Cytokine Receptors—Regulators of Antimycobacterial Immune Response
Abstract
1. Introduction
2. Cytokine Receptors Involved in Antimycobacterial Immune Response
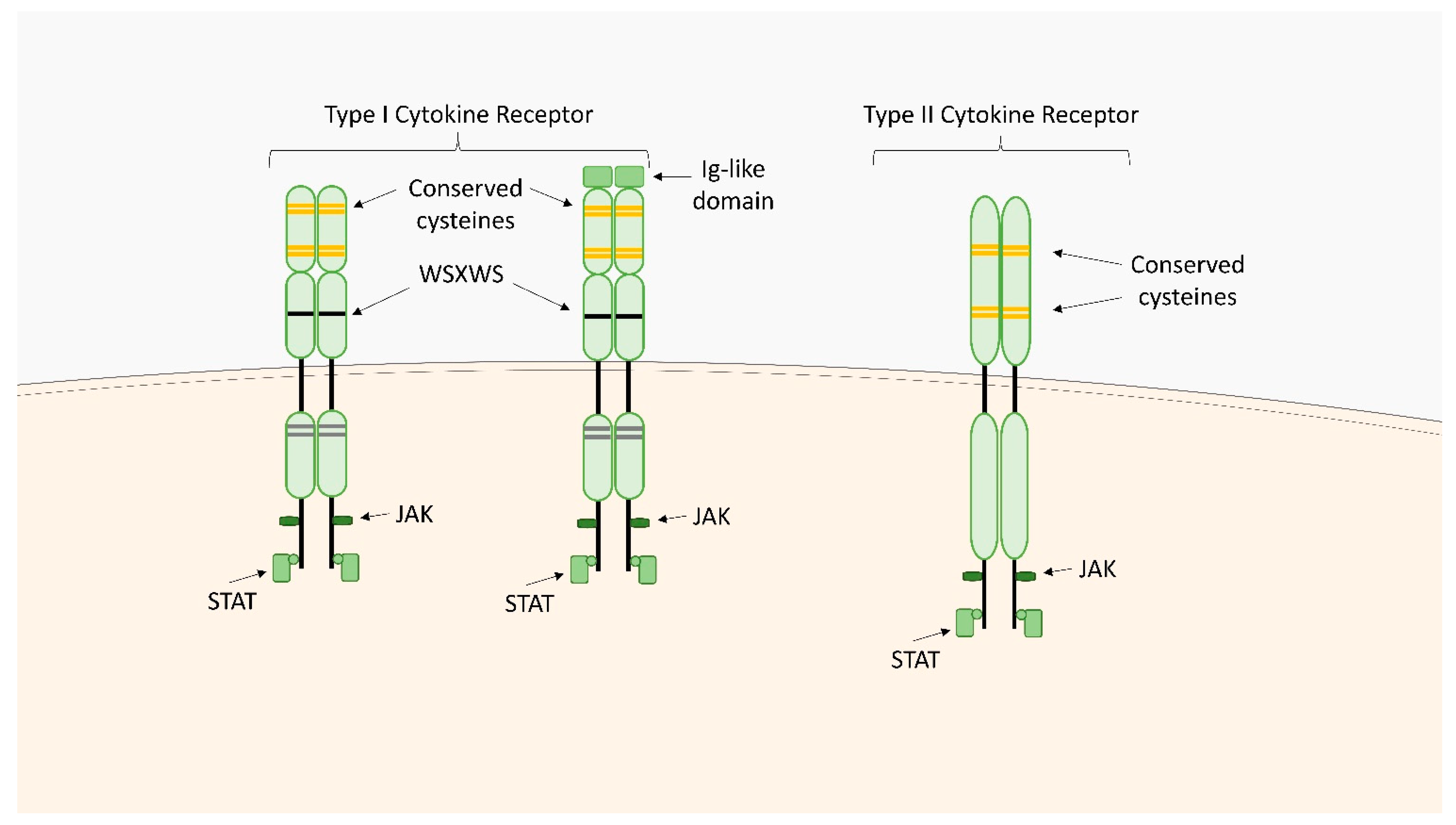
2.1. Type I Cytokine Receptors
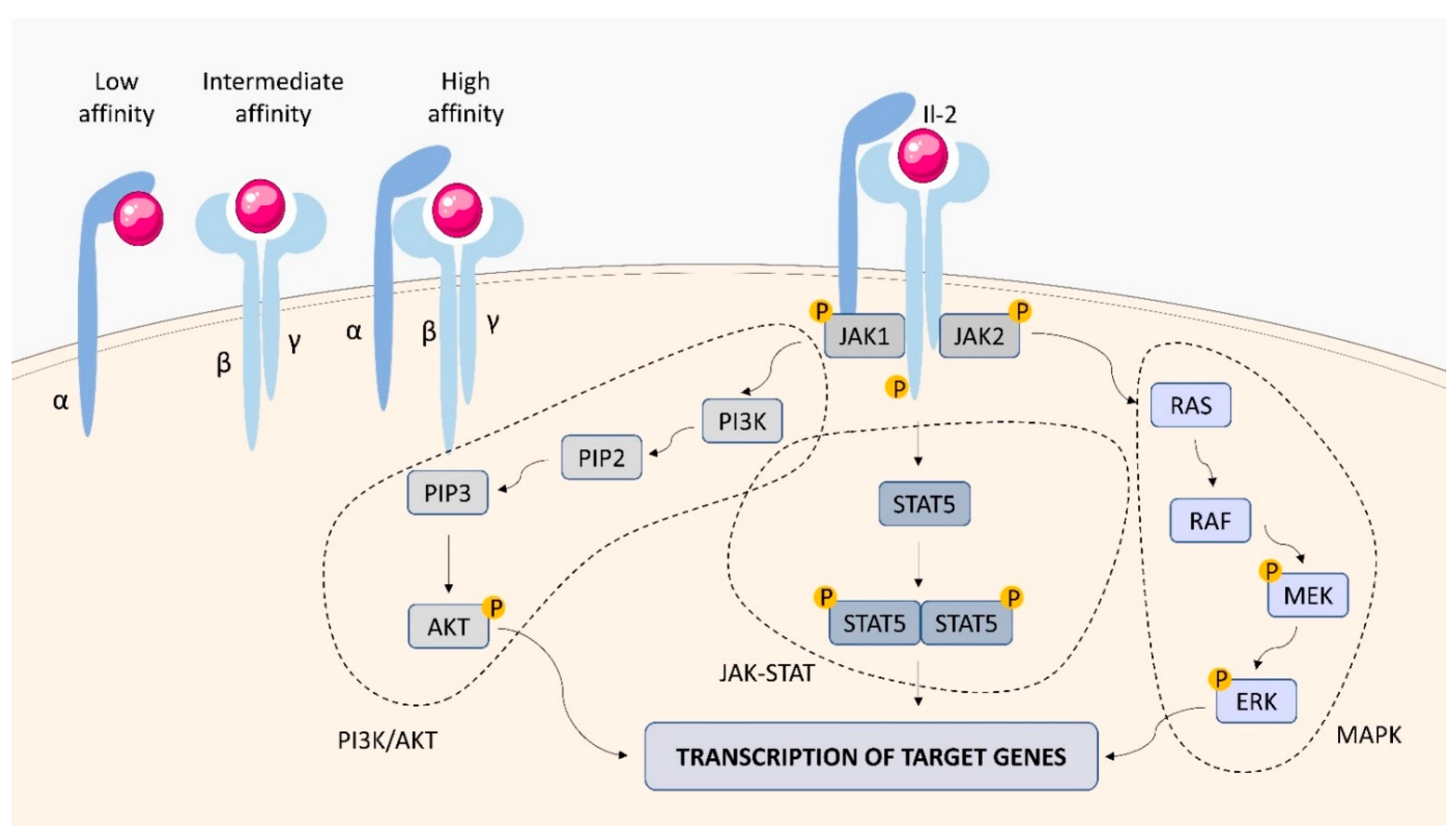
2.1.1. IL-2 Receptor (IL-2R)
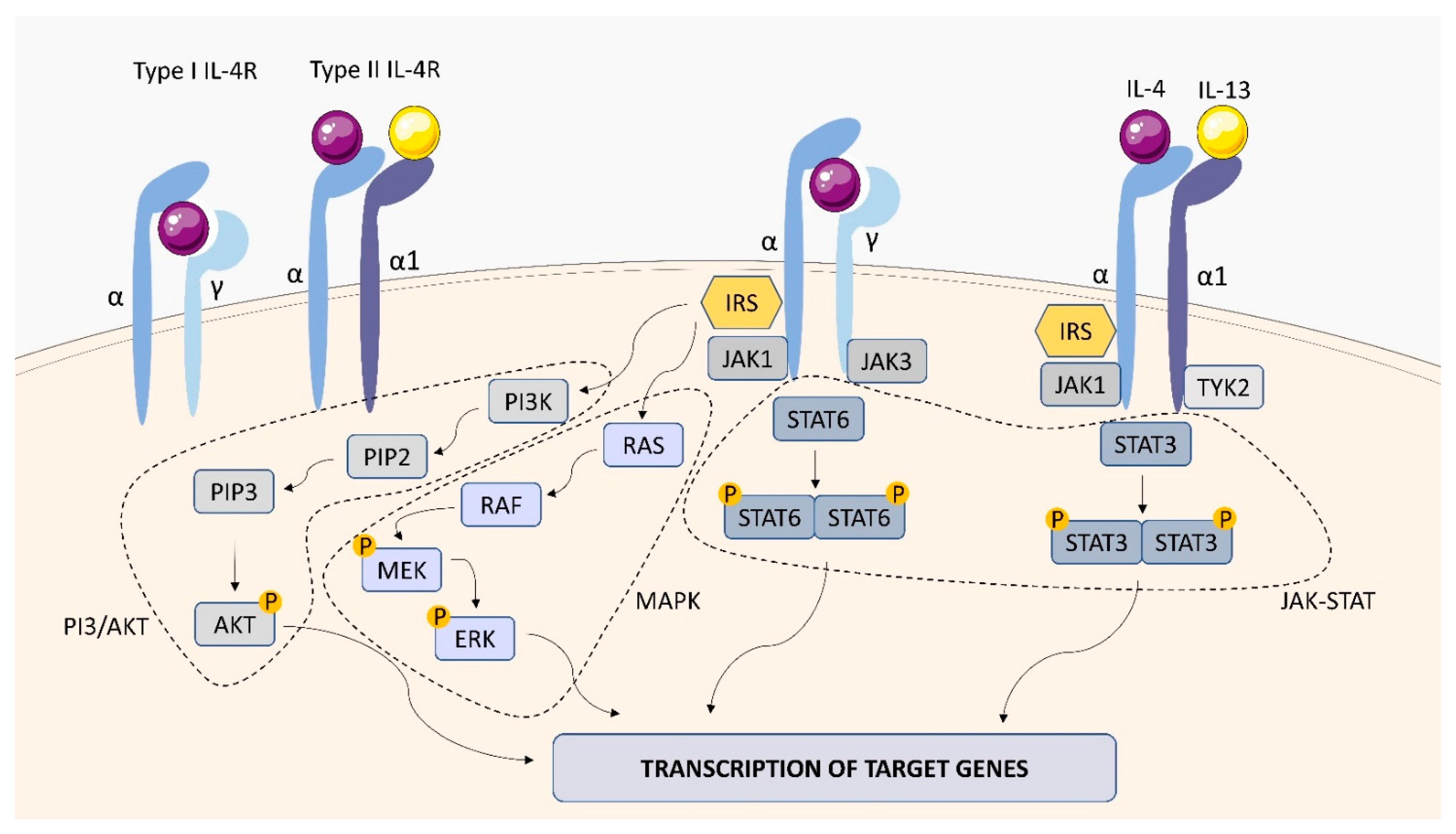
2.1.2. IL-4 Receptor (IL-4R)
2.1.3. IL-6 Receptor (IL-6R)
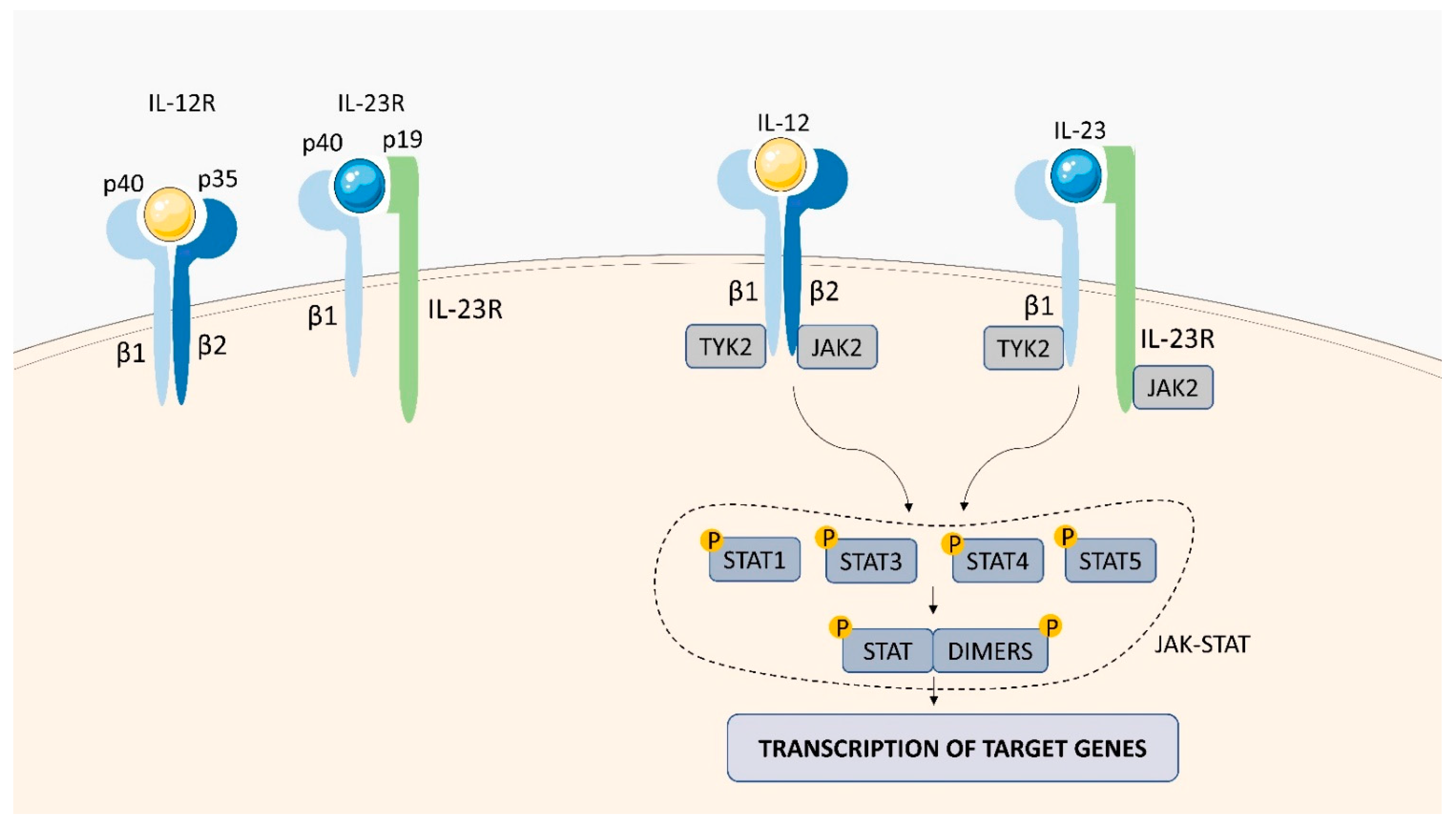
2.1.4. IL-12 Receptor (IL-12R)
2.1.5. IL-23 Receptor (IL-23R)
2.2. Type II Cytokine Receptors
2.2.1. Interferon (IFN)-Gamma (γ) Receptor (IFN-γR)
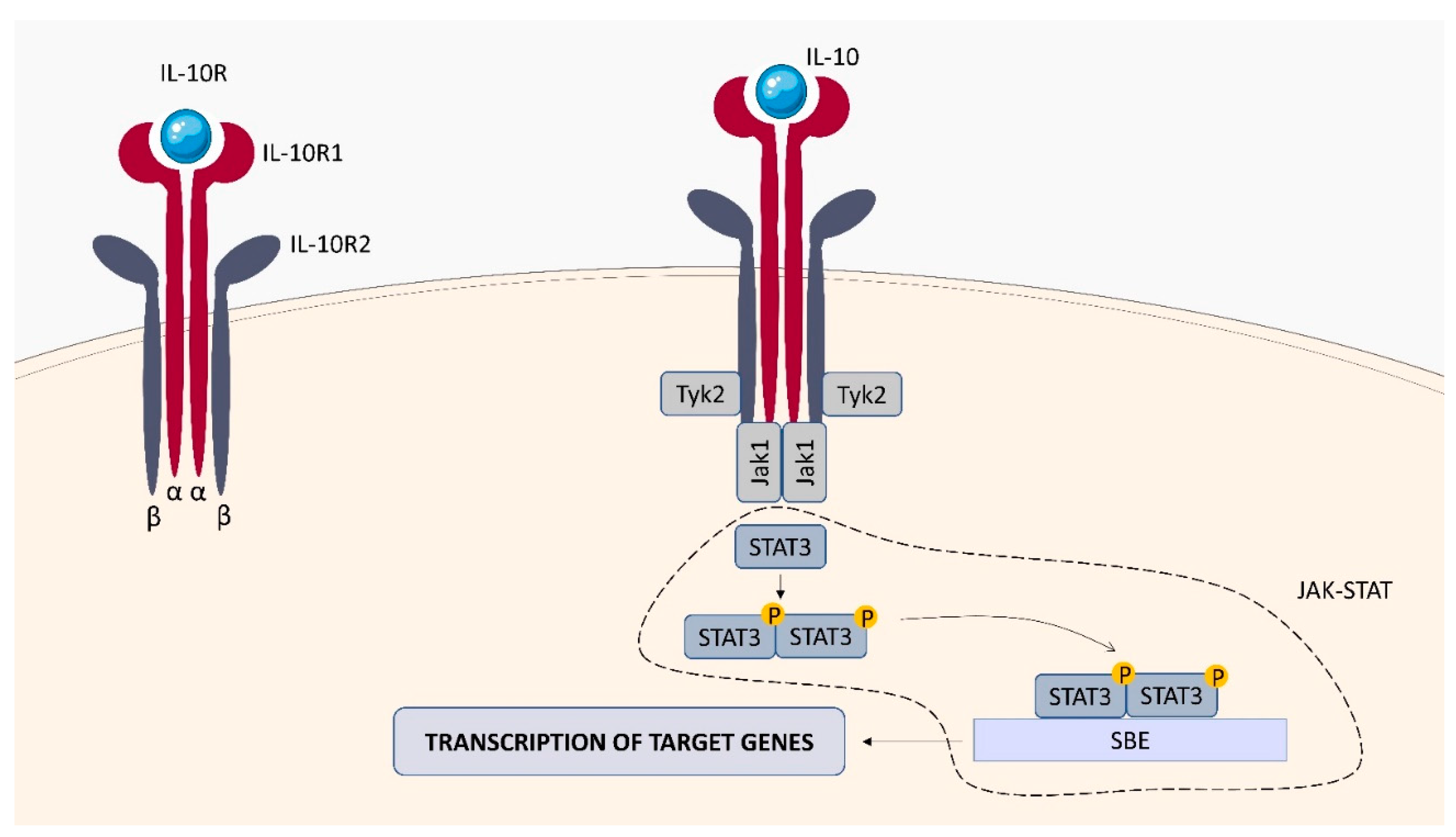
2.2.2. IL-10 Receptor (IL-10R)
2.2.3. IL-22 Receptor (IL-22R)
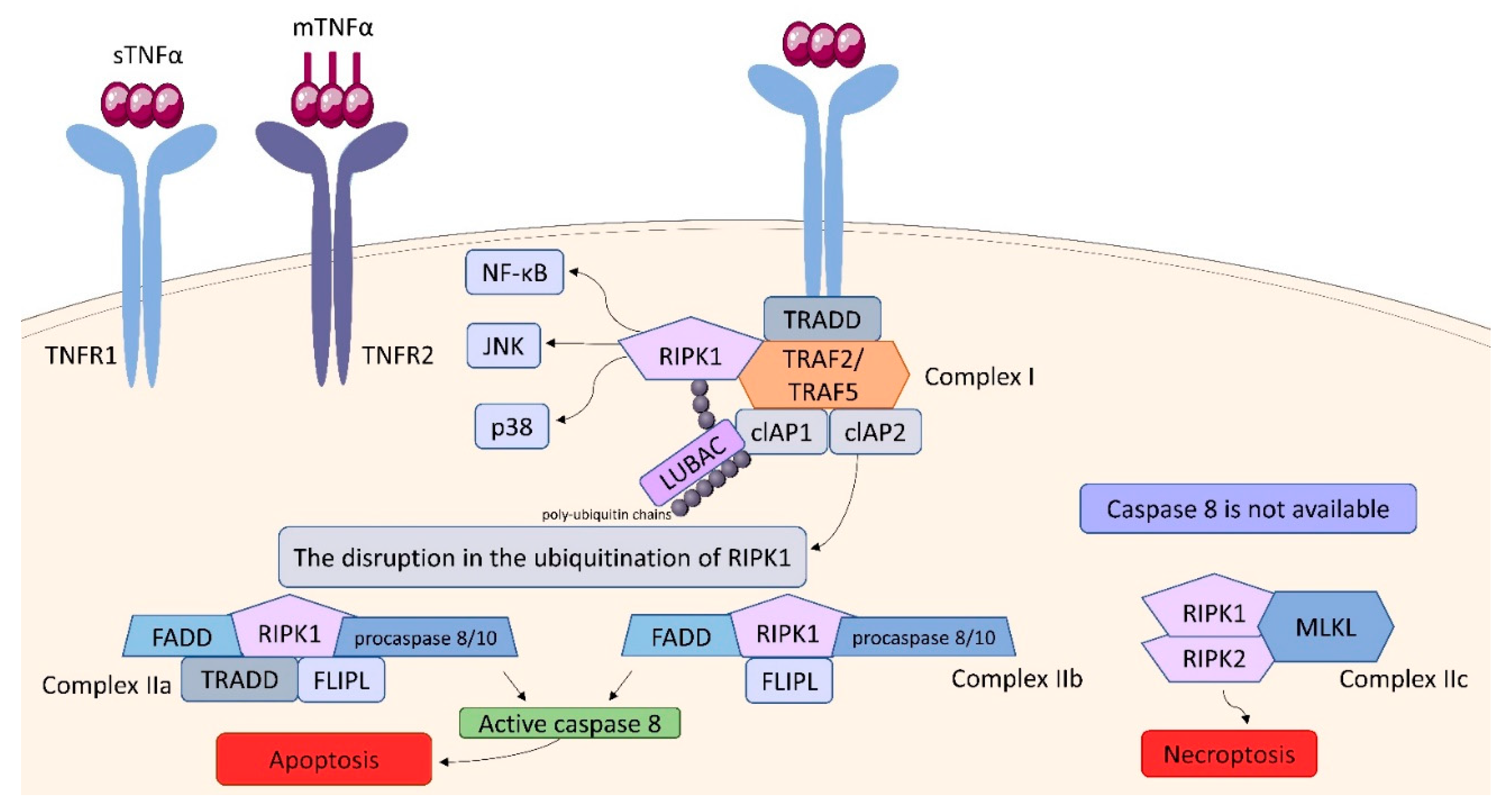
2.3. Tumor Necrosis Factor (TNF) Receptor (TNFR) Superfamily
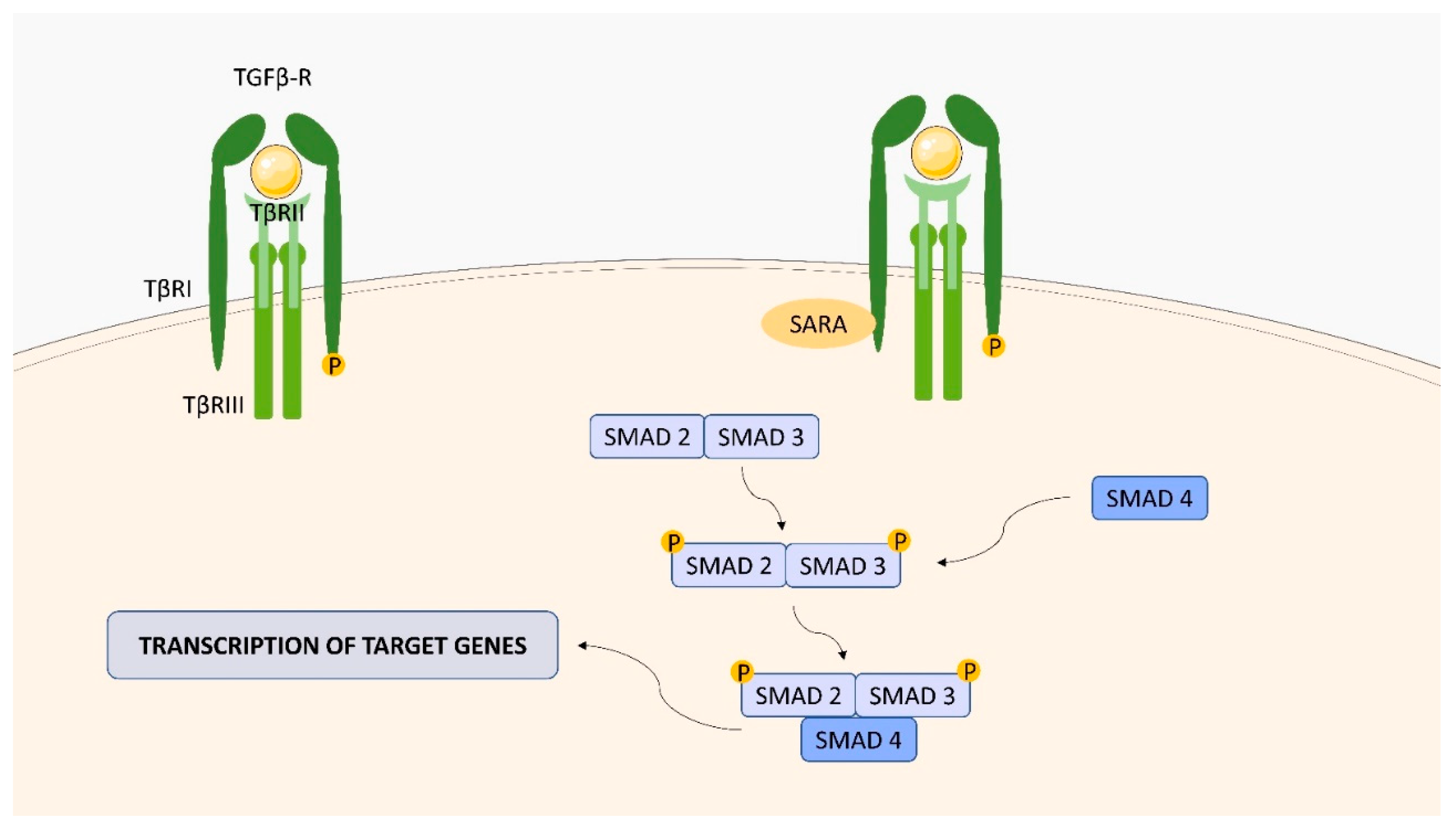
2.4. Transforming Growth Factor (TGF)-Beta (β) Receptor (TGFBR) Family
2.5. IL-1 Receptor (IL-1R) Family
2.6. Chemokine Receptors
2.6.1. CXCR3
2.6.2. CXCR1 and CXCR2
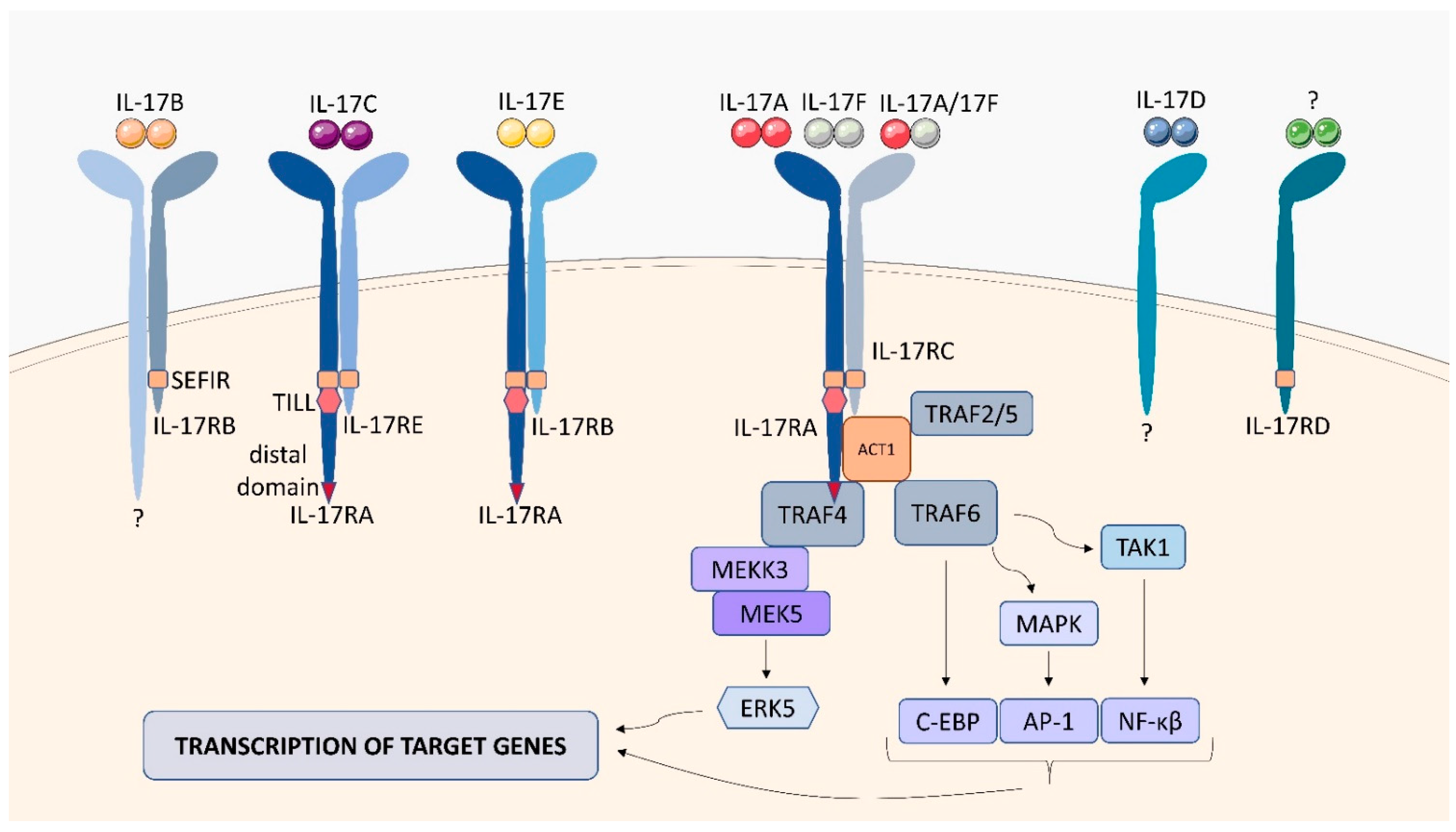
2.7. IL-17 Receptor Family (IL-17R)
3. Implications of Cytokine Receptor Gene Defects for the Course of Mycobacterial Infection
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Harriff, M.J.; Purdy, G.E.; Lewinsohn, D.M. Escape from the Phagosome: The Explanation for MHC-I Processing of Mycobacterial Antigens? Front. Immunol. 2012, 3, 40. [Google Scholar] [CrossRef]
- Abrahams, K.A.; Besra, G.S. Synthesis and recycling of the mycobacterial cell envelope. Curr. Opin. Microbiol. 2021, 60, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Domingo-Gonzalez, R.; Prince, O.; Cooper, A.; Khader, S.A. Cytokines and Chemokines in Mycobacterium tuberculosis infection. Microbiol. Spectr. 2016, 4, 4–5. [Google Scholar] [CrossRef]
- Torrelles, J.B.; Schlesinger, L.S. Integrating Lung Physiology, Immunology, and Tuberculosis. Trends Microbiol. 2017, 25, 688–697. [Google Scholar] [CrossRef]
- Etna, M.P.; Giacomini, E.; Severa, M.; Coccia, E.M. Pro- and anti-inflammatory cytokines in tuberculosis: A two-edged sword in TB pathogenesis. Semin. Immunol. 2014, 26, 543–551. [Google Scholar] [CrossRef]
- Ndlovu, H.; Marakalala, M.J. Granulomas and inflammation: Host-directed therapies for tuberculosis. Front. Immunol. 2016, 7, 1. [Google Scholar] [CrossRef] [PubMed]
- Harding, C.V.; Boom, W.H. Regulation of antigen presentation by Mycobacterium tuberculosis: A role for Toll-like receptors. Nat. Rev. Microbiol. 2010, 8, 296. [Google Scholar] [CrossRef] [PubMed]
- Shiromizu, C.M.; Jancic, C.C. γδ T Lymphocytes: An Effector Cell in Autoimmunity and Infection. Front. Immunol. 2018, 9, 2389. [Google Scholar] [CrossRef]
- Orme, I.M.; Henao-Tamayo, M.I. Trying to see the forest through the trees: Deciphering the nature of memory immunity to Mycobacterium tuberculosis. Front. Immunol. 2018, 9, 461. [Google Scholar] [CrossRef]
- Redford, P.S.; Murray, P.J.; O’Garra, A. The role of IL-10 in immune regulation during M. tuberculosis infection. Mucosal Immunol. 2011, 4, 261–270. [Google Scholar] [CrossRef]
