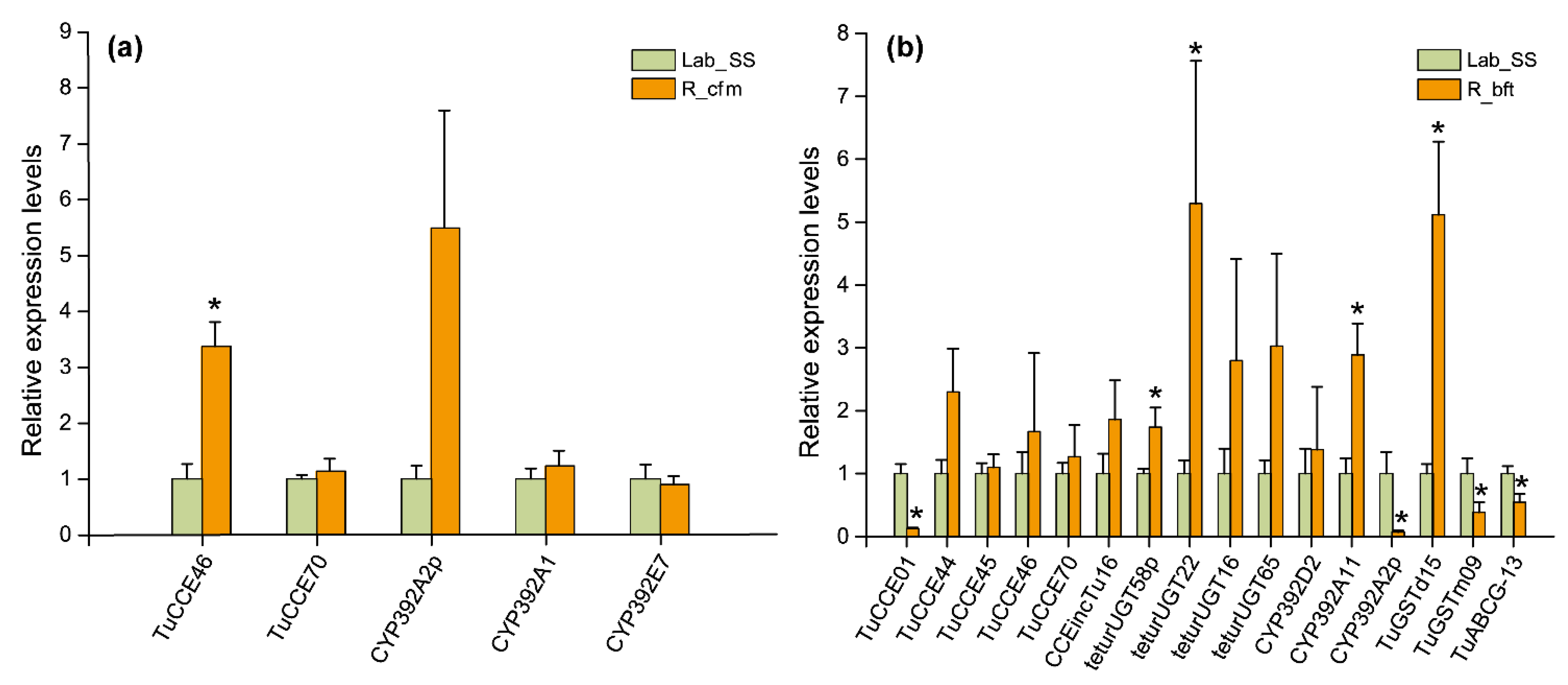
2.6. Selected DEGs of Detoxification Enzymes of Lab_SS vs R_cfm and Lab_SS vs R_bft
2.7. Validation of Detoxification Enzyme Genes
3. Discussion
4. Materials and Methods
4.1. Mites
4.2. Reagents
4.3. Resistance Selection
4.4. Bioassays of Cyflumetofen- and Bifenthrin-Resistant Strains of T. urticae
4.5. Determination of the Activity of the Detoxification Enzymes UGTs, GSTs, CarEs and P450s
4.6. Transcriptome Sequencing
4.6.1. Total RNA Extraction, Library Construction and RNA-Seq
4.6.2. Transcriptome Sequencing Data Analysis
4.6.3. Transcriptome Sequencing Data Analysis
4.7. Quantitative Real-Time PCR (qRT-PCR) Analysis
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Migeon, A.; Nouguier, E.; Dorkeld, F. Spider mites web: A comprehensive database for the Tetranychidae. In Trends in Acarology; Sabelis, M.W., Bruin, J., Eds.; Springer: Dordrecht, The Netherlands; Heidelberg, Germany; London, UK; New York, NY, USA, 2010; pp. 557–560. [Google Scholar]
- Adesanya, A.W.; Lavine, M.D.; Moural, T.W.; Lavine, L.C.; Zhu, F.; Walsh, D.B. Mechanisms and management of acaricide resistance for Tetranychus urticae in agroecosystems. J. Pest Sci. 2021, 94, 639–663. [Google Scholar] [CrossRef]
- Padilha, G.; Fiorin, R.A.; Cargnelutti Filho, A.; Pozebon, H.; Rogers, J.; Marques, R.P.; Castilhos, L.B.; Donatti, A.; Stefanelo, L.; Burtet, L.M.; et al. Damage assessment and economic injury level of the two-spotted spider mite Ttetranychus urticae in soybean. Pesq. Agropec. Bras. 2020, 55, e01836. [Google Scholar] [CrossRef]
- Ndiaye, B.; Joutei, A.B.; Lahlali, R. Managing spider mites in corn: A review. Moroccan. J. Agr. Sci. 2022, 1, 19–28. [Google Scholar]
- Sood, A.K.; Pathania, N.; Sharma, P. Annual Progress Report of RKVY Project: Technological Interventions for Protected Cultivation of Vegetable Crops in Himachal Pradesh; CSK Himachal Pradesh Krishi Vishvavidyalya: Palampur, India, 2015. [Google Scholar]
- Ferreira, C.B.S.; Andrade, F.H.N.; Rodrigues, A.R.S.; Siqueira, H.A.A.; Gondim, M.G.C. Resistance in field populations of Tetranychus urticae to acaricides and characterization of the inheritance of abamectin resistance. Crop Prot. 2015, 67, 77–83. [Google Scholar] [CrossRef]
- Campbell, L.; Euston, S.R.; Ahmed, M.A. Effect of addition of thermally modified cowpea protein on sensory acceptability and textural properties of wheat bread and sponge cake. Food Chem. 2016, 194, 1230–1237. [Google Scholar] [CrossRef]
