Integration of the Metabolomic and Transcriptome Analysis Reveals the Remarkable Compounds of G. bicolor Young and Mature Leaves under Different Iron Nutrient Conditions
Abstract
:1. Introduction
2. Results
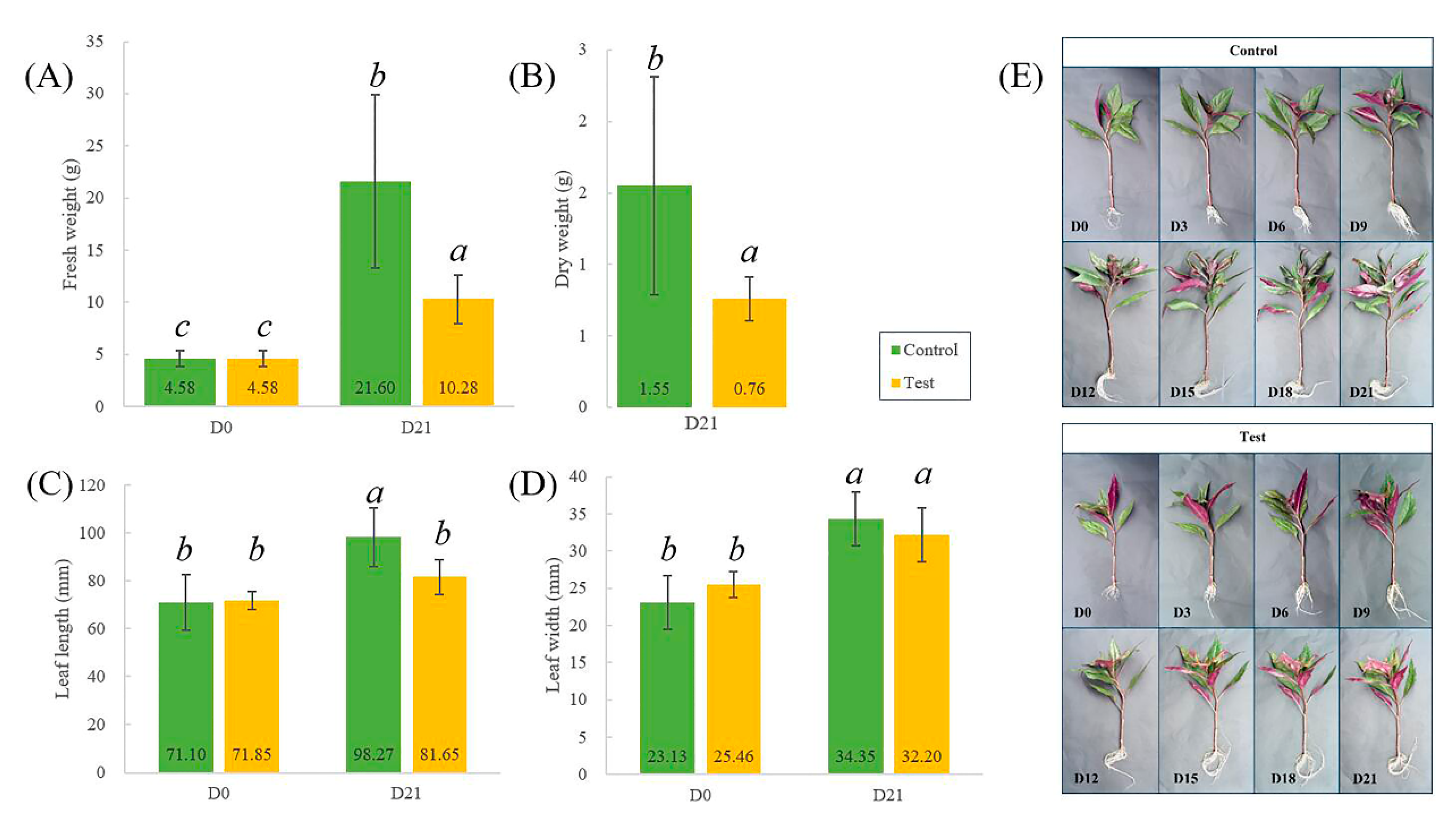
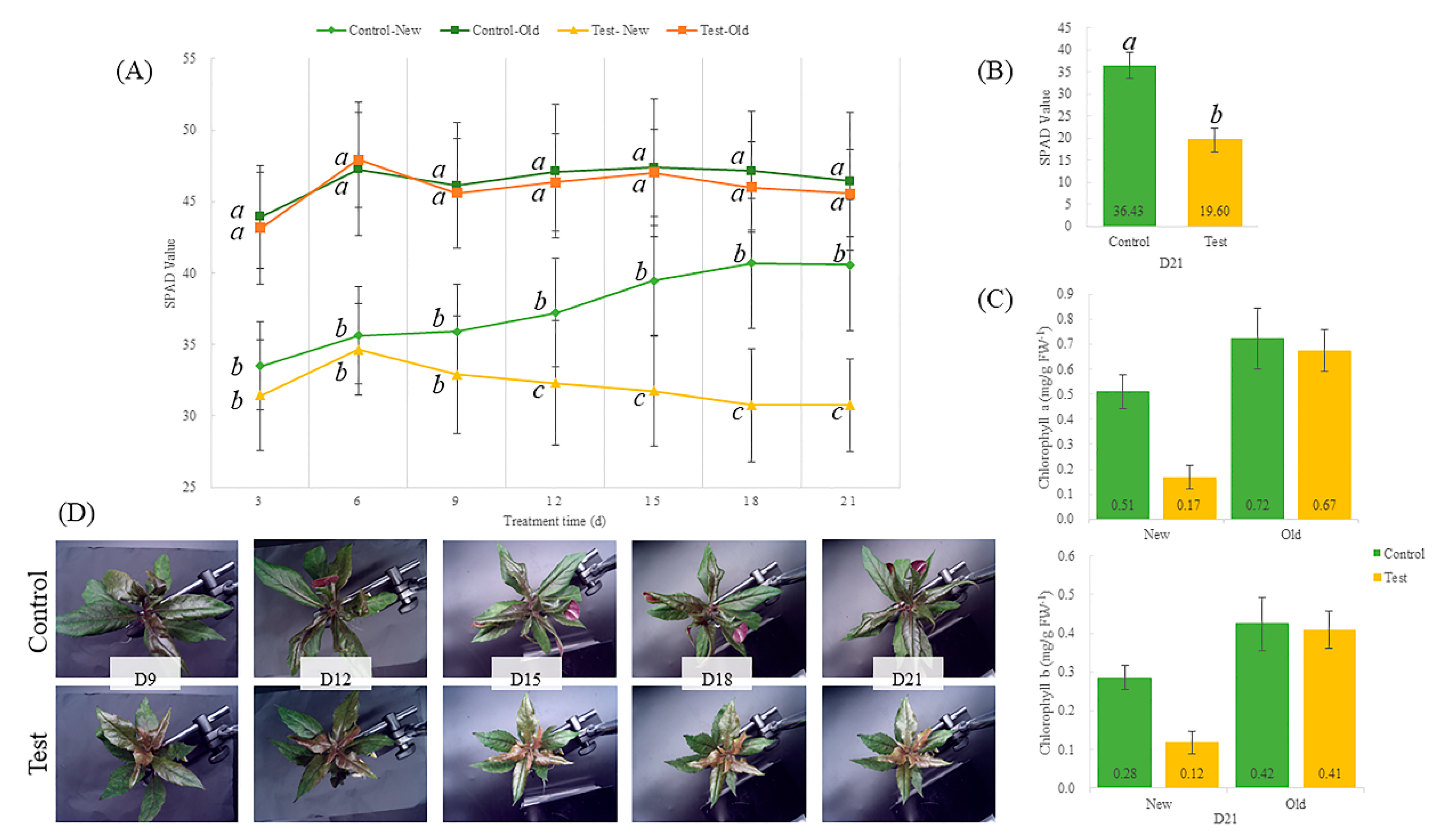
2.1. Phenotypic Analysis of G. bicolor under Fe-Deficiency Stress
2.2. Effect of Fe-Deficiency on Element Content of Leaves
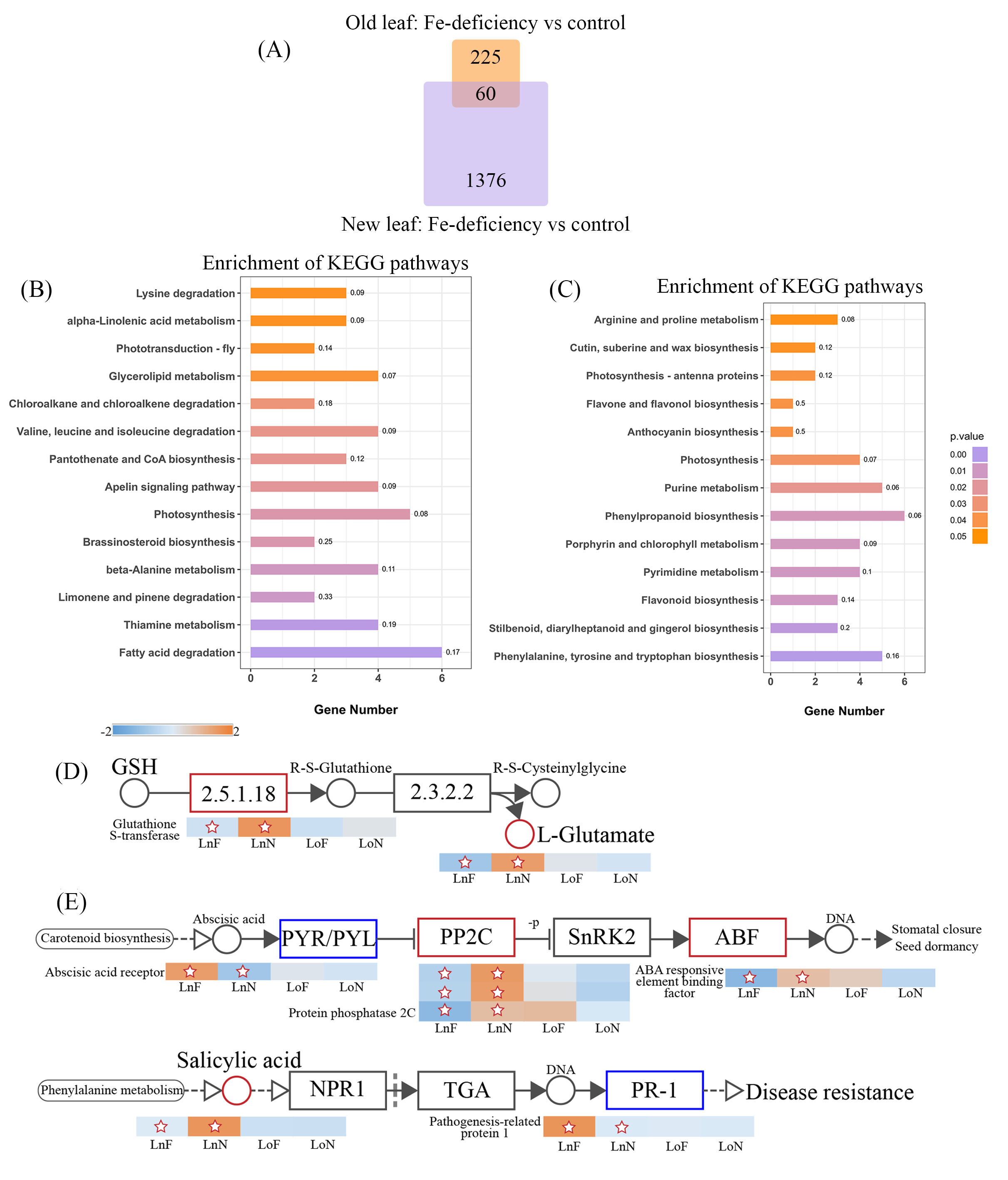
2.3. Transcriptome Analysis of the G. bicolor Response to Fe Deficiency
2.4. The Correlation between the Metabolome and Transcriptome
3. Discussion
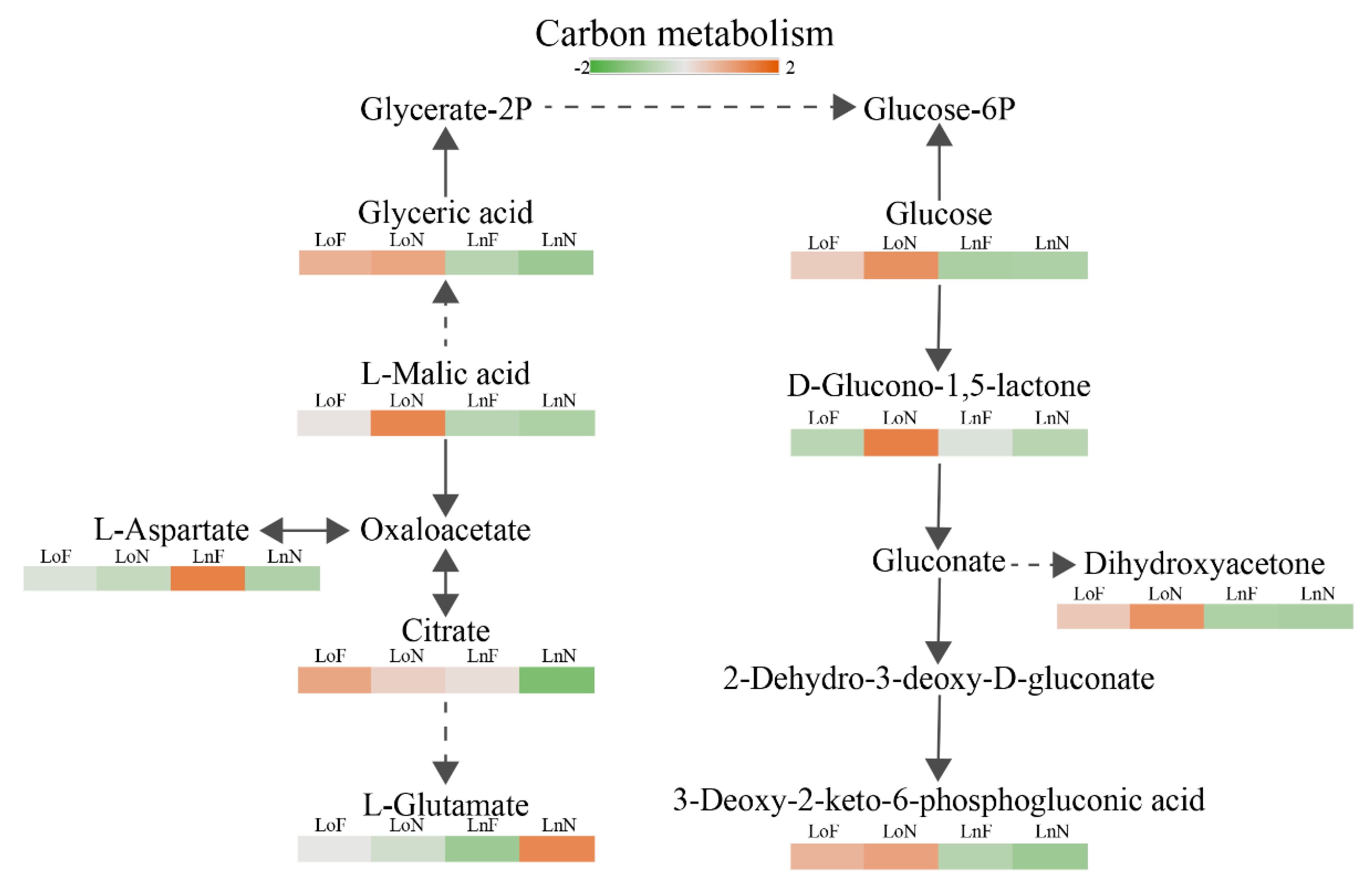
3.1. Impacts of Fe Deficiency on Metabolism
3.2. Impacts of Fe Deficiency on Transcriptome
4. Materials and Methods
4.1. Plant Materials
4.2. Determination of Chlorophyll Content
4.3. Transcriptome Data Acquisition
4.4. Metabolomics Data Acquisition
4.5. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cohen, C.K.; Fox, T.C.; Garvin, D.F.; Kochian, L.V. The Role of Iron-Deficiency Stress Responses in Stimulating Heavy-Metal Transport in Plants. Plant Physiol. 1998, 116, 1063–1072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rotaru, V.; Sinclair, T.R. Interactive influence of phosphorus and iron on nitrogen fixation by soybean. Environ. Exp. Bot. 2009, 66, 94–99. [Google Scholar] [CrossRef]
- Wang, Y.; Hu, Y.; Zhu, Y.; Baloch, A.W.; Jia, X.; Guo, A. Transcriptional and physiological analyses of short-term iron deficiency response in apple seedlings provide insight into the regulation involved in photosynthesis. BMC Genom. 2018, 19, 461. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Zhang, D.; Sun, W.; Wang, T. The Adaptive Mechanism of Plants to Iron Deficiency via Iron Uptake, Transport, and Homeostasis. Int. J. Mol. Sci. 2019, 20, 2424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kathpalia, R.; Bhatla, S.C. Plant Mineral Nutrition. In Plant Physiology, Development and Metabolism; Bhatla, S.C., Lal, M.A., Eds.; Springer: Singapore, 2018; pp. 37–81. ISBN 9789811320231. [Google Scholar]