- Mogues, T.; Goodrich, M.E.; Ryan, L.; LaCourse, R.; North, R.J. The relative importance of T cell subsets in immunity and immunopathology of airborne Mycobacterium tuberculosis infection in mice. J. Exp. Med. 2001, 193, 271–280. [Google Scholar] [CrossRef]
- Behar, S.M.; Carpenter, S.M.; Booty, M.G.; Barber, D.L.; Jayaraman, P. Orchestration of pulmonary T cell immunity during Mycobacterium tuberculosis infection: Immunity interruptus. Semin. Immunol. 2014, 26, 559–577. [Google Scholar] [CrossRef]
- Ehlers, S.; Schaible, U.E. The granuloma in tuberculosis: Dynamics of a host-pathogen collusion. Front. Immunol. 2012, 3, 411. [Google Scholar] [CrossRef]
- Domingues, J.D.S.; Ducasse, L.; Peixoto, A. Exploring the mechanisms of granuloma formation in vivo to prevent dissemination of a respiratory mycobacterium tuberculosis infection: A live imaging approach. Eur. Respir. J. 2016, 48, PA2696. [Google Scholar] [CrossRef]
- Verduzco-Sierra, O.A.; Rosas-Taraco, A.G. Cytokine Environment in Tuberculoid Lung Granuloma. Austin Tuberc. Res. Treat. 2016, 1, 1004. [Google Scholar] [CrossRef]
- Pagán, A.J.; Ramakrishnan, L. Immunity and Immunopathology in the Tuberculous Granuloma. Cold Spring Harb. Perspect. Med. 2014, 5, a018499. [Google Scholar] [CrossRef]
- Gideon, H.P.; Phuah, J.; Junecko, B.A.; Mattila, J.T. Neutrophils express pro- and anti-inflammatory cytokines in granulomas from Mycobacterium tuberculosis-infected cynomolgus macaques. Mucosal Immunol. 2019, 12, 1370–1381. [Google Scholar] [CrossRef]
- Millar, J.A.; Butler, J.R.; Evans, S.; Mattila, J.T.; Linderman, J.J.; Flynn, J.A.L.; Kirschner, D.E. Spatial Organization and Recruitment of Non-Specific T Cells May Limit T Cell-Macrophage Interactions Within Mycobacterium tuberculosis Granulomas. Front. Immunol. 2021, 11, 3496. [Google Scholar] [CrossRef] [PubMed]
- Hulme, M.A.; Wasserfall, C.H.; Atkinson, M.A.; Brusko, T.M. Central Role for Interleukin-2 in Type 1 Diabetes. Diabetes 2012, 61, 14. [Google Scholar] [CrossRef] [PubMed]
- Keegan, A.D.; Nelms, K.; Wang, L.M.; Pierce, J.H.; Paul, W.E. Interleukin 4 receptor: Signaling mechanisms. Immunol. Today 1994, 15, 423. [Google Scholar] [CrossRef]
- Gadani, S.P.; Cronk, J.C.; Norris, G.T.; Kipnis, J. IL-4 in the brain: A cytokine to remember. J. Immunol. 2012, 189, 4213. [Google Scholar] [CrossRef] [PubMed]
- Beekman, R.; Touw, I.P. G-CSF and its receptor in myeloid malignancy. Blood 2010, 115, 5131. [Google Scholar] [CrossRef] [PubMed]
- Neurath, M.F.; Finotto, S. IL-6 signaling in autoimmunity, chronic inflammation and inflammation-associated cancer. Cytokine Growth Factor Rev. 2011, 22, 83. [Google Scholar] [CrossRef] [PubMed]
- Sansone, P.; Bromberg, J. Targeting the interleukin-6/Jak/stat pathway in human malignancies. J. Clin. Oncol. 2012, 30, 1005. [Google Scholar] [CrossRef]
