Complete Characterization of the O-Antigen from the LPS of Aeromonas bivalvium
Abstract
:1. Introduction
2. Results
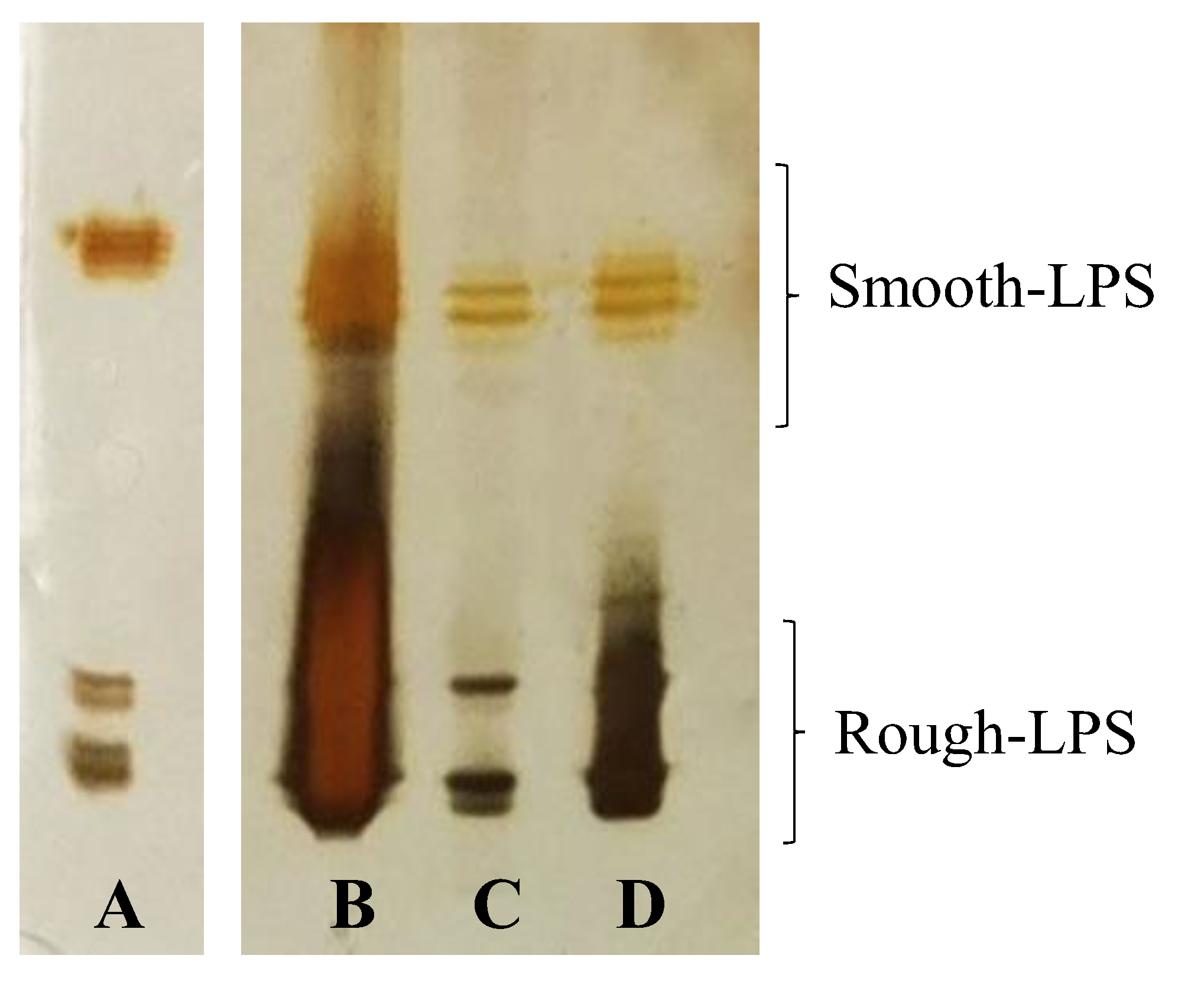
2.1. LPS Isolation and DOC-PAGE Analysis
2.2. Compositional Analysis
2.3. Isolation and Chemical Analysis of O-Chain
2.4. NMR Analysis of the O-Chain (OPS)
3. Materials and Methods
3.1. Bacterial Characteristics, Isolation and Growth
3.2. LPS Isolation
3.3. DOC-PAGE Analysis
3.4. Chemical Analyses
3.5. Isolation of O-Chain
3.6. NMR Spectroscopy
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Janda, J.M.; Abbott, S.L. The Genus Aeromonas: Taxonomy, Pathogenicity, and Infection. Clin. Microbiol. Rev. 2010, 23, 35–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernández-Bravo, A.; Figueras, M.J. An Update on the Genus Aeromonas: Taxonomy, Epidemiology, and Pathogenicity. Microorganisms 2020, 8, 129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janda, J.M.; Abbott, S.L. Evolving concepts regarding the genus Aeromonas: An expanding panorama of species, disease presentations, and unanswered questions. Clin. Infect. Dis. 1998, 27, 332–344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mendoza-Barberá, E.; Merino, S.; Tomás, J. Surface Glucan Structures in Aeromonas spp. Mar. Drugs 2021, 19, 649. [Google Scholar] [CrossRef]
- Raetz, C.R.H.; Whitfield, C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002, 71, 635–700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forn-Cuní, G.; Merino, S.; Tomás, J.M. Comparative Genomics of the Aeromonadaceae Core Oligosaccharide Biosynthetic Regions. Int. J. Mol. Sci. 2017, 18, 519. [Google Scholar] [CrossRef] [Green Version]
- Knirel, Y. A Structure of O-Antigens. In Bacterial Lipopolysaccharides; Knirel, Y., Valvano, M., Eds.; Springer: Vienna, Austria, 2011; pp. 41–115. [Google Scholar]
- Heiss, C.; Wang, Z.; Thurlow, C.M.; Hossain, M.J.; Sun, D.; Liles, M.R.; Saper, M.A.; Azadi, P. Structure of the capsule and lipopolysaccharide O-antigen from the channel catfish pathogen, Aeromonas hydrophila. Carbohydr. Res. 2019, 486, 107858–107864. [Google Scholar] [CrossRef]
- Dworaczek, K.; Kurzylewska, M.; Karaś, M.A.; Janczarek, M.; Pękala-Safińska, A.; Turska-Szewczuk, A. A Unique Sugar l-Perosamine (4-Amino-4,6-dideoxy-l-mannose) Is a Compound Building Two O-Chain Polysaccharides in the Lipopolysaccharide of Aeromonas hydrophila Strain JCM 3968, Serogroup O6. Mar. Drugs 2019, 17, 254–271. [Google Scholar] [CrossRef] [Green Version]