- Adu, M.O.; Asare, P.A.; Yawson, D.O.; Dzidzienyo, D.K.; Nyadanu, D.; Asare-Bediako, E.; Afutu, E.; Tachie-Menson, J.W.; Amoah, M.N. Identifying key contributing root system traits to genetic diversity in field-grown cowpea (Vigna unguiculata L. Walp.) genotypes. Field Crop Res. 2019, 232, 106–118. [Google Scholar] [CrossRef]
- Grbić, M.; Van Leeuwen, T.; Clark, R.M.; Rombauts, S.; Rouzé, P.; Grbić, V.; Osborne, E.J.; Dermauw, W.; Thi Ngoc, P.C.; Ortego, F.; et al. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature 2011, 479, 487–492. [Google Scholar] [CrossRef]
- Van Leeuwen, T.; Tirry, L.; Yamamoto, A.; Nauen, R.; Dermauw, W. The economic importance of acaricides in the control of phytophagous mites and an update on recent acaricide mode of action research. Pestic. Biochem. Phys. 2015, 121, 12–21. [Google Scholar] [CrossRef]
- Hayashi, N.; Sasama, Y.; Takahashi, N.; Ikemi, N. Cyflumetofen, a novel acaricide-its mode of action and selectivity. Pest Manag. Sci. 2013, 69, 1080–1084. [Google Scholar] [CrossRef]
- Pavlidi, N.; Khalighi, M.; Myridakis, A.; Dermauw, W.; Wybouw, N.; Tsakireli, D.; Stephanou, E.G.; Labrou, N.E.; Vontas, J.; Van Leeuwen, T. A glutathione-S-transferase (TuGSTd05) associated with acaricide resistance in Tetranychus urticae directly metabolizes the complex II inhibitor cyflumetofen. Insect Biochem. Molec. 2017, 80, 101–115. [Google Scholar] [CrossRef]
- Van Leeuwen, T.; Dermauw, W. The molecular evolution of xenobiotic metabolism and resistance in chelicerate mites. Annu. Rev. Entomol. 2016, 61, 475–498. [Google Scholar] [CrossRef] [PubMed]
- Wei, P.; Chen, M.; Nan, C.; Feng, K.; Shen, G.; Cheng, J.; He, L. Downregulation of carboxylesterase contributes to cyflumetofen resistance in Tetranychus cinnabarinus (Boisduval). Pest Manag. Sci. 2019, 75, 2166–2173. [Google Scholar] [CrossRef] [PubMed]
- Khalighi, M.; Tirry, L.; Van Leeuwen, T. Cross-resistance risk of the novel complex II inhibitors cyenopyrafen and cyflumetofen in resistant strains of the two-spotted spider mite Tetranychus urticae. Pest Manag. Sci. 2014, 70, 365–368. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, N.; Takahashi, A.; Ihara, R.; Itoh, Y.; Jouraku, A.; Van Leeuwen, T.; Osakabe, M. QTL mapping using microsatellite linkage reveals target-site mutations associated with high levels of resistance against three mitochondrial complex II inhibitors in Tetranychus urticae. Insect Biochem. Molec. 2020, 123, 103410. [Google Scholar] [CrossRef] [PubMed]