- Vainchenker, W.; Constantinescu, S.N. JAK/STAT signaling in hematological malignancies. Oncogene 2013, 32, 2601. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Pardoll, D.; Jove, R. STATs in cancer inflammation and immunity: A leading role for STAT3. Nat. Rev. Cancer 2009, 9, 798. [Google Scholar] [CrossRef]
- Hamza, T.; Barnett, J.B.; Li, B. Interleukin 12 a key immunoregulatory cytokine in infection applications. Int. J. Mol. Sci. 2010, 11, 789–806. [Google Scholar] [CrossRef]
- Guo, Y.; Cao, W.; Zhu, Y. Immunoregulatory Functions of the IL-12 Family of Cytokines in Antiviral Systems. Viruses 2019, 11, 772. [Google Scholar] [CrossRef] [PubMed]
- Pastor-Fernández, G.; Mariblanca, I.R.; Navarro, M.N. Decoding IL-23 Signaling Cascade for New Therapeutic Opportunities. Cells 2020, 9, 2044. [Google Scholar] [CrossRef] [PubMed]
- Furmento, V.A.; Marino, J.; Blank, V.C.; Roguin, L.P. The granulocyte colony-stimulating factor (G-CSF) upregulates metalloproteinase-2 and VEGF through PI3K/Akt and Erk1/2 activation in human trophoblast Swan 71 cells. Placenta 2014, 35, 937. [Google Scholar] [CrossRef]
- Yang, J.Z.; Zhang, J.Q.; Sun, L.X. Mechanisms for T cell tolerance induced with granulocyte colony-stimulating factor. Mol. Immunol. 2016, 70, 56. [Google Scholar] [CrossRef]
- Na, Y.R.; Gu, G.J.; Jung, D.; Kim, Y.W.; Na, J.; Woo, J.S.; Cho, J.Y.; Youn, H.; Seok, S.H. GM-CSF Induces Inflammatory Macrophages by Regulating Glycolysis and Lipid Metabolism. J. Immunol. 2016, 197, 4101. [Google Scholar] [CrossRef]
- Schülke, S. Induction of interleukin-10 producing dendritic cells as a tool to suppress allergen-specific T helper 2 responses. Front. Immunol. 2018, 9, 455. [Google Scholar] [CrossRef] [PubMed]
- Thompson, C.D.; Zurko, J.C.; Hanna, B.F.; Hellenbrand, D.J.; Hanna, A. The therapeutic role of interleukin-10 after spinal cord injury. J. Neurotrauma 2013, 30, 1311. [Google Scholar] [CrossRef]
- Dhama, K.; Latheef, S.K.; Dadar, M.; Samad, H.A.; Munjal, A.; Khandia, R.; Karthik, K.; Tiwari, R.; Yatoo, M.I.; Bhatt, P.; et al. Biomarkers in stress related diseases/disorders: Diagnostic, prognostic, and therapeutic values. Front. Mol. Biosci. 2019, 6, 91. [Google Scholar] [CrossRef]
- DiNardo, A.R.; Rajapakshe, K.; Gandhi, T.; Grimm, S.L.; Nishiguchi, T.; Heyckendorf, J.; Kahari, J.; Dlamini, Q.; Lange, C. Discerning divergent tuberculosis endotypes: A meta-analysis and systematic review of individual patient data. medRxiv 2021. [Google Scholar] [CrossRef]
- Bhat, M.Y.; Solanki, H.S.; Advani, J.; Khan, A.A.; Keshava Prasad, T.S.; Gowda, H.; Thiyagarajan, S.; Chatterjee, A. Comprehensive network map of interferon gamma signaling. J. Cell Commun. Signal. 2018, 12, 745. [Google Scholar] [CrossRef]
- Zhou, A.; Scoggin, S.; Gaynor, R.B.; Williams, N.S. Identification of NF-kappa B-regulated genes induced by TNF-alpha utilizing expression profiling and RNA interference. Oncogene 2003, 22, 2054. [Google Scholar] [CrossRef] [PubMed]
- Kandasamy, K.; Mohan, S.S.; Raju, R.; Keerthikumar, S.; Kumar, G.S.; Venugopal, A.K.; Telikicherla, D.; Navarro, J.D.; Mathivanan, S.; Pecquet, C.; et al. NetPath: A public resource of curated signal transduction pathways. Genome Biol. 2010, 11, R3. [Google Scholar] [CrossRef]