- Briguglio, M.; Hrelia, S.; Malaguti, M.; Lombardi, G.; Riso, P.; Porrini, M.; Perazzo, P.; Banfi, G. The Central Role of Iron in Human Nutrition: From Folk to Contemporary Medicine. Nutrients 2020, 12, 1761. [Google Scholar] [CrossRef]
- Abbaspour, N.; Hurrell, R.; Kelishadi, R. Review on iron and its importance for human health. J. Res. Med. Sci. 2014, 19, 164–174. [Google Scholar]
- Li, J.; Cao, X.; Jia, X.; Liu, L.; Cao, H.; Qin, W.; Li, M. Iron Deficiency Leads to Chlorosis through Impacting Chlorophyll Synthesis and Nitrogen Metabolism in Areca catechu L. Front. Plant Sci. 2021, 12, 710093. [Google Scholar] [CrossRef]
- Kour, J.; Kohli, S.K.; Khanna, K.; Bakshi, P.; Sharma, P.; Singh, A.D.; Ibrahim, M.; Devi, K.; Sharma, N.; Ohri, P.; et al. Brassinosteroid Signaling, Crosstalk and, Physiological Functions in Plants under Heavy Metal Stress. Front. Plant Sci. 2021, 12, 608061. [Google Scholar] [CrossRef]
- Zhang, X.; Jia, X.; Zhang, R.; Zhu, Z.; Liu, B.; Gao, L.; Wang, Y. Metabolic analysis in Malus halliana leaves in response to iron deficiency. Sci. Hortic. 2019, 258, 108792. [Google Scholar] [CrossRef]
- Qiu, X.-L.; Guo, Y.-X.; Zhang, Q.-F. Chemical profile and antioxidant activity of Gynura bicolor DC. ethanolic extract. Int. J. Food Prop. 2018, 21, 407–415. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.; Chang, W.; Lu, C.; Chang, Y.; Wang, J.; Hsieh, S. Effects of extracts from Gynura bicolor (Roxb. & Willd.) DC. on iron bioavailability in rats. J. Food Drug Anal. 2015, 23, 425–432. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Zhou, M.; Liu, Y.; Ye, Q.; Gu, J.; Luo, G. Isolation and Identification of Antioxidant Compounds from Gynura Bicolor Stems and Leaves. Int. J. Food Prop. 2016, 19, 233–241. [Google Scholar] [CrossRef]
- ShangQuan, R.; Yue, Y.; JianHong, W.; Di, R. Distribution of mineral elements in Gynura bicolor D. C. and its correlation with soil mineral elements. Med. Plant. 2010, 1, 5–7. [Google Scholar]