- Dworaczek, K.; Drzewiecka, D.; Pękala-Safińska, A.; Turska-Szewczuk, A. Structural and Serological Studies of the O6-Related Antigen of Aeromonas veronii bv. sobria Strain K557 Isolated from Cyprinus carpio on a Polish Fish Farm, which Contains l-perosamine (4-amino-4,6-dideoxy-l-mannose), a Unique Sugar Characteristic for Aeromonas Serogroup O6. Mar. Drugs 2019, 17, 399–418. [Google Scholar]
- Dworaczek, K.; Kurzylewska, M.; Laban, M.; Drzewiecka, D.; Pękala-Safińska, A.; Turska-Szewczuk, A. Structural Studies of the Lipopolysaccharide of Aeromonas veronii bv. sobria Strain K133 Which Represents New Provisional Serogroup PGO1 Prevailing among Mesophilic Aeromonads on Polish Fish Farms. Int. J. Mol. Sci. 2021, 22, 4272–4291. [Google Scholar] [CrossRef]
- Dworaczek, K.; Kurzylewska, M.; Laban, M.; Pękala-Safińska, A.; Marczak, M.; Turska-Szewczuk, A. Structure of the disaccharide repeating unit of O-specific polysaccharide isolated from Aeromonas veronii strain Bs8 pathogenic to common carp (Cyprinus carpio). Carbohydr. Res. 2021, 500, 108210–108214. [Google Scholar] [CrossRef] [PubMed]
- Miñana-Galbis, D.; Farfán, M.; Fusté, M.C.; Lorén, J.G. Aeromonas bivalvium sp. nov., isolated from bivalve molluscs. Int. J. Syst. Evol. Microbiol. 2007, 57, 582–587. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, P.K. NMR spectroscopy in the structural elucidation of the oligosaccharides and glycosides. Phytochemistry 1992, 31, 3307–3330. [Google Scholar] [CrossRef]
- Jansson, P.E.; Kenne, L.; Widmalm, G. Computer-assisted structural analysis of polysaccharides with an extended version of CASPER using 1H- and 13C-NMR data. Carbohydr. Res. 1989, 188, 169–171. [Google Scholar] [CrossRef]
- Bock, K.; Pedersen, C. Carbon-13 Nuclear Magnetic Resonance Spectroscopy of Monosaccharides. Adv. Carbohydr. Chem. Biochem. 1983, 41, 27–66. [Google Scholar]
- Turska-Szewczuk, A.; Kozinska, A.; Russa, R.; Holst, O. The structure of the O-specific polysaccharide from the lipopolysaccharide of Aeromonas bestiarum strain 207. Carbohydr. Res. 2010, 345, 680–684. [Google Scholar] [CrossRef] [PubMed]