- Ranganathan, P.; Agrawal, A.; Bhushan, R.; Chavalmane, A.K.; Kalathur, R.K.; Takahashi, T.; Kondaiah, P. Expression profiling of genes regulated by TGF-beta: Differential regulation in normal and tumour cells. BMC Genom. 2007, 8, 98. [Google Scholar] [CrossRef]
- Manna, S.K.; Ramesh, G.T. Interleukin-8 induces nuclear transcription factor-kappaB through a TRAF6-dependent pathway. J. Biol. Chem. 2005, 280, 7010. [Google Scholar] [CrossRef]
- Onishi, R.M.; Gaffen, S.L. Interleukin-17 and its target genes: Mechanisms of interleukin-17 function in disease. Immunology 2010, 129, 311. [Google Scholar] [CrossRef]
- Morris, R.; Kershaw, N.J.; Babon, J.J. The molecular details of cytokine signaling via the JAK/STAT pathway. Protein Sci. 2018, 27, 1984. [Google Scholar] [CrossRef] [PubMed]
- Fujii, H. Mechanisms of Signal Transduction from Receptors of Type I and Type II Cytokines. J. Immunotoxicol. 2007, 4, 69–76. [Google Scholar] [CrossRef]
- Esen, İ. The Structure and Signaling Mechanisms of Type 1 Cytokine Receptors: A Brief Overview Tip 1 Sitokin Reseptörlerinin Yapısı ve Sinyal Mekanizmaları: Kısa Bir Genel Bakış. Turk. J. Immunol. 2015, 3, 121–124. [Google Scholar] [CrossRef]
- Malek, T.R.; Castro, I. Interleukin-2 receptor signaling: At the interface between tolerance and immunity. Immunity 2010, 33, 153–165. [Google Scholar] [CrossRef]
- Damoiseaux, J. The IL-2 - IL-2 receptor pathway in health and disease: The role of the soluble IL-2 receptor. Clin. Immunol. 2020, 218, 108515. [Google Scholar] [CrossRef]
- Lawn, S.D.; Rudolph, D.; Ackah, A.; Coulibaly, D.; Wiktor, S.; Lal, R.B. Lack of induction of interleukin-2-receptor-α in patients with tuberculosis and human immunodeficiency virus co-infection: Implications for pathogenesis. Trans. R. Soc. Trop. Med. Hyg. 2001, 95, 449–452. [Google Scholar] [CrossRef]
- Ryu, Y.J.; Ryu, K.H.; Kim, S.H.; Lee, J.S.; Cheon, S.H.; Seoh, J.Y. Soluble IL-2R, IFN-γ and neopterin as immunologic markers in patients with tuberculosis. Tuberc. Respir. Dis. 2002, 53, 294–308. [Google Scholar] [CrossRef]
- Lee, S.-I.; Jin, H.-S.; Park, S. Association of genetic polymorphism of IL-2 receptor subunit and tuberculosis case [Internet]. Biomedical Science Letters. Korean Soc. Biomed. Lab. Sci. 2018, 24, 94–101. [Google Scholar] [CrossRef]
- Chendi, B.H.; Tveitenc, H.; Snyders, C.I.; Tonbya, K.; Jenumd, S.; Dam Nielsene, S.; Hove-Skovsgaarde, M.; Walzl, G.; Chegoub, N.N.; Dyrhol-Riise, A.M. CCL1 and IL-2Ra differentiate Tuberculosis disease from latent infection Irrespective of HIV infection in low TB burden countries. J. Infect. 2021, 83, 433–443. [Google Scholar] [CrossRef]
- Cheung, L.S. Therapeutic Potential of an IL-2 Fusion Toxin in Tuberculosis and Melanoma. Doctoral Dissertations, Johns Hopkins University, Baltimore, Maryland, 2018. Available online: http://jhir.library.jhu.edu/handle/1774.2/61021 (accessed on 28 December 2021).