- Kubes, J.; Skalicky, M.; Tumova, L.; Martin, J.; Hejnak, V.; Martinkova, J. Vanadium elicitation of Trifolium pratense L. cell culture and possible pathways of produced isoflavones transport across the plasma membrane. Plant Cell Rep. 2019, 38, 657–671. [Google Scholar] [CrossRef] [PubMed]
- Chu, Q.; Sha, Z.; Maruyama, H.; Yang, L.; Pan, G.; Xue, L.; Watanabe, T. Metabolic reprogramming in nodules, roots, and leaves of symbiotic soybean in response to iron deficiency. Plant. Cell Environ. 2019, 42, 3027–3043. [Google Scholar] [CrossRef] [PubMed]
- Bouain, N.; Krouk, G.; Lacombe, B.; Rouached, H. Getting to the Root of Plant Mineral Nutrition: Combinatorial Nutrient Stresses Reveal Emergent Properties. Trends Plant Sci. 2019, 24, 542–552. [Google Scholar] [CrossRef]
- Cui, J.; Pottosin, I.; Lamade, E.; Tcherkez, G. What is the role of putrescine accumulated under potassium deficiency? Plant. Cell Environ. 2020, 43, 1331–1347. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, S.; Naldrett, M.J. Mass spectrometry based untargeted metabolomics for plant systems biology. Emerg. Top. Life Sci. 2021, 5, 189–201. [Google Scholar] [CrossRef]
- Zhang, K.; Chen, L.; Wei, M.; Qiao, H.; Zhang, S.; Li, Z.; Fang, Y.; Chen, K. Metabolomic profile combined with transcriptomic analysis reveals the value of UV-C in improving the utilization of waste grape berries. Food Chem. 2021, 363, 130288. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Feng, M.; Yang, K.; Cao, Y.; Zhang, J.; Xu, J.; Hernández, S.H.; Wei, X.; Fan, M. Transcriptomic and metabolomic analyses reveal antibacterial mechanism of astringent persimmon tannin against Methicillin-resistant Staphylococcus aureus isolated from pork. Food Chem. 2020, 309, 125692. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.; Guerinot, M. Lou Homing in on iron homeostasis in plants. Trends Plant Sci. 2009, 14, 280–285. [Google Scholar] [CrossRef]
- Jain, A.; Wilson, G.; Connolly, E. The diverse roles of FRO family metalloreductases in iron and copper homeostasis. Front. Plant Sci. 2014, 5, 100. [Google Scholar] [CrossRef] [Green Version]