- Susurluk, H.; Gürkan, M.O. Mode of inheritance and biochemical mechanisms underlying lambda-cyhalothrin and bifenthrin resistance in the laboratory-selected two-spotted spider mite, Tetranychus urticae. Crop Prot. 2020, 137, 105280. [Google Scholar] [CrossRef]
- Tsagkarakou, A.; Van Leeuwen, T.; Khajehali, J.; Ilias, A.; Grispou, M.; Williamson, M.S.; Tirry, L.; Vontas, J. Identification of pyrethroid resistance associated mutations in the para sodium channel of the two-spotted spider mite Tetranychus urticae (Acari: Tetranychidae). Insect Mol. Biol. 2009, 18, 583–593. [Google Scholar] [CrossRef]
- Van Leeuwen, T.; Tirry, L. Esterase-mediated bifenthrin resistance in a multiresistant strain of the two-spotted spider mite, Tetranychus urticae. Pest Manag. Sci. 2007, 63, 150–156. [Google Scholar] [CrossRef]
- Ay, R.; Gürkan, M.O. Resistance to bifenthrin and resistance mechanisms of different strains of the two-spotted spider mite (Tetranychus urticae) from turkey. Phytoparasitica 2005, 33, 237–244. [Google Scholar] [CrossRef]
- Xu, D.; Zhang, Y.; Zhang, Y.; Wu, Q.; Guo, Z.; Xie, W.; Zhou, X.; Wang, S. Transcriptome profiling and functional analysis suggest that the constitutive overexpression of four cytochrome P450s confers resistance to abamectin in Tetranychus urticae from China. Pest Manag. Sci. 2021, 77, 1204–1213. [Google Scholar] [CrossRef]
- Papapostolou, K.M.; Riga, M.; Charamis, J.; Skoufa, E.; Souchlas, V.; Ilias, A.; Dermauw, W.; Ioannidis, P.; Van Leeuwen, T.; Vontas, J. Identification and characterization of striking multiple-insecticide resistance in a Tetranychus urticae field population from Greece. Pest Manag. Sci. 2021, 77, 666–676. [Google Scholar] [CrossRef]
- Liu, J.; Jiang, Z.; Feng, K.; Lu, W.; Wen, X.; Sun, J.; Li, J.; Liu, J.; He, L. Transcriptome analysis revealed that multiple genes were related to the cyflumetofen resistance of Tetranychus cinnabarinus (Boisduval). Pestic. Biochem. Phys. 2021, 173, 104799. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.; Ou, S.; Zhang, P.; Wen, X.; Shi, L.; Yang, Y.; Hu, Y.; Zhang, Y.; Shen, G.; Xu, Z.; et al. The cytochrome P450 CYP389C16 contributes to the cross-resistance between cyflumetofen and pyridaben in Tetranychus cinnabarinus (Boisduval). Pest Manag. Sci. 2020, 76, 665–675. [Google Scholar] [CrossRef] [PubMed]
- Alpkent, Y.N.; İnak, E.; Ulusoy, S.; Ay, R. Acaricide resistance and mechanisms in Tetranychus urticae populations from greenhouses in Turkey. Syst. Appl. Acarol. 2020, 25, 155–168. [Google Scholar]