- Junttila, I.S. Tuning the Cytokine Responses: An Update on Interleukin (IL)-4 and IL-13 Receptor Complexes. Front. Immunol. 2018, 9, 888. [Google Scholar] [CrossRef]
- Nelms, K.; Keegan, A.D.; Zamorano, J.; Ryan, J.J.; Paul, W.E. The IL-4 receptor: Signaling mechanisms and biologic functions. Annu. Rev. Immunol. 1999, 17, 701–738. [Google Scholar] [CrossRef]
- Andrews, A.-L.; Holloway, J.W.; Holgate, S.T.; Davies, D.E. IL-4 receptor alpha is an important modulator of IL-4 and IL-13 receptor binding: Implications for the development of therapeutic targets. J. Immunol. 2006, 176, 7456–7461. [Google Scholar] [CrossRef]
- Hölscher, C.; Heitmann, L.; Owusu-Dabo, E.; Horstmann, R.D.; Meyer, C.G.; Ehlers, S.; Thye, T. A mutation in IL4RA is associated with the degree of pathology in human TB patients. Mediat. Inflamm. 2016, 2016, 4245028. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Heitmann, L.; Abad Dar, M.; Schreiber, T.; Erdmann, H.; Behrends, J.; Mckenzie, A.N.; Brombacher, F.; Ehlers, S.; Hölscher, C. The IL-13/IL-4Rα axis is involved in tuberculosis-associated pathology. J. Pathol. 2014, 234, 338–350. [Google Scholar] [CrossRef] [PubMed]
- Parihar, S.P.; Ozturk, M.; Höft, M.A.; Chia, J.E.; Guler, R.; Keeton, R.; van Rensburg, I.C.; Loxton, A.G.; Brombacher, F. IL-4-responsive B cells are detrimental during chronic tuberculosis infection in mice. Front. Immunol. 2021, 12, 611673. [Google Scholar] [CrossRef]
- Mihara, M.; Hashizume, M.; Yoshida, H.; Suzuki, M.; Shiina, M. IL-6/IL-6 receptor system and its role in physiological and pathological conditions. Clin. Sci. 2012, 122, 143–159. [Google Scholar] [CrossRef]
- Simpson, R.J.; Hammacher, A.; Smith, D.K.; Matthews, J.M.; Ward, L.D. Interleukin-6: Structure-function relationships. Protein Sci. 1997, 6, 929–955. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Narazaki, M.; Kishimoto, T. IL-6 in inflammation, immunity, and disease. Cold Spring Harb. Perspect. Biol. 2014, 6, a016295. [Google Scholar] [CrossRef] [PubMed]
- Boni, F.G.; Hamdi, I.; Koundi, L.M.; Shrestha, K.; Xie, J. Cytokine storm in tuberculosis and IL-6 involvement. Infect. Genet. Evol. 2021, 97, 105166. [Google Scholar] [CrossRef]
- Domingo-Gonzalez, R.; Prince, O.; Cooper, A.; Khader, S.A. Cytokines and chemokines in Mycobacterium tuberculosis infection. In Tuberculosis and the Tubercle Bacillus, 2nd ed.; William, R.J., Jr., Helen, M., Valerie, M., Ian, M.O., Eds.; ASM Press: Washington, DC, USA, 2017; pp. 33–72. [Google Scholar]
- Rantala, A.; Lajunen, T.; Juvonen, R.; Silvennoinen-Kassinen, S.; Peitso, A.; Vainio, O.; Saikku, P.; Leinonen, M. Association of IL-6 and IL-6R gene polymorphisms with susceptibility to respiratory tract infections in young Finnish men. Hum. Immunol. 2011, 72, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Ritter, K.; Sodenkamp, J.C.; Hölscher, A.; Behrends, J.; Hölscher, C. IL-6 Is Not Absolutely Essential for the Development of a TH17 Immune Response after an Aerosol Infection with Mycobacterium tuberculosis H37RV. Cells 2021, 10, 9. [Google Scholar] [CrossRef]
- Delgobo, M.; Mendes, D.A.; Kozlova, E.; Rocha, E.L.; Rodrigues-Luiz, G.F.; Mascarin, L.; Dias, G.; Patrício, D.O.; Dierckx, T.; Bicca, M.A.; et al. An evolutionary recent IFN/IL-6/CEBP axis is linked to monocyte expansion and tuberculosis severity in humans. eLife 2019, 8, e47013. [Google Scholar] [CrossRef]
- Sinlgaglia, F.; Ambrosio, D.D.; Panina-Bordignon, P.; Rogse, L. Regulation of the IL-12/IL-12R axis: A critical step in T-helper cell differentiation and effector function. Immunol. Rev. 1999, 170, 65–72. [Google Scholar] [CrossRef]
- Wu, C.Y.; Warrier, R.R.; Carvajal, D.M.; Chua, A.O.; Minetti, L.J.; Chizzonite, R.; Mongini, P.K.A.; Stern, A.S.; Gubler, U.; Presky, D.H.; et al. Biological function and distribution of human interleukin-12 receptor β chain. Eur. J. Immunol. 1996, 26, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Gong, J.; Presky, D.H.; Xue, W.; Barnes, P.F. Expression of the IL-12 Receptor β1 and β2 Subunits in Human Tuberculosis. J. Immunol. 1999, 162, 2441–2447. [Google Scholar]
- Bloch, Y.; Bouchareychas, L.; Merceron, R.; Składanowska, K.; Van den Bossche, L.; Detry, S.; Govindarajan, S.; Elewaut, D.; Haerynck, F.; Dullaers, M.; et al. Structural activation of pro-inflammatory human cytokine IL-23 by cognate IL-23 receptor enables recruitment of the shared receptor IL-12Rβ1. Immunity 2018, 48, 45. [Google Scholar] [CrossRef]
- Tang, C.; Chen, S.; Qian, H.; Huang, W. Interleukin-23: As a drug target for autoimmune inflammatory diseases. Immunology 2012, 135, 112–124. [Google Scholar] [CrossRef]
- Shen, H.; Chen, Z.W. The crucial roles of Th17-related cytokines/signal pathways in M. tuberculosis infection. Cell. Mol. Immunol. 2018, 15, 216–225. [Google Scholar] [CrossRef]
- Khader, S.A.; Bell, G.K.; Pearl, J.E.; Fountain, J.J.; Rangel-Moreno, J.; Cilley, G.E.; Shen, F.; Eaton, S.M.; Gaffen, S.L.; Swain, S.L.; et al. IL-23 and IL-17 in the establishment of protective pulmonary CD4+ T cell responses after vaccination and during Mycobacterium tuberculosis challenge. Nat. Immunol. 2007, 84, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Wang, Y.; Chen, C.Y.; Frencher, J.; Huang, D.; Yang, E.; Ryan-Payseur, B.; Chen, Z.W. Th17-related cytokines contribute to recall-like expansion/effector function of HMBPP-specific Vγ2Vδ2 T cells after Mycobacterium tuberculosis infection or vaccination. Eur. J. Immunol. 2015, 45, 442–451. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, B.S.; Kastelein, R.A.; Cua, D.J. Understanding the IL-23–IL-17 immune pathway. Trends Immunol. 2006, 27, 17–23. [Google Scholar] [CrossRef]
- Shen, H.; Gu, J.; Xiao, H.; Liang, S.; Yang, E.; Yang, R.; Huang, D.; Chen, C.; Wang, F.; Shen, L.; et al. Selective Destruction of Interleukin 23–Induced Expansion of a Major Antigen–Specific γδ T-Cell Subset in Patients With Tuberculosis. J. Infect. Dis. 2017, 215, 420. [Google Scholar] [CrossRef] [PubMed]
- Renauld, J.C. Class II cytokine receptors and their ligands: Key antiviral and inflammatory modulators. Nat. Rev. Immunol. 2003, 38, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Ivashkiv, L.B. IFNγ: Signalling, epigenetics and roles in immunity, metabolism, disease and cancer immunotherapy. Nat. Rev. Immunol. 2018, 18, 545. [Google Scholar] [CrossRef]
- Tau, G.; Rothman, P. Biologic functions of the IFN-gamma receptors. Allergy 1999, 54, 1233–1251. [Google Scholar] [CrossRef] [PubMed]
- Kak, G.; Tiwari, B.K.; Singh, Y.; Natarajan, K. Regulation of Interferon-γreceptor (IFN-γR) expression in macrophages during Mycobacterium tuberculosis infection. Biomol. Concepts 2020, 11, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Cooke, G.S.; Campbell, S.J.; Sillah, J.; Gustafson, P.; Bah, B.; Sirugo, G.; Bennett, S.; McAdam, K.P.W.J.; Sow, O.; Lienhardt, C.; et al. Polymorphism within the Interferon-γ/Receptor Complex Is Associated with Pulmonary Tuberculosis. Am. J. Respir. Crit. Care Med. 2006, 174, 339–343. [Google Scholar] [CrossRef]
- Singhal, A.; Jaiswal, A.; Arora, V.K.; Prasad, H.K. Modulation of gamma interferon receptor 1 by Mycobacterium tuberculosis: A potential immune response evasive mechanism. Infect. Immun. 2007, 75, 2500–2510. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gupta, D.; Sharma, S.; Singhal, J.; Satsangi, A.T.; Antony, C.; Natarajan, K. Suppression of TLR2-induced IL-12, reactive oxygen species, and inducible nitric oxide synthase expression by Mycobacterium tuberculosis antigens expressed inside macrophages during the course of infection. J. Immunol. 2010, 184, 5444–5455. [Google Scholar] [CrossRef] [PubMed]
- Couper, K.N.; Blount, D.G.; Riley, E.M. IL-10: The master regulator of immunity to infection. J. Immunol. 2008, 180, 5771–5777. [Google Scholar] [CrossRef] [PubMed]
- Iyer, S.S.; Cheng, G. Role of Interleukin 10 Transcriptional Regulation in Inflammation and Autoimmune Disease. Crit. Rev. Immunol. 2012, 32, 23. [Google Scholar] [CrossRef] [PubMed]
- Shouval, D.S.; Ouahed, J.; Biswas, A.; Goettel, J.A.; Horwitz, B.H.; Klein, C.; Muise, A.M.; Snapper, S.B. Interleukin 10 Receptor Signaling: Master Regulator of Intestinal Mucosal Homeostasis in Mice and Humans. Adv. Immunol. 2014, 122, 177. [Google Scholar] [CrossRef]
- Murray, P.J. The primary mechanism of the IL-10-regulated antiinflammatory response is to selectively inhibit transcription. Proc. Natl. Acad. Sci. USA 2005, 102, 8686–8691. [Google Scholar] [CrossRef] [PubMed]