- Muhammad, I.; Jing, X.; Shalmani, A.; Ali, M.; Yi, S.; Gan, P.; Li, W.; Liu, W.; Chen, K. Comparative In Silico Analysis of Ferric Reduction Oxidase (FRO) Genes Expression Patterns in Response to Abiotic Stresses, Metal and Hormone Applications. Molecules 2018, 23, 1163. [Google Scholar] [CrossRef] [Green Version]
- Fan, W.; Lou, H.Q.; Gong, Y.L.; Liu, M.Y.; Wang, Z.Q.; Yang, J.L.; Zheng, S.J. Identification of early Al-responsive genes in rice bean (Vigna umbellata) roots provides new clues to molecular mechanisms of Al toxicity and tolerance. Plant. Cell Environ. 2014, 37, 1586–1597. [Google Scholar] [CrossRef]
- Dai, J.; Qiu, W.; Wang, N.; Nakanishi, H.; Zuo, Y. Comparative transcriptomic analysis of the roots of intercropped peanut and maize reveals novel insights into peanut iron nutrition. Plant Physiol. Biochem. 2018, 127, 516–524. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, B.; Li, M.; Feng, D.; Jin, H.; Wang, P.; Liu, J.; Xiong, F.; Wang, J.; Wang, H. The bHLH transcription factor bHLH104 interacts with IAA-LEUCINE RESISTANT3 and modulates iron homeostasis in Arabidopsis. Plant Cell 2015, 27, 787–805. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Yang, C.; Chen, H.; Luo, L.; Leng, Q.; Li, S.; Han, Z.; Li, X.; Song, C.; Zhang, X.; et al. Exploring transcription factors reveals crucial members and regulatory networks involved in different abiotic stresses in Brassica napus L. BMC Plant Biol. 2018, 18, 202. [Google Scholar] [CrossRef] [Green Version]
- Kabir, A.H.; Akther, M.S.; Skalicky, M.; Das, U.; Gohari, G.; Brestic, M.; Hossain, M.M. Downregulation of Zn-transporters along with Fe and redox imbalance causes growth and photosynthetic disturbance in Zn-deficient tomato. Sci. Rep. 2021, 11, 6040. [Google Scholar] [CrossRef]
- Kerry, R.G.; Mahapatra, G.P.; Patra, S.; Sahoo, S.L.; Pradhan, C.; Padhi, B.K.; Rout, J.R. Proteomic and genomic responses of plants to nutritional stress. BioMetals 2018, 31, 161–187. [Google Scholar] [CrossRef]
- Zeng, J.; He, X.; Quan, X.; Cai, S.; Han, Y.; Nadira, U.A.; Zhang, G. Identification of the proteins associated with low potassium tolerance in cultivated and Tibetan wild barley. J. Proteom. 2015, 126, 1–11. [Google Scholar] [CrossRef]