- Yang, X.; Zhu, K.Y.; Buschman, L.L.; Margolies, D.C. Comparative susceptibility and possible detoxification mechanisms for selected miticides in banks grass mite and two-spotted spider mite (Acari: Tetranychidae). Exp. Appl. Acarol. 2001, 25, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Luan, Z.; Cheng, X.; Xu, J. Molecular dynamics investigation of the substrate binding mechanism in carboxylesterase. Biochemistry 2015, 54, 1841–1848. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.L.; Huang, Y.; Lu, X.P.; Jiang, X.Z.; Smagghe, G.; Feng, Z.J.; Yuan, G.R.; Wei, D.; Wang, J.J. Overexpression of two α-esterase genes mediates metabolic resistance to malathion in the oriental fruit fly, Bactrocera dorsalis (Hendel). Insect Mol. Biol. 2015, 24, 467–479. [Google Scholar] [CrossRef]
- Devonshire, A.L.; Moores, G.D. A carboxylesterase with broad substrate specificity causes organophosphorus, carbamate and pyrethroid resistance in peach-potato aphids (Myzus persicae). Pestic. Biochem. Phys. 1982, 18, 235–246. [Google Scholar] [CrossRef]
- Shi, L.; Wei, P.; Wang, X.; Shen, G.; Zhang, J.; Xiao, W.; Xu, Z.; Xu, Q.; He, L. Functional analysis of esterase TCE2 gene from Tetranychus cinnabarinus (Boisduval) involved in acaricide resistance. Sci Rep. 2016, 6, 18646. [Google Scholar] [CrossRef]
- Xu, Z.S.; Lin, Y.Q.; Xu, J.; Zhu, B.; Zhao, W.; Peng, R.H.; Yao, Q.H. Selective detoxification of phenols by Pichia pastoris and Arabidopsis thaliana heterologously expressing the PtUGT72B1 from Populus trichocarpa. PLoS ONE 2013, 8, e66878. [Google Scholar] [CrossRef]
- Tiwari, P.; Sangwan, R.S.; Sangwan, N.S. Plant secondary metabolism linked glycosyltransferases: An update on expanding knowledge and scopes. Biotechnol. Adv. 2016, 34, 714–739. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, S.; Yang, J.; Kang, C.; Huang, L.; Guo, L. Glycosylation of plant secondary metabolites: Regulating from chaos to harmony. Environ. Exp. Bot. 2022, 194, 104703. [Google Scholar] [CrossRef]
- Bock, K.W. Vertebrate UDP-glucuronosyltransferases: Functional and evolutionary aspects. Biochem. Pharmacol. 2003, 66, 691–696. [Google Scholar] [CrossRef] [PubMed]
- Després, L.; David, J.; Gallet, C. The evolutionary ecology of insect resistance to plant chemicals. Trends Ecol. Evol. 2007, 22, 298–307. [Google Scholar] [CrossRef]
- Daborn, P.J.; Yen, J.L.; Bogwitz, M.R.; Le Goff, G.; Feil, E.; Jeffers, S.; Tijet, N.; Perry, T.; Heckel, D.; Batterham, P.; et al. A single P450 allele associated with insecticide resistance in Drosophila. Science 2002, 297, 2253–2256. [Google Scholar] [CrossRef] [PubMed]