- Turner, J.; Gonzalez-Juarrero, M.; Ellis, D.L.; Basaraba, R.J.; Kipnis, A.; Orme, I.M.; Cooper, A.M. In vivo IL-10 production reactivates chronic pulmonary tuberculosis in C57BL/6 mice. J. Immunol. 2002, 169, 6343–6351. [Google Scholar] [CrossRef]
- Redford, P.S.; Boonstra, A.; Read, S.; Pitt, J.; Graham, C.; Stavropoulos, E.; Bancroft, G.J.; O’Garra, A. Enhanced protection to Mycobacterium tuberculosis infection in IL-10-deficient mice is accompanied by early and enhanced Th1 responses in the lung. Eur. J. Immunol. 2010, 40, 2200–2210. [Google Scholar] [CrossRef] [PubMed]
- Beamer, G.L.; Flaherty, D.K.; Assogba, B.D.; Stromberg, P.; Gonzalez-Juarrero, M.; de Waal Malefyt, R.; Vesosky, B.; Turner, J. Interleukin-10 promotes Mycobacterium tuberculosis disease progression in CBA/J mice. J. Immunol. 2008, 181, 5545–5550. [Google Scholar] [CrossRef]
- Cyktor, J.C.; Carruthers, B.; Kominsky, R.A.; Beamer, G.L.; Stromberg, P.; Turner, J. IL-10 inhibits mature fibrotic granuloma formation during Mycobacterium tuberculosis infection. J. Immunol. 2013, 190, 2778–2790. [Google Scholar] [CrossRef]
- Pitt, J.M.; Stavropoulos, E.; Redford, P.S.; Beebe, A.M.; Bancroft, G.J.; Young, D.B.; O’Garra, A. Blockade of IL-10 signaling during BCG vaccination enhances and sustains Th1, Th17, and innate lymphoid IFN-γ and IL-17 responses and increases protection to Mycobacterium tuberculosis infection. J. Immunol. 2012, 189, 4079. [Google Scholar] [CrossRef]
- Dwivedi, V.; Gautam, S.; Headley, C.A.; Piergallini, T.; Torrelles, J.B.; Turner, J. IL-10 receptor blockade delivered simultaneous with BCG vaccination sustains long term protection against Mycobacterium tuberculosis infection in mice. BioRxiv. 2021, 458995. [Google Scholar] [CrossRef]
- Zenewicz, L.A. IL-22: There Is a Gap in Our Knowledge. ImmunoHorizons 2018, 2, 198–207. [Google Scholar] [CrossRef]
- Dudakov, J.A.; Hanash, A.M.; Van Den Brink, M.R.M. Interleukin-22: Immunobiology and pathology. Annu. Rev. Immunol. 2015, 33, 747–785. [Google Scholar] [CrossRef] [PubMed]
- Parks, O.B.; Pociask, D.A.; Hodzic, Z.; Kolls, J.K.; Good, M. Interleukin-22 signaling in the regulation of intestinal health and disease. Front. Cell Dev. Biol. 2016, 3, 85. [Google Scholar] [CrossRef] [PubMed]
- Arshad, T.; Mansur, F.; Palek, R.; Manzoor, S.; Liska, V. A Double Edged Sword Role of Interleukin-22 in Wound Healing and Tissue Regeneration. Front. Immunol. 2020, 11, 2148. [Google Scholar] [CrossRef]
- Aujla, S.J.; Chan, Y.R.; Zheng, M.; Fei, M.; Askew, D.J.; Pociask, D.A.; Reinhart, T.A.; McAllister, F.; Edeal, J.; Gaus, K.; et al. IL-22 mediates mucosal host defense against Gram-negative bacterial pneumonia. Nat. Med. 2008, 14, 275. [Google Scholar] [CrossRef] [PubMed]
- Zeng, G.; Chen, C.Y.; Huang, D.; Yao, S.; Wang, R.C.; Chen, Z.W. Membrane-bound IL-22 after de novo production in tuberculosis and anti-M.tuberculosis effector function of IL-22+CD4+ T cells. J. Immunol. 2011, 187, 190. [Google Scholar] [CrossRef] [PubMed]
- Ronacher, K.; Sinha, R.; Cestari, M. IL-22: An Underestimated Player in Natural Resistance to Tuberculosis? Front. Immunol. 2018, 9, 2209. [Google Scholar] [CrossRef]
- Whittington, H.A.; Armstrong, L.; Uppington, K.M.; Millar, A.B. Interleukin-22: A potential immunomodulatory molecule in the lung. Am. J. Respir. Cell Mol. Biol. 2004, 31, 220–226. [Google Scholar] [CrossRef]
- Treerat, P.; Prince, O.; Cruz-Lagunas, A.; Muñoz-Torrico, M.; Salazar-Lezama, M.A.; Selman, M.; Fallert-Junecko, B.; Reinhardt, T.A.; Alcorn, J.F.; Kaushal, D.; et al. Novel role for IL-22 in protection during chronic Mycobacterium tuberculosis HN878 infection. Mucosal Immunol. 2017, 10, 1069–1081. [Google Scholar] [CrossRef]
- Chu, W.M. Tumor necrosis factor. Cancer Lett. 2013, 328, 222. [Google Scholar] [CrossRef] [PubMed]
- Segueni, N.; Benmerzoug, S.; Rose, S.; Gauthier, A.; Bourigault, M.L.; Reverchon, F.; Philippeau, A.; Erard, F.; Le Bert, M.; Bouscayrol, H.; et al. Innate myeloid cell TNFR1 mediates first line defence against primary Mycobacterium tuberculosis infection. Sci. Rep. 2016, 6, 22454. [Google Scholar] [CrossRef] [PubMed]
- Dostert, C.; Grusdat, M.; Letellier, E.; Brenner, D. The TNF family of ligands and receptors: Communication modules in the immune system and beyond. Physiol. Rev. 2019, 99, 115–160. [Google Scholar] [CrossRef] [PubMed]
- Holbrook, J.; Lara-Reyna, S.; Jarosz-Griffiths, H.; McDermott, M. Tumour necrosis factor signalling in health and disease. F1000Res 2019, 8, 111. [Google Scholar] [CrossRef] [PubMed]
- Idriss, H.T.; Naismith, J.H. TNF alpha and the TNF receptor superfamily: Structure-function relationship(s). Microsc. Res. Tech. 2000, 50, 184–195. [Google Scholar] [CrossRef]
- Tartaglia, L.A.; Pennica, D.; GoeddelS, D.V. Ligand Passing: The 75-kDa Tumor Necrosis Factor (TNF) Receptor Recruits TNF for Signaling by the 55-kDa TNF’ Receptor*. J. Biol. Chem. 1993, 268, 18542–18548. [Google Scholar] [CrossRef]
- Keeton, R.; du Toit, J.P.; Hsu, N.J.; Dube, F.; Jacobs, M. Immune control of Mycobacterium tuberculosis is dependent on both soluble TNFRp55 and soluble TNFRp75. Immunology 2021, 164, 524–540. [Google Scholar] [CrossRef] [PubMed]
- Hijdra, D.; Vorselaars, A.D.; Grutters, J.C.; Claessen, A.M.; Rijkers, G.T. Differential expression of TNFR1 (CD120a) and TNFR2 (CD120b) on subpopulations of human monocytes. J. Inflamm. 2012, 9, 38. [Google Scholar] [CrossRef] [PubMed]
- Gough, P.; Myles, I.A. Tumor Necrosis Factor Receptors: Pleiotropic Signaling Complexes and Their Differential Effects. Front. Immunol. 2020, 11, 585880. [Google Scholar] [CrossRef]
- Lavrik, I.; Golks, A.; Krammer, P.H. Death receptor signaling. J. Cell Sci. 2005, 118, 265–267. [Google Scholar] [CrossRef]
- Micheau, O.; Tschopp, J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell 2003, 114, 181–190. [Google Scholar] [CrossRef]
- Rossol, M.; Meusch, U.; Pierer, M.; Kaltenhäuser, S.; Häntzschel, H.; Hauschildt, S.; Wagner, U. Interaction between transmembrane TNF and TNFR1/2 mediates the activation of monocytes by contact with T cells. J. Immunol. 2007, 179, 4239–4248. [Google Scholar] [CrossRef]
- Aspalter, R.M.; Eibl, M.M.; Wolf, H.M. Regulation of TCR-mediated T cell activation by TNF-RII. J. Leukoc. Biol. 2003, 74, 572–582. [Google Scholar] [CrossRef]
- Chen, X.; Oppenheim, J.J. The phenotypic and functional consequences of tumour necrosis factor receptor type 2 expression on CD4+ FoxP3+ regulatory T cells. Immunology 2011, 133, 426. [Google Scholar] [CrossRef] [PubMed]
- Maney, N.J.; Reynolds, G.; Krippner-Heidenreich, A.; Hilkens, C.M.U. Dendritic cell maturation and survival are differentially regulated by TNFR1 and TNFR2. J. Immunol. 2014, 193, 4914–4923. [Google Scholar] [CrossRef] [PubMed]
- Wajant, H.; Siegmund, D. TNFR1 and TNFR2 in the Control of the Life and Death Balance of Macrophages. Front. Cell Dev. Biol. 2019, 7, 91. [Google Scholar] [CrossRef]
- Yang, S.; Wang, J.; Brand, D.D.; Zheng, S.G. Role of TNF-TNF Receptor 2 Signal in Regulatory T Cells and Its Therapeutic Implications. Front. Immunol. 2018, 9, 784. [Google Scholar] [CrossRef] [PubMed]
- Ticha, O.; Slanina, P.; Moos, L.; Stichova, J.; Vlkova, M.; Bekeredjian-Ding, I. TNFR2 expression is a hallmark of human memory B cells with suppressive function. Eur. J. Immunol. 2021, 51, 1195–1205. [Google Scholar] [CrossRef]
- Téllez-Navarrete, N.A.; Ramon-Luing, L.A.; Muñoz-Torrico, M.; Preciado-García, M.; Medina-Quero, K.; Hernandez-Pando, R.; Chavez-Galan, L. Anti-tuberculosis chemotherapy alters TNFR2 expression on CD4+ lymphocytes in both drug-sensitive and -resistant tuberculosis: However, only drug-resistant tuberculosis maintains a pro-inflammatory profile after a long time. Mol. Med. 2021, 27, 76. [Google Scholar] [CrossRef] [PubMed]
- Flynn, J.A.L.; Goldstein, M.M.; Chan, J.; Triebold, K.J.; Pfeffer, K.; Lowenstein, C.J.; Schrelber, R.; Mak, T.W.; Bloom, B.R. Tumor necrosis factor-alpha is required in the protective immune response against Mycobacterium tuberculosis in mice. Immunity 1995, 2, 561–572. [Google Scholar] [CrossRef]
- Keeton, R.; Allie, N.; Dambuza, I.; Abel, B.; Hsu, N.J.; Sebesho, B.; Randall, P.; Burger, P.; Fick, E.; Quesniaux, V.F.J.; et al. Soluble TNFRp75 regulates host protective immunity against Mycobacterium tuberculosis. J. Clin. Investig. 2014, 124, 1537. [Google Scholar] [CrossRef] [PubMed]
- Olleros, M.L.; Guler, R.; Corazza, N.; Vesin, D.; Eugster, H.-P.; Marchal, G.; Chavarot, P.; Mueller, C.; Garcia, I. Transmembrane TNF Induces an Efficient Cell-Mediated Immunity and Resistance to Mycobacterium bovis Bacillus Calmette-Guérin Infection in the Absence of Secreted TNF and Lymphotoxin-α. J. Immunol. 2002, 168, 3394–3401. [Google Scholar] [CrossRef]