- Yang, Q.; Liu, Y.; Zeng, Q. Overexpression of three orthologous glutathione S-transferases from Populus increased salt and drought resistance in Arabidopsis. Biochem. Syst. Ecol. 2019, 83, 57–61. [Google Scholar] [CrossRef]
- Forde, B.G.; Lea, P.J. Glutamate in plants: Metabolism, regulation, and signalling. J. Exp. Bot. 2007, 58, 2339–2358. [Google Scholar] [CrossRef]
- Wang, W.; Wang, J.; Wei, Q.; Li, B.; Zhong, X.; Hu, T.; Hu, H.; Bao, C. Transcriptome-Wide Identification and Characterization of Circular RNAs in Leaves of Chinese Cabbage (Brassica rapa L. ssp. pekinensis) in Response to Calcium Deficiency-Induced Tip-burn. Sci. Rep. 2019, 9, 14544. [Google Scholar] [CrossRef] [Green Version]
- Dossa, K.; Li, D.; Wang, L.; Zheng, X.; Liu, A.; Yu, J.; Wei, X.; Zhou, R.; Fonceka, D.; Diouf, D.; et al. Transcriptomic, biochemical and physio-anatomical investigations shed more light on responses to drought stress in two contrasting sesame genotypes. Sci. Rep. 2017, 7, 8755. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, P.L. Protein phosphatase 2C (PP2C) function in higher plants. Plant Mol. Biol. 1998, 38, 919–927. [Google Scholar] [CrossRef]
- Singh, A.; Yadav, A.K.; Kaur, K.; Sanyal, S.K.; Jha, S.K.; Fernandes, J.L.; Sharma, P.; Tokas, I.; Pandey, A.; Luan, S.; et al. A protein phosphatase 2C, AP2C1, interacts with and negatively regulates the function of CIPK9 under potassium-deficient conditions in Arabidopsis. J. Exp. Bot. 2018, 69, 4003–4015. [Google Scholar] [CrossRef] [Green Version]
- Tiwari, J.K.; Buckseth, T.; Zinta, R.; Saraswati, A.; Singh, R.K.; Rawat, S.; Dua, V.K.; Chakrabarti, S.K. Transcriptome analysis of potato shoots, roots and stolons under nitrogen stress. Sci. Rep. 2020, 10, 1152. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Ge, H.; Wang, X.; Tang, R.; Wang, Y.; Zhao, F.; Lan, W.; Luan, S.; Yang, L. Overexpression of Pyrabactin Resistance-Like Abscisic Acid Receptors Enhances Drought, Osmotic, and Cold Tolerance in Transgenic Poplars. Front. Plant Sci. 2017, 8, 1752. [Google Scholar] [CrossRef]