- Djouaka, R.F.; Bakare, A.A.; Coulibaly, O.N.; Akogbeto, M.C.; Ranson, H.; Hemingway, J.; Strode, C. Expression of the cytochrome P450s, CYP6P3 and CYP6M2 are significantly elevated in multiple pyrethroid resistant populations of Anopheles gambiae s.s. from Southern Benin and Nigeria. BMC Genom. 2008, 9, 538. [Google Scholar] [CrossRef]
- Xing, X.; Yan, M.; Pang, H.; Wu, F.A.; Wang, J.; Sheng, S. Cytochrome P450s are essential for insecticide tolerance in the endoparasitoid wasp Meteorus pulchricornis (Hymenoptera: Braconidae). Insects 2021, 12, 651. [Google Scholar] [CrossRef]
- Antony, B.; Johny, J.; Abdelazim, M.M.; Jakše, J.; Al-Saleh, M.A.; Pain, A. Global transcriptome profiling and functional analysis reveal that tissue-specific constitutive overexpression of cytochrome P450s confers tolerance to imidacloprid in palm weevils in date palm fields. BMC Genom. 2019, 20, 440. [Google Scholar] [CrossRef]
- Shi, Y.; Qu, Q.; Wang, C.; He, Y.; Yang, Y.; Wu, Y. Involvement of CYP2 and mitochondrial clan P450s of Helicoverpa armigera in xenobiotic metabolism. Insect Biochem. Molec. 2022, 140, 103696. [Google Scholar] [CrossRef]
- Yang, T.; Liu, N. Genome analysis of cytochrome P450s and their expression profiles in insecticide resistant mosquitoes, Culex quinquefasciatus. PLoS ONE 2011, 6, e29418. [Google Scholar] [CrossRef]
- Riga, M.; Myridakis, A.; Tsakireli, D.; Morou, E.; Stephanou, E.G.; Nauen, R.; Van Leeuwen, T.; Douris, V.; Vontas, J. Functional characterization of the Tetranychus urticae CYP392A11, a cytochrome P450 that hydroxylates the METI acaricides cyenopyrafen and fenpyroximate. Insect Biochem. Molec. 2015, 65, 91–99. [Google Scholar] [CrossRef]
- Samra, A.I.; Kamita, S.G.; Yao, H.; Cornel, A.J.; Hammock, B.D. Cloning and characterization of two glutathione S-transferases from pyrethroid-resistant Culex pipiens. Pest Manag. Sci. 2012, 68, 764–772. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Xie, H.; Li, M.; Xu, X.; Lei, Z. Highly virulent Beauveria bassiana strains against the two-spotted spider mite, Tetranychus urticae, show no pathogenicity against five phytoseiid mite species. Exp. Appl. Acarol. 2016, 70, 421–435. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhou, L.; Yao, Q.; Liu, Y.; Bi, X.; Huang, J. Laboratory selection, resistance risk assessment, multi-resistance, and management of Tetranychus urticae Koch to bifenthrin, bifenazate and cyflumetofen on cowpea. Pest Manag. Sci. 2020, 76, 1912–1919. [Google Scholar] [CrossRef] [PubMed]