- Robertson, I.B.; Horiguchi, M.; Zilberberg, L.; Dabovic, B.; Hadjiolova, K.; Rifkin, D.B. Latent TGF-β-binding proteins. Matrix Biol. 2015, 47, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Vander Ark, A.; Cao, J.; Li, X. TGF-β receptors: In and beyond TGF-β signaling. Cell. Signal. 2018, 52, 112–120. [Google Scholar] [CrossRef]
- Goumans, M.J.; Valdimarsdottir, G.; Itoh, S.; Lebrin, F.; Larsson, J.; Mummery, C.; Karlsson, S.; Ten Dijke, P. Activin receptor-like kinase (ALK)1 is an antagonistic mediator of lateral TGF beta/ALK5 signaling. Mol. Cell 2003, 12, 817–828. [Google Scholar] [CrossRef]
- Santibañez, J.F.S.; Quintanilla, M.; Bernabeu, C. TGF-β/TGF-β receptor system and its role in physiological and pathological conditions. Clin. Sci. 2011, 121, 233–251. [Google Scholar] [CrossRef]
- Huang, T.; David, L.; Mendoza, V.; Yang, Y.; Villarreal, M.; De, K.; Sun, L.; Fang, X.; López-Casillas, F.; Wrana, J.L.; et al. TGF-β signalling is mediated by two autonomously functioning TβRI:TβRII pairs. EMBO J. 2011, 30, 1263–1276. [Google Scholar] [CrossRef] [PubMed]
- Gatza, C.E.; Oh, S.Y.; Blobe, G.C. Roles for the type III TGF-beta receptor in human cancer. Cell. Signal. 2010, 22, 1163–1174. [Google Scholar] [CrossRef] [PubMed]
- Warsinske, H.C.; Pienaar, E.; Linderman, J.J.; Mattila, J.T.; Kirschner, D.E. Deletion of TGF-β1 increases bacterial clearance by cytotoxic t cells in a tuberculosis granuloma model. Front. Immunol. 2017, 8, 1843. [Google Scholar] [CrossRef]
- Valentini, D.; Rao, M.; Rane, L.; Rahman, S.; Axelsson-Robertson, R.; Heuchel, R.; Löhr, M.; Hoft, D.; Brighenti, S.; Zumla, A.; et al. Peptide microarray-based characterization of antibody responses to host proteins after bacille Calmette–Guérin vaccination. Int. J. Infect. Dis. 2017, 56, 140–154. [Google Scholar] [CrossRef]
- Bonecini-Almeida, M.G.; Ho, J.L.; Boéchat, N.; Huard, R.C.; Chitale, S.; Doo, H.; Geng, J.; Rego, L.; Lazzarini, L.C.O.; Kritski, A.L.; et al. Down-modulation of lung immune responses by interleukin-10 and transforming growth factor beta (TGF-beta) and analysis of TGF-beta receptors I and II in active tuberculosis. Infect. Immun. 2004, 72, 2628–2634. [Google Scholar] [CrossRef]
- Adams, K.N.; Moguche, A.O.; Plumlee, C.R.; Urdahl, K.B. TGF-β-mediated inhibition of IFN-γ production by Mycobacterium tuberculosis-specific T cells in the infected lung. J. Immunol. 2016, 196, 1. [Google Scholar]
- Dinarello, C.A. Overview of the IL-1 family in innate inflammation and acquired immunity. Immunol. Rev. 2018, 281, 8. [Google Scholar] [CrossRef]
- Dinarello, C.A. The IL-1 family of cytokines and receptors in rheumatic diseases. Nat. Rev. Rheumatol. 2019, 15, 612–632. [Google Scholar] [CrossRef] [PubMed]
- Boraschi, D.; Italiani, P.; Weil, S.; Martin, M.U. The family of the interleukin-1 receptors. Immunol. Rev. 2018, 281, 197–232. [Google Scholar] [CrossRef]
- Buhl, A.L.; Wenzel, J. Interleukin-36 in Infectious and Inflammatory Skin Diseases. Front. Immunol. 2019, 10, 1162. [Google Scholar] [CrossRef] [PubMed]
- Fremond, C.M.; Togbe, D.; Doz, E.; Rose, S.; Vasseur, V.; Maillet, I.; Jacobs, M.; Ryffel, B.; Quesniaux, V.F.J. IL-1 Receptor-Mediated Signal Is an Essential Component of MyD88-Dependent Innate Response to Mycobacterium tuberculosis Infection. J. Immunol. 2007, 179, 1178–1189. [Google Scholar] [CrossRef] [PubMed]
- Juffermans, N.P.; Florquin, S.; Camoglio, L.; Verbon, A.; Kolk, A.H.; Speelman, P.; Van Deventer, S.J.H.; Van der Poll, T. Interleukin-1 signaling is essential for host defense during murine pulmonary tuberculosis. J. Infect. Dis. 2000, 182, 902–908. [Google Scholar] [CrossRef] [PubMed]
- Rosas, L.E.; Barbi, J.; Lu, B.; Fujiwara, Y.; Gerard, C.; Sanders, V.M.; Satoskar, A.R. CXCR3−/− mice mount an efficient Th1 response but fail to control Leishmania major infection. Eur. J. Immunol. 2005, 35, 515–523. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Gu, Z.; Fu, Y.; Wang, J. CXCR3 from chemokine receptor family correlates with immune infiltration and predicts poor survival in osteosarcoma. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef]
- Sun, H.; Fan, J.; Shang, X.; Tuohetaerbaike, B.; Li, Y.; Lv, J.; Wang, Y.; Wang, L.; Wang, J.; Ma, X. Study on the relationship between CXCR3 and its ligands and tubal tuberculosis. Life Sci. 2021, 272, 119047. [Google Scholar] [CrossRef]
- Michlmayr, D.; McKimmie, C.S. Role of CXCL10 in central nervous system inflammation. Int. J. Interf. Cytokine Mediat. Res. 2014, 6, 1–18. [Google Scholar] [CrossRef]
- Kauffman, K.D.; Sallin, M.A.; Sakai, S.; Kamenyeva, O.; Kabat, J.; Weiner, D.; Sutphin, M.; Schimel, D.; Via, L.; Barry, C.E.; et al. Defective positioning in granulomas but not lung-homing limits CD4 T-cell interactions with Mycobacterium tuberculosis-infected macrophages in rhesus macaques. Mucosal Immunol. 2018, 11, 462–473. [Google Scholar] [CrossRef] [PubMed]
- Shanmugasundaram, U.; Bucsan, A.N.; Ganatra, S.R.; Ibegbu, C.; Quezada, M.; Blair, R.V.; Alvarez, X.; Velu, V.; Kaushal, D.; Rengarajan, J. Pulmonary Mycobacterium tuberculosis control associates with CXCR3- and CCR6-expressing antigen-specific Th1 and Th17 cell recruitment. JCI Insight 2020, 5, e137858. [Google Scholar] [CrossRef] [PubMed]
- Seiler, P.; Aichele, P.; Bandermann, S.; Hauser, A.E.; Lu, B.; Gerard, N.P.; Gerard, C.; Ehlers, S.; Mollenkopf, H.J.; Kaufmann, S.H.E. Early granuloma formation after aerosol Mycobacterium tuberculosis infection is regulated by neutrophils via CXCR3-signaling chemokines. Eur. J. Immunol. 2003, 33, 2676–2686. [Google Scholar] [CrossRef] [PubMed]
- Chakravarty, S.D.; Xu, J.; Lu, B.; Gerard, C.; Flynn, J.; Chan, J. The Chemokine Receptor CXCR3 Attenuates the Control of Chronic Mycobacterium tuberculosis Infection in BALB/c Mice. J. Immunol. 2007, 178, 1723–1735. [Google Scholar] [CrossRef] [PubMed]
- Bromley, S.K.; Peterson, D.A.; Gunn, M.D.; Dustin, M.L. Cutting edge: Hierarchy of chemokine receptor and TCR signals regulating T cell migration and proliferation. J. Immunol. 2000, 165, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Shang, X.; Wang, L.; Liu, Y.; Liu, X.; Lv, J.; Zhou, X.; Wang, H.; Nazierhan, S.; Wang, J.; Ma, X. Diagnostic value of CXCR3 and its ligands in spinal tuberculosis. Exp. Ther. Med. 2021, 21, 73. [Google Scholar] [CrossRef]
- Yu, S.; Shen, J.; Lao, S.; Yang, B.; Wu, C. Distinct functions of CXCR3+ and CCR4+CD4+ T-cells accumulated in human tuberculosis pleural fluid. Int. J. Tuberc. Lung Dis. 2018, 22, 1514–1522. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Chung, W.; Jung, Y.; Kim, Y.; Park, J.; Sheen, S.; Park, K. CXCR3 ligands as clinical markers for pulmonary tuberculosis. Int. J. Tuberc. Lung Dis. 2015, 19, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Krupa, A.; Fol, M.; Dziadek, B.R.; Kepka, E.; Wojciechowska, D.; Brzostek, A.; Torzewska, A.; Dziadek, J.; Baughman, R.P.; Griffith, D.; et al. Binding of CXCL8/IL-8 to Mycobacterium tuberculosis Modulates the Innate Immune Response. Mediat. Inflamm. 2015, 2015, 124762. [Google Scholar] [CrossRef] [PubMed]
- Horuk, R. The Duffy antigen receptor for chemokines DARC/ACKR1. Front. Immunol. 2015, 6, 279. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Das, B.B.; Casagrande, F.; Tian, Y.; Nothnagel, H.J.; Chu, M.; Kiefer, H.; Maier, K.; De Angelis, A.A.; Marassi, F.M.; et al. Structure of the chemokine receptor CXCR1 in phospholipid bilayers. Nature 2012, 491, 779–783. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, S.K.; Özçelik, T.; Milatovitch, A.; Francke, U.; Murphy, P.M. Molecular evolution of the human interleukin–8 receptor gene cluster. Nat. Genet. 1992, 2, 31–36. [Google Scholar] [CrossRef][Green Version]
- Ha, H.; Debnath, B.; Neamati, N. Role of the CXCL8-CXCR1/2 Axis in Cancer and Inflammatory Diseases. Theranostics 2017, 7, 1543–1588. [Google Scholar] [CrossRef] [PubMed]
- Murphy, P. Neutrophil receptors for interleukin-8 and related CXC chemokines. Semin. Hematol. 1997, 34, 311–318. [Google Scholar] [PubMed]
- Hammond, M.E.; Lapointe, G.R.; Feucht, P.H.; Hilt, S.; Gallegos, C.A.; Gordon, C.A.; Giedlin, M.A.; Mullenbach, G.; Tekamp-Olson, P. IL-8 induces neutrophil chemotaxis predominantly via type I IL-8 receptors. J. Immunol. 1995, 155, 1428–1433. [Google Scholar] [PubMed]
- Alaridah, N.; Winqvist, N.; Håkansson, G.; Tenland, E.; Rönnholm, A.; Sturegård, E.; Björkman, P.; Godaly, G. Impaired CXCR1-dependent oxidative defence in active tuberculosis patients. Tuberculosis 2015, 95, 744–750. [Google Scholar] [CrossRef] [PubMed]