- Arrivault, S.; Senger, T.; Krämer, U. The Arabidopsis metal tolerance protein AtMTP3 maintains metal homeostasis by mediating Zn exclusion from the shoot under Fe deficiency and Zn oversupply. Plant J. 2006, 46, 861–879. [Google Scholar] [CrossRef]
- Vert, G.; Grotz, N.; Dédaldéchamp, F.; Gaymard, F.; Guerinot, M.L.; Briat, J.-F.; Curie, C. IRT1, an Arabidopsis Transporter Essential for Iron Uptake from the Soil and for Plant Growth. Plant Cell 2002, 14, 1223–1233. [Google Scholar] [CrossRef] [Green Version]
- Eprintsev, A.T.; Selivanova, N.V.; Igamberdiev, A.U. Enzymatic Conversions of Glutamate and γ-Aminobutyric Acid as Indicators of Plant Stress Response. In Nitrogen Metabolism in Plants; Gupta, K.J., Ed.; Springer: New York, NY, USA, 2020; Volume 2057, pp. 71–78. ISBN 9781493997909. [Google Scholar]
- Shen, C.; Yang, Y.; Liu, K.; Zhang, L.; Guo, H.; Sun, T.; Wang, H. Involvement of endogenous salicylic acid in iron-deficiency responses in Arabidopsis. J. Exp. Bot. 2016, 67, 4179–4193. [Google Scholar] [CrossRef] [Green Version]
- Urbanczyk-Wochniak, E.; Fernie, A.R. Metabolic profiling reveals altered nitrogen nutrient regimes have diverse effects on the metabolism of hydroponically-grown tomato (Solanum lycopersicum) plants. J. Exp. Bot. 2005, 56, 309–321. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Lin, L. Effect of Heat Stress on Sargassum fusiforme Leaf Metabolome. J. Plant Biol. 2020, 63, 229–241. [Google Scholar] [CrossRef] [Green Version]
- Popp, M.; Kinzel, H. Changes in the Organic Acid Content of some Cultivated Plants Induced by Mineral Ion Deficiency. J. Exp. Bot. 1981, 32, 1–8. [Google Scholar] [CrossRef]
- Yoneyama, T. Iron delivery to the growing leaves associated with leaf chlorosis in mugineic acid family phytosiderophores-generating graminaceous crops. Soil Sci. Plant Nutr. 2021, 67, 415–426. [Google Scholar] [CrossRef]