- Dittrich, V.; Cranham, J.; Jepson, L. Revised method for spider mites and their eggs (eg Tetranychus spp. and Panonychus ulmi koch). FAO Plant Prod. Prot. Pap. 1980, 21, 49–53. [Google Scholar]
- Wang, Y.; Zhao, S.; Shi, L.; Xu, Z.; He, L. Resistance selection and biochemical mechanism of resistance against cyflumetofen in Tetranychus cinnabarinus (Boisduval). Pestic. Biochem. Phys. 2014, 111, 24–30. [Google Scholar] [CrossRef]
- Chen, A.; Zhang, H.; Shan, T.; Shi, X.; Gao, X. The overexpression of three cytochrome P450 genes CYP6CY14, CYP6CY22 and CYP6UN1 contributed to metabolic resistance to dinotefuran in melon/cotton aphid, Aphis gossypii Glover. Pestic. Biochem. Phys. 2020, 167, 104601. [Google Scholar] [CrossRef]
- Wang, Q.; Rui, C.; Wang, L.; Nahiyoon, S.A.; Huang, W.; Zhu, J.; Ji, X.; Yang, Q.; Yuan, H.; Cui, L. Field-evolved resistance to 11 insecticides and the mechanisms involved in Helicoverpa armigera (Lepidoptera: Noctuidae). Pest Manag. Sci. 2021, 77, 5086–5095. [Google Scholar] [CrossRef]
- Zhang, Y.; Huang, D.; Lv, N.; Zhu, G.; Peng, J.; Chou, T.; Zhu, Z.; Wang, J.; Chen, Y.; Fang, X.; et al. Global quantification of glutathione S-transferases in human serum using LC-MS/MS coupled with affinity enrichment. J. Proteome Res. 2022, 21, 1311–1320. [Google Scholar] [CrossRef]
- Yin, Z.; Dong, X.; Kang, K.; Chen, H.; Dai, X.; Wu, G.; Zheng, L.; Yu, Y.; Zhai, Y. FoxO transcription factor regulate hormone mediated signaling on nymphal diapause. Front. Physiol. 2018, 9, 1654. [Google Scholar] [CrossRef]
- Zhang, W.; Chen, W.; Li, Z.; Ma, L.; Yu, J.; Wang, H.; Liu, Z.; Xu, B. Identification and characterization of three new cytochrome P450 genes and the use of RNA interference to evaluate their roles in antioxidant defense in Apis cerana cerana Fabricius. Front. Physiol. 2018, 9, 1608. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Morales, M.A.; Mendoza, B.M.; Lavine, L.C.; Lavine, M.D.; Walsh, D.B.; Zhu, F. Selection of reference genes for expression studies of xenobiotic adaptation in Tetranychus urticae. Int. J. Biol. Sci. 2016, 12, 1129–1139. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
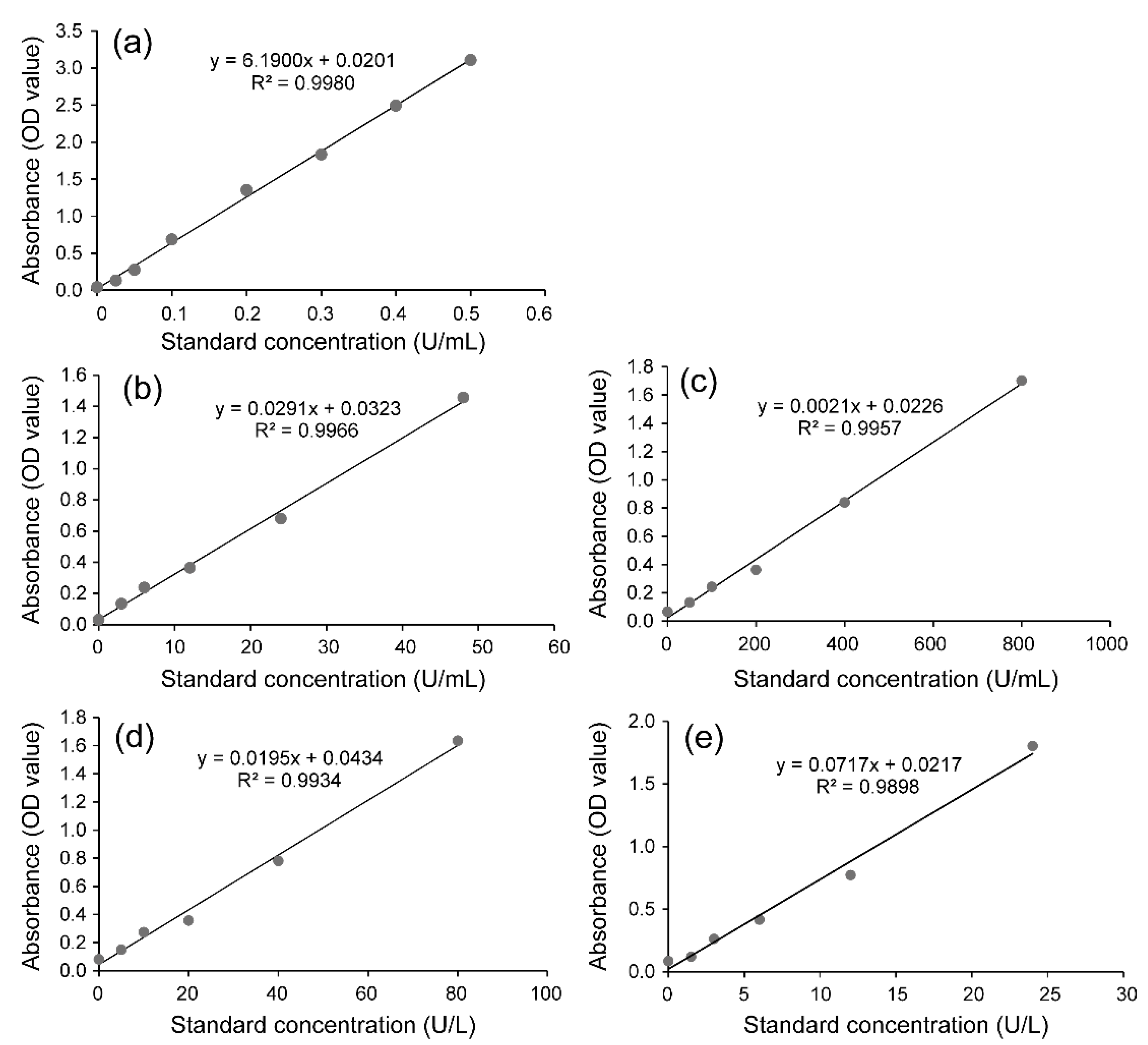
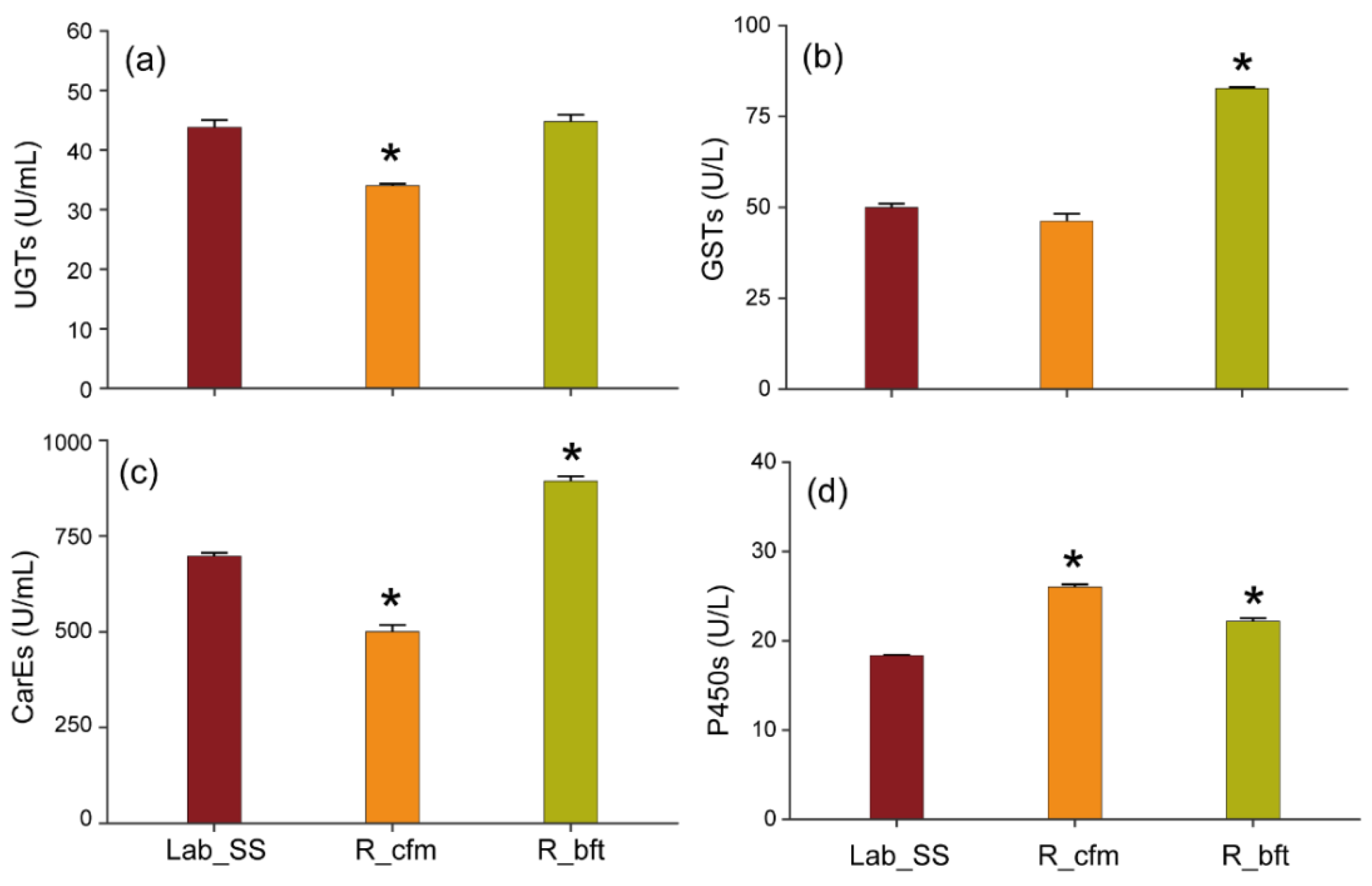
 :Lab_SS;
:Lab_SS;  : R_bft;
: R_bft;  : R_cfm).
: R_cfm).
 :Lab_SS;
:Lab_SS;  : R_bft;
: R_bft;  : R_cfm).
: R_cfm).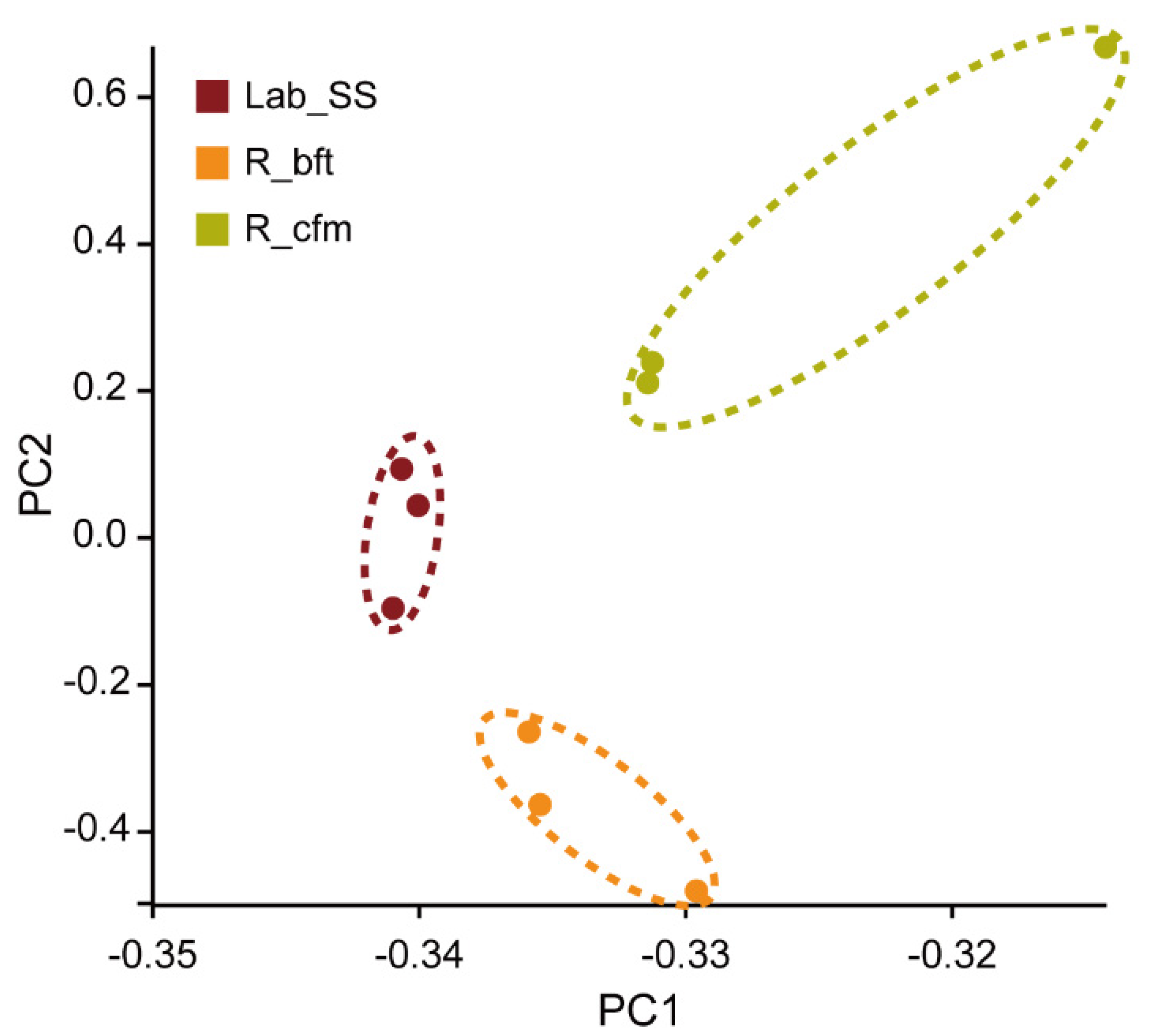


| Acaricide | Generation | Regression | LC50 (mg/L) (95% CI) | Resistance Ratio |
|---|---|---|---|---|
| Cyflumetofen | F0 | y = 4.14 + 1.10x | 6.02 (4.29~8.43) | - |