- Turska-Szewczuk, A.; Guz, L.; Lindner, B.; Pietras, H.; Russa, R.; Holst, O. Structural characterization of the O-specific polysaccharide from the lipopolysaccharide of the fish pathogen Aeromonas bestiarum strain P1S. Carbohydr. Res. 2011, 346, 815–821. [Google Scholar] [CrossRef]
- Pakiet, K.; Turska-Szewczuk, A.; Karas, M.A.; Pekala, A.; Pietras, H. Structure of the O-specific polysaccharide from the lipopolysaccharide of Aeromonas hydrophila strain K691 containing 4-acetamido-4,6-dideoxy-D-glucose. Carbohydr. Res. 2017, 439, 23–29. [Google Scholar] [CrossRef]
- Galanos, C.; Lüderitz, O.; Westphal, O. A new method for the extraction of R lipopolysaccharides. Eur. J. Biochem. 1969, 9, 245–249. [Google Scholar] [CrossRef]
- Westphal, O.; Jann, K. Bacterial lipopolysaccharides extraction with phenol-water and further applications of the procedure, Methods. Carbohydr. Chem. 1965, 5, 83–91. [Google Scholar]
- Laemmli, U.K. Most commonly used discontinuous buffer system for SDS electrophoresis. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.M.; Frasch, C.E. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 1982, 119, 115–119. [Google Scholar] [CrossRef]
- Abid, Y.; Azabou, S.; Joulak, I.; Casillo, A.; Lanzetta, R.; Corsaro, M.M.; Gharsallaoui, A.; Attia, H. Potential biotechnological properties of an exopolysaccharide produced by newly isolated Bacillus tequilensis-GM from spontaneously fermented goat milk. LWT 2019, 105, 135–141. [Google Scholar] [CrossRef]
- Leontein, K.; Lindberg, B.; Lonngren, J. Assignment of absolute configuration of sugars by G.L.C. of their acetylated glycosides formed from chiral alcohols. Carbohydr. Res. 1978, 62, 359–362. [Google Scholar] [CrossRef]
- Ciucanu, I.; Kerek, F. A simple and rapid method for the permethylation of carbohydrates. Carbohydr. Res. 1984, 131, 209–217. [Google Scholar] [CrossRef]
- Fresno, S.; Jiménez, N.; Canals, R.; Merino, S.; Corsaro, M.M.; Lanzetta, R.; Parrilli, M.; Pieretti, G.; Regué, M.; Tomás, J.M. A Second Galacturonic Acid Transferase Is Required for Core Lipopolysaccharide Biosynthesis and Complete Capsule Association with the Cell Surface in Klebsiella pneumoniae. J. Bacteriol. 2007, 189, 1128–1137. [Google Scholar] [CrossRef] [Green Version]