- Juffermans, N.P.; Dekkers, P.E.P.; Peppelenbosch, M.P.; Speelman, P.; Van Deventer, S.J.H.; Van der Poll, T. Expression of the Chemokine Receptors CXCR1 and CXCR2 on Granulocytes in Human Endotoxemia and Tuberculosis: Involvement of the p38 Mitogen—Activated Protein Kinase Pathway. J. Infect. Dis. 2000, 182, 888–894. [Google Scholar] [CrossRef] [PubMed]
- Meddows-Taylor, S.; Martin, D.J.; Tiemessen, C.T. Reduced expression of interleukin-8 receptors A and B on polymorphonuclear neutrophils from persons with human immunodeficiency virus type 1 disease and pulmonary tuberculosis. J. Infect. Dis. 1998, 177, 921–930. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Alaridah, N.; Lutay, N.; Tenland, E.; Rönnholm, A.; Hallgren, O.; Puthia, M.; Westergren-Thorsson, G.; Godaly, G. Mycobacteria Manipulate G-Protein-Coupled Receptors to Increase Mucosal Rac1 Expression in the Lungs. J. Innate Immun. 2017, 9, 318–329. [Google Scholar] [CrossRef] [PubMed]
- Antas, P.; Holland, S.; Sterling, T. Abnormal spontaneous interleukin 8 receptor expression: A brief report of two cases. Rev. Soc. Bras. Med. Trop. 2012, 45, 134. [Google Scholar] [CrossRef] [PubMed]
- Zenobia, C.; Hajishengallis, G. Basic biology and role of interleukin-17 in immunity and inflammation. J. Periodontol. 2000, 69, 142. [Google Scholar] [CrossRef] [PubMed]
- Monin, L.; Gaffen, S.L. Interleukin 17 Family Cytokines: Signaling Mechanisms, Biological Activities, and Therapeutic Implications. Cold Spring Harb. Perspect. Biol. 2018, 10, a028522. [Google Scholar] [CrossRef] [PubMed]
- Amatya, N.; Garg, A.V.; Gaffen, S.L. IL-17 Signaling: The Yin and the Yang. Trends Immunol. 2017, 38, 310–322. [Google Scholar] [CrossRef] [PubMed]
- Freches, D.; Korf, H.; Denis, O.; Havaux, X.; Huygen, K.; Romano, M. Mice genetically inactivated in interleukin-17A receptor are defective in long-term control of Mycobacterium tuberculosis infection. Immunology 2013, 140, 220–231. [Google Scholar] [CrossRef]
- Lombard, R.; Doz, E.; Carreras, F.; Epardaud, M.; Le Vern, Y.; Buzoni-Gatel, D.; Winter, N. IL-17RA in Non-Hematopoietic Cells Controls CXCL-1 and 5 Critical to Recruit Neutrophils to the Lung of Mycobacteria-Infected Mice during the Adaptive Immune Response. PLoS ONE 2016, 11, e0149455. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kamijo, R.; Le, J.; Shapiro, D.; Havell, E.A.; Huang, S.; Aguet, M.; Bosland, M.; Vilček, J. Mice that lack the interferon-gamma receptor have profoundly altered responses to infection with Bacillus Calmette-Guérin and subsequent challenge with lipopolysaccharide. J. Exp. Med. 1993, 178, 1435–1440. [Google Scholar] [CrossRef] [PubMed]
- de Souza, T.L.; Fernandes, R.C.d.S.C.; da Silva, J.A.; Alves, V.G.; Coelho, A.G.; Faria, A.C.S.; Simão, N.M.M.S.; Filho, J.T.S.; Deswarte, C.; Boisson-Dupuis, S.; et al. Microbial Disease Spectrum Linked to a Novel IL-12Rβ1 N-Terminal Signal Peptide Stop-Gain Homozygous Mutation with Paradoxical Receptor Cell-Surface Expression. Front. Microbiol. 2017, 8, 616. [Google Scholar] [CrossRef]
- Taur, P.D.; Gowri, V.; Pandrowala, A.A.; Iyengar, V.V.; Chougule, A.; Golwala, Z.; Chandak, S.; Agarwal, R.; Keni, P.; Dighe, N.; et al. Clinical and Molecular Findings in Mendelian Susceptibility to Mycobacterial Diseases: Experience From India. Front. Immunol. 2021, 12, 426. [Google Scholar] [CrossRef] [PubMed]
- Bustamante, J.; Boisson-Dupuis, S.; Abel, L.; Casanova, J.L. Mendelian susceptibility to mycobacterial disease: Genetic, immunological, and clinical features of inborn errors of IFN-γ immunity. Semin. Immunol. 2014, 26, 454–470. [Google Scholar] [CrossRef] [PubMed]
- Ottenhoff, T.H.M.; Verreck, F.A.W.; Lichtenauer-Kaligis, E.G.R.; Hoeve, M.A.; Sanal, O.; van Dissel, J.T. Genetics, cytokines and human infectious disease: Lessons from weakly pathogenic mycobacteria and salmonellae. Nat. Genet. 2002, 32, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Alejo, N.; Santos-Argumedo, L. Innate Defects of the IL-12/IFN-γ Axis in Susceptibility to Infections by Mycobacteria and Salmonella. J. Interf. Cytokine Res. 2014, 34, 307. [Google Scholar] [CrossRef]
- Altare, F.; Ensser, A.; Breiman, A.; Reichenbach, J.; El Baghdadi, J.; Fischer, A.; Emile, J.F.; Gaillard, J.L.; Meinl, E.; Casanova, J.L. Interleukin-12 receptor beta1 deficiency in a patient with abdominal tuberculosis. J. Infect. Dis. 2001, 184, 231–236. [Google Scholar] [CrossRef]
- Casanova, J.L.; Abel, L. Genetic Dissection of Immunity to Mycobacteria: The Human Model. Annu. Rev. Immunol. 2002, 20, 581. [Google Scholar] [CrossRef] [PubMed]
- Alinejad Dizaj, M.; Mortaz, E.; Mahdaviani, S.A.; Mansouri, D.; Mehrian, P.; Verhard, E.M.; Varahram, M.; Babaie, D.; Adcock, I.M.; Garssen, J.; et al. Susceptibility to mycobacterial disease due to mutations in IL-12Rβ1 in three Iranian patients. Immunogenetics 2018, 70, 373. [Google Scholar] [CrossRef] [PubMed]







| Family | Cytokine Receptors | Features | Mechanism of Action | Targeting Genes/Responsive Genes |
|---|---|---|---|---|
| type I cytokine receptors | IL-2R, IL-4R, IL-6R, IL-12R, IL-23R, G-CSFR, GM-CSFR |
| transduction of the signaling pathway through the involvement of the non-receptor Janus kinases (JAKs) and the signal transducer and activator of transcription (STATs) factors | IL-2R: IL-2, CTLA-4, Bcl-2, cyclins, FoxP3, FasL, CD25 [19] IL-4R: CD23, MHC class II, IL-4α, GATA3, BDNF/NGF, YM-1 [20,21] IL-6R: CIS, SOCS, Mcl-1, TIMP-1, Pim-1, c-Myc, cytokines, TFs [22,23,24,25,26] IL-12R: IFN-γ, TNF-α, T-bet, PD-1 [27,28] IL-23R: RIG-I, IL-23, IL-23R, IL-17, GM-CSF [28,29] G-CSFR : MMP2, VEGF, GATA-3, ATG2A, and PIAS3 [30,31] GM-CSFR: c-Myc, IL-1β, IL-12p70, IL-6, TNF-α, iNOS [32] |
| type II cytokine receptors | IL-10R, IL-22R, IFN-γR |
| transduction of the signaling pathway through the involvement of the non-receptor Janus kinases (JAKs) and the signal transducer and activator of transcription (STATs) factors | IL-10R: IL-4, IL-13, IL-1β, IL-6, TNF-α, CD80,CD83, CD86, Bcl-2, Bcl-xL, caspase-3 [33,34] IL-22R: Bcl-2, Bcl-xL, MCL-1, Cyclin D1, CDK4, CCL2, RANKL2, CXCL5, S100A7, S100A8, S100A9, lipocalin 2, MMP1, MMP3 [35] IFN-γR : S100A family, IL-7, TAP1, SERPING1, VAMP5, TNFAIP2, TNFSF10, PARP-1, CD274, CCL2, NOX1, NOX4, IRF2 [36,37] |
| tumor necrosis factor (TNF) receptor family | TNFR |
| transduction of the signaling pathways through IKK/NF-κB, JNK/AP-1 and p38 MAP signaling cascades. TNF also can trigger apoptosis via caspase-8 or necroptosis by activating intracellular receptor-interacting serine/threonine-protein (RIPK) kinases | MKP-1, MCP-1, COX-2, IL-6, SOD-2 [38] |
| transforming growth factor (TGF)-β receptor family | TGFBR |
| ligand-bound type II receptors activate type I receptors by phosphorylation, which then autophosphorylate and bind intracellular Sma- and Mad-related proteins (SMAD) | PDGF-B, FoxP3, Bcl-xL, Beclin 1 [39,40] |
| chemokine receptors | IL-8R CXCR3 |
| transduction of the signal by G-protein coupled receptors, which dissociate to activate diverse downstream pathways resulting in cellular polarization and actin reorganization | ICAM-1, VCAM-1, Cox-2 [41] |
| immunoglobulin (Ig) receptor superfamily | IL-1R family |
| signal transduction through the toll/interleukin-1 receptor (TIR) domain, which recruits MyD88 adaptor protein activating the NF-κB pathway. | IL-6, CCL2, IL-8 [39] |
| interleukin (IL)-17 receptor family | IL-17RA-E |
| signaling cascade activates the extracellular signal-regulated protein kinase (ERK), the c-jun N-terminal kinase (JNK), and the p38/MAPK pathway | IL-6, IL-1β, CXCL1, CXCL2, G-CSF [42] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Druszczyńska, M.; Godkowicz, M.; Kulesza, J.; Wawrocki, S.; Fol, M. Cytokine Receptors—Regulators of Antimycobacterial Immune Response. Int. J. Mol. Sci. 2022, 23, 1112. https://doi.org/10.3390/ijms23031112
Druszczyńska M, Godkowicz M, Kulesza J, Wawrocki S, Fol M. Cytokine Receptors—Regulators of Antimycobacterial Immune Response. International Journal of Molecular Sciences. 2022; 23(3):1112. https://doi.org/10.3390/ijms23031112
Chicago/Turabian StyleDruszczyńska, Magdalena, Magdalena Godkowicz, Jakub Kulesza, Sebastian Wawrocki, and Marek Fol. 2022. "Cytokine Receptors—Regulators of Antimycobacterial Immune Response" International Journal of Molecular Sciences 23, no. 3: 1112. https://doi.org/10.3390/ijms23031112
APA StyleDruszczyńska, M., Godkowicz, M., Kulesza, J., Wawrocki, S., & Fol, M. (2022). Cytokine Receptors—Regulators of Antimycobacterial Immune Response. International Journal of Molecular Sciences, 23(3), 1112. https://doi.org/10.3390/ijms23031112