| F16 | y = −6.65 + 4.09x | 707.95 (646.64~775.09) | 117.60 | |
| Bifenthrin | F0 | y = −0.21 + 2.46x | 125.04 (103.19~151.52) | - |
| F20 | - | >20000 | >159.95 |
| Samples | Total Raw Reads (Mb) | Total Clean Reads (Mb) | Clean Reads Ratio (%) | Mapping Rate (%) | Clean Reads Q20 (%) | Clean Reads Q30 (%) |
|---|---|---|---|---|---|---|
| Lab_SS1 | 45.57 | 42.24 | 92.68 | 63.45 | 94.94 | 88.86 |
| Lab_SS2 | 47.33 | 42.83 | 90.50 | 62.02 | 94.55 | 88.10 |
| Lab_SS3 | 45.57 | 42.26 | 92.72 | 64.88 | 94.67 | 88.30 |
| R_cfm1 | 47.33 | 43.11 | 91.10 | 67.04 | 94.72 | 88.42 |
| R_cfm2 | 42.65 | 38.51 | 90.30 | 70.27 | 94.80 | 88.53 |
| R_cfm3 | 47.33 | 43.10 | 91.07 | 66.80 | 94.84 | 88.66 |
| R_bft1 | 49.08 | 42.81 | 87.22 | 49.29 | 95.17 | 89.34 |
| R_bft2 | 47.33 | 43.10 | 91.08 | 54.89 | 94.85 | 88.69 |
| R_bft3 | 45.57 | 42.22 | 92.65 | 69.80 | 94.70 | 88.36 |
| Detoxification Enzymes | Gene ID | log2 (R_cfm or R_bft /Lab_SS) | Symbol | |
|---|---|---|---|---|
| Lab_SS vs R_cfm | CCEs | tetur17g00350 | 1.77 | TuCCE46 |
| tetur207g00010 | 1.28 | TuCCE70 | ||
| P450s | tetur07g06440 | 1.64 | CYP392A2p | |
| tetur07g06410 | 1.10 | CYP392A1 | ||
| tetur27g00340 | −1.32 | CYP392E7 | ||
| Lab_SS vs R_bft | CCEs | tetur17g00350 | 3.68 | TuCCE46 |
| tetur207g00010 | 3.09 | TuCCE70 | ||
| tetur207g00020 | 2.65 | CCEincTu16 | ||
| tetur17g00300 | 2.63 | TuCCE45 | ||
| tetur17g00080 | 2.21 | TuCCE44 | ||
| tetur17g00360 | 2.04 | CCEincTu08 | ||
| tetur35g00180 | 1.89 | CCEincTu13 | ||
| tetur35g00200 | 1.77 | TuCCE65 | ||
| tetur04g06770 | 1.53 | TuCCE25 | ||
| tetur01g14180 | 1.26 | TuCCE12 | ||
| tetur11g05770 | 1.15 | TuCCE35 | ||
| tetur16g02380 | −1.01 | TuCCE40 | ||
| tetur04g02550 | −1.04 | TuCCE22 | ||
| tetur30g01290 | −1.26 | TuCCE61 | ||
| tetur01g08680 | −1.46 | TuCCE01 | ||
| UGTs | tetur04g07630 | 3.31 | UGT16 | |
| tetur05g00080 | 2.83 | UGT22 | ||
| tetur21g01400 | 2.29 | UGT58p | ||
| tetur22g00420 | 1.75 | UGT65 | ||
| tetur08g00190 | 1.73 | UGT40 | ||
| tetur09g01660 | 1.69 | UGT | ||
| tetur05g09325 | 1.56 | UGT | ||
| tetur05g00090 | 1.30 | UGT23 | ||
| tetur22g00510 | 1.17 | UGT69 | ||
| P450s | tetur03g04990 | 2.04 | CYP392D2 | |
| tetur03g00970 | 1.75 | CYP392A11 | ||
| tetur03g09961 | −1.14 | CYP392D7 | ||
| tetur46g00150 | −1.44 | CYP385C4v2 | ||
| tetur07g06440 | −1.89 | CYP392A2p | ||
| GSTs | tetur31g01330 | 1.88 | TuGSTd15 | |
| tetur05g05260 | 1.65 | TuGSTm09 | ||
| tetur26g01510 | 1.49 | TuGSTd13 | ||
| tetur05g05250 | 1.48 | TuGSTm08 | ||