- Almeida-Silva, F.; Venancio, T.M. Pathogenesis-related protein 1 (PR-1) genes in soybean: Genome-wide identification, structural analysis and expression profiling under multiple biotic and abiotic stresses. bioRxiv 2021, 809, 146013. [Google Scholar] [CrossRef]
- Schwarz, B.; Bauer, P. FIT, a regulatory hub for iron deficiency and stress signaling in roots, and FIT-dependent and -independent gene signatures. J. Exp. Bot. 2020, 71, 1694–1705. [Google Scholar] [CrossRef] [Green Version]
- Toprak Aktas, E.; Yildiz, H. Effects of electroplasmolysis treatment on chlorophyll and carotenoid extraction yield from spinach and tomato. J. Food Eng. 2011, 106, 339–346. [Google Scholar] [CrossRef]
- Cao, J.; Li, F.; Li, Y.; Chen, H.; Liao, X.; Zhang, Y. Hydrophobic interaction driving the binding of soybean protein isolate and chlorophyll: Improvements to the thermal stability of chlorophyll. Food Hydrocoll. 2021, 113, 106465. [Google Scholar] [CrossRef]


| Tissue | Group | Element Content (mg/g FW-1) | ||||
|---|---|---|---|---|---|---|
| Fe | Ca | Mg | Zn | P | ||
| New leaf | Control | 0.04 ± 0.022 | 16.25 ± 2.56 | 7.57 ± 0.94 | 0.06 ± 0.00 | 4.94 ± 0.40 |
| Test | 0.02 ± 0.006 * | 18.21 ± 1.81 ns | 7.49 ± 0.52 ns | 0.16 ± 0.02 ** | 6.46 ± 0.52 ** | |
| Old leaf | Control | 0.06 ± 0.017 | 11.85 ± 1.16 | 5.46 ± 0.36 | 0.06 ± 0.01 | 4.16 ± 0.20 |
| Test | 0.04 ± 0.021 ns | 12.82 ± 2.62 ns | 5.52 ± 1.12 ns | 0.11 ± 0.03 ** | 4.90 ± 1.08 ns | |
| Iron Mode | Tissue | Metabolites | VIP | Fold Change | p-Value | m/z | RT(s) |
|---|---|---|---|---|---|---|---|
| Positive | New leaf | 4-Hydroxybutanoic acid lactone | 1.57 | 0.11 | 0.0000 | 87.0432 | 402.47 |
| Phe-Phe | 1.00 | 0.23 | 0.0000 | 313.1533 | 154.17 | ||
| Pyridoxal (Vitamin B6) | 2.10 | 0.36 | 0.0000 | 168.0639 | 95.96 | ||
| 4-Aminobutyric acid | 2.00 | 0.37 | 0.0000 | 104.0705 | 368.79 | ||
| S-Methyl-5’-thioadenosine | 3.65 | 0.38 | 0.0000 | 298.0953 | 98.92 | ||
| Adenosine | 13.85 | 0.47 | 0.0001 | 268.1026 | 168.71 | ||
| Nicotinate | 1.04 | 0.56 | 0.0415 | 124.0373 | 215.08 | ||
| Choline | 1.58 | 0.59 | 0.0458 | 104.1059 | 390.71 | ||
| 20-Hydroxyarachidonic acid | 1.19 | 0.64 | 0.0000 | 343.2246 | 34.76 | ||