- Di Guida, R.; Casillo, A.; Corsaro, M.M. O-specific polysaccharide structure isolated from the LPS of the Antarctic bacterium Pseudomonas ANT_J38B. Carbohydr. Res. 2020, 497, 108125. [Google Scholar] [CrossRef]
- Ståhle, J.; Fontana, C.; Weintraub, A.; Widmalm, G. Elucidation of the O-antigen structure of Escherichia coli O63. Glycobiology 2019, 29, 179–187. [Google Scholar] [CrossRef]




| Residue | H1 C1 1JC1,H1 | H2 C2 | H3 C3 | H4 C4 | H5 C5 | H6 C6 | CH3CO | CO |
|---|---|---|---|---|---|---|---|---|
| α-2-L-Rha A | 5.15 102.0 181 Hz | 4.05 79.0 | 3.92 71.1 | 3.46 73.4 | 3.77 70.3 | 1.30 17.9 | ||
| α-t-L-Rha B | 5.01 100.6 179 Hz | 3.83 71.6 | 3.91 70.4 | 3.39 73.6 | 4.49 69.8 | 1.34 18.0 | ||
| α-2,3-L-Rha C | 4.94 100.7 178 Hz | 4.17 77.3 | 3.95 78.6 | 3.54 72.8 | 3.89 70.4 | 1.30 17.9 | ||
| α-3,4-D-GalNAc D | 4.92 97.6 180 Hz | 4.28 49.5 | 3.88 78.2 | 3.66 76.1 | 3.91 72.2 | 3.77, 3.86 62.2 | 2.04 23.4 | 175.3 |
| α-3-L-Rha E | 4.90 103.3 179 Hz | 3.87 71.5 | 3.87 76.0 | 3.54 70.7 | 3.74 70.5 | 1.30 17.8 | ||
| β-t-D-GlcNAc F | 4.74 103.6 164 Hz | 3.96 57.2 | 3.87 74.8 | 3.81 71.6 | 3.50 76.2 | 3.74, 3.93 61.1 | 2.04 23.4 | 176.0 |
| β-3-D-FucNAc G | 4.46 102.4 163 Hz | 3.94 53.7 | 3.92 78.5 | 3.70 71.9 | 4.47 68.5 | 1.29 16.8 | 2.04 23.4 | 176.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Guida, R.; Casillo, A.; Tomás, J.M.; Merino, S.; Corsaro, M.M. Complete Characterization of the O-Antigen from the LPS of Aeromonas bivalvium. Int. J. Mol. Sci. 2022, 23, 1204. https://doi.org/10.3390/ijms23031204
Di Guida R, Casillo A, Tomás JM, Merino S, Corsaro MM. Complete Characterization of the O-Antigen from the LPS of Aeromonas bivalvium. International Journal of Molecular Sciences. 2022; 23(3):1204. https://doi.org/10.3390/ijms23031204
Chicago/Turabian StyleDi Guida, Rossella, Angela Casillo, Juan M. Tomás, Susana Merino, and Maria Michela Corsaro. 2022. "Complete Characterization of the O-Antigen from the LPS of Aeromonas bivalvium" International Journal of Molecular Sciences 23, no. 3: 1204. https://doi.org/10.3390/ijms23031204
APA StyleDi Guida, R., Casillo, A., Tomás, J. M., Merino, S., & Corsaro, M. M. (2022). Complete Characterization of the O-Antigen from the LPS of Aeromonas bivalvium. International Journal of Molecular Sciences, 23(3), 1204. https://doi.org/10.3390/ijms23031204