| ABCs | tetur09g01970 | 1.35 | TuABCG-13 | |
| tetur18g00230 | 1.14 | TuABCH-13 | ||
| tetur11g02120 | 1.06 | TuABCC-28 |
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| tetur03g04990 | TCTAAAGGACCAAGAGGAG | ACGATGGGTTTGATAATGT |
| tetur04g07630 | ATCAGGACCTCCACCATTT | AATCTATTCGGCACCAACC |
| tetur05g00080 | AACATCGCTTCATCGTCTC | CAATCAATCTTGCCACTTC |
| tetur21g01400 | GGACCAAGAGGAGACGAAC | TTCAGCATACGATGGGTTT |
| tetur17g00350 | TTCCTTATGCGAAGCCAACG | AACGGACCAAGATCCAGCA |
| tetur207g00010 | AAAAGGACCAGCGAAGACT | AACGGACCAAGATCCAGCA |
| tetur207g00020 | TATGTTTTCGGTTTACCTC | TTTCCCCAATCATCTATG |
| tetur17g00300 | CAAGAAGAACCAGCGAAGA | CTGACCAAGATCCAGCAGA |
| tetur17g00080 | AACATCGCTTCATCGTCTC | CAATCAATCTTGCCACTTC |
| tetur17g00360 | TCAAACAAACTACGACCAG | CTCCGAAGTAGATGAAACC |
| tetur01g08680 | TTTTGATAATTCGGTTGACG | ACGATATTGGCTTAGGAGG |
| tetur22g00420 | ATGGTCCACCTCTTCGTTC | TTCCCATTGAGACATAGATAAGC |
| tetur03g00970 | AGGAAGCAATTCGCTGATA | GCCAACAATAGGAAGACCC |
| tetur07g06440 | CTCGTCCAATATCCTCAAT | AGCACTAAATGGCACTAAA |
| tetur31g01330 | GATTGGTTTACGCTGTCCT | GTTTCGTTTCGATAAGATTG |
| tetur05g05260 | AGGTGCTGTTGAGCCTATT | TTTCTTGGTCCTAAATCGT |
| tetur09g01970 | ATGTTGCTGGCTGAGGGTC | ATGGCTTCTTGATTGTAGTCCTG |
| tetur27g00340 | GGCTTAGGAAAGAGTGAAA | CGAAATACGGAAACAGACC |
| tetur01g12670 | CTTCAAGGCGGTGACTTTACC | CCATTGAAAGGATACCTGGTCC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Wu, F.; Liang, W.; Zhou, L.; Huang, J. Molecular Mechanisms Underlying Metabolic Resistance to Cyflumetofen and Bifenthrin in Tetranychus urticae Koch on Cowpea. Int. J. Mol. Sci. 2022, 23, 16220. https://doi.org/10.3390/ijms232416220
Liu Z, Wu F, Liang W, Zhou L, Huang J. Molecular Mechanisms Underlying Metabolic Resistance to Cyflumetofen and Bifenthrin in Tetranychus urticae Koch on Cowpea. International Journal of Molecular Sciences. 2022; 23(24):16220. https://doi.org/10.3390/ijms232416220
Chicago/Turabian StyleLiu, Zhenxiu, Fuxing Wu, Weikang Liang, Lijuan Zhou, and Jiguang Huang. 2022. "Molecular Mechanisms Underlying Metabolic Resistance to Cyflumetofen and Bifenthrin in Tetranychus urticae Koch on Cowpea" International Journal of Molecular Sciences 23, no. 24: 16220. https://doi.org/10.3390/ijms232416220
APA StyleLiu, Z., Wu, F., Liang, W., Zhou, L., & Huang, J. (2022). Molecular Mechanisms Underlying Metabolic Resistance to Cyflumetofen and Bifenthrin in Tetranychus urticae Koch on Cowpea. International Journal of Molecular Sciences, 23(24), 16220. https://doi.org/10.3390/ijms232416220