| (4Z,7Z,10Z,13Z,16Z,19Z)-4,7,10,13,16,19-Docosahexaenoic acid | 1.41 | 0.66 | 0.0137 | 328.2463 | 68.75 | ||
| D-Proline | 1.57 | 1.92 | 0.0361 | 116.0693 | 308.51 | ||
| L-Asparagine | 1.86 | 3.30 | 0.0001 | 133.0592 | 396.60 | ||
| 1-Indanone | 1.24 | 3.58 | 0.0035 | 193.0843 | 32.62 | ||
| 1-Aminocyclopropanecarboxylic acid | 1.14 | 3.62 | 0.0002 | 84.0433 | 376.98 | ||
| 1-Palmitoyl-sn-glycero-3-phosphocholine | 1.02 | 7.70 | 0.0085 | 496.3372 | 188.96 | ||
| L-Glutamate | 1.99 | 12.71 | 0.0000 | 148.0591 | 394.20 | ||
| Old leaf | Adenosine | 4.36 | 0.46 | 0.0025 | 268.1026 | 168.71 | |
| Cytidine | 2.74 | 2.10 | 0.0074 | 244.0922 | 236.66 | ||
| 2’-O-methyladenosine | 1.46 | 1.48 | 0.0154 | 282.1183 | 130.78 | ||
| Uracil | 1.02 | 1.22 | 0.0226 | 113.0331 | 162.04 | ||
| Negative | New leaf | Salicylic acid | 1.00 | 2.76 | 0.0001 | 137.0237 | 78.27 |
| Citrate | 3.58 | 0.06 | 0.0004 | 191.0203 | 470.66 | ||
| L-Glutamine | 1.08 | 3.71 | 0.0009 | 145.0609 | 386.60 | ||
| Old leaf | D-Lyxose | 1.66 | 1.51 | 0.0072 | 149.0445 | 153.51 | |
| D-Glucono-1,5-lactone | 1.28 | 1.94 | 0.0185 | 177.0394 | 158.68 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, Z.; Ji, S.; Cui, D. Integration of the Metabolomic and Transcriptome Analysis Reveals the Remarkable Compounds of G. bicolor Young and Mature Leaves under Different Iron Nutrient Conditions. Int. J. Mol. Sci. 2022, 23, 1160. https://doi.org/10.3390/ijms23031160
Feng Z, Ji S, Cui D. Integration of the Metabolomic and Transcriptome Analysis Reveals the Remarkable Compounds of G. bicolor Young and Mature Leaves under Different Iron Nutrient Conditions. International Journal of Molecular Sciences. 2022; 23(3):1160. https://doi.org/10.3390/ijms23031160
Chicago/Turabian StyleFeng, Zhe, Shuyu Ji, and Di Cui. 2022. "Integration of the Metabolomic and Transcriptome Analysis Reveals the Remarkable Compounds of G. bicolor Young and Mature Leaves under Different Iron Nutrient Conditions" International Journal of Molecular Sciences 23, no. 3: 1160. https://doi.org/10.3390/ijms23031160





