Protein Modification with Ribose Generates Nδ-(5-hydro-5-methyl-4-imidazolone-2-yl)-ornithine
Abstract
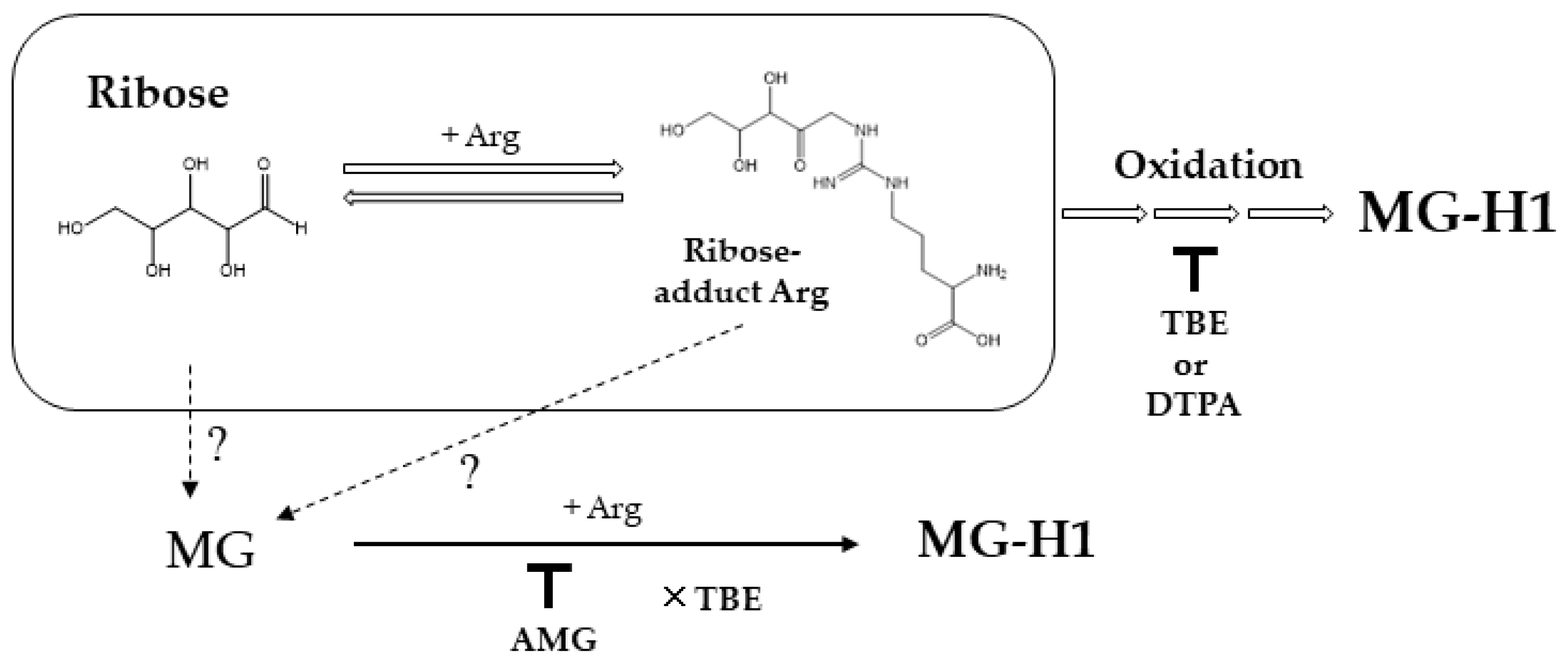
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
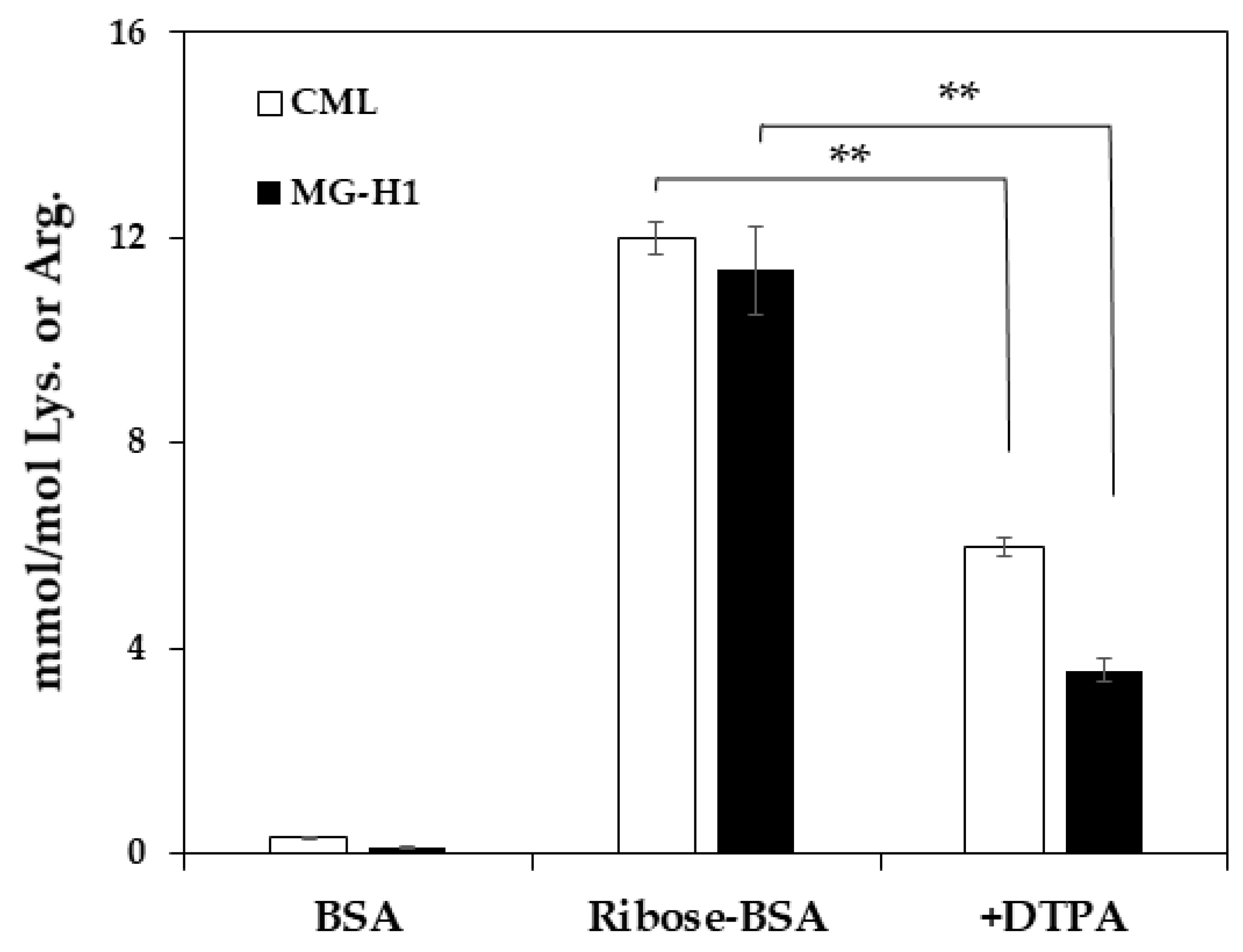
4.1. Measurement of AGE Contents in Ribose-BSA
4.2. Quantification of MG-H1 in Ribated-BSA or Glycated-BSA
4.3. Detection of MG in Ribated-BSA
4.4. LC-QTOF Condition
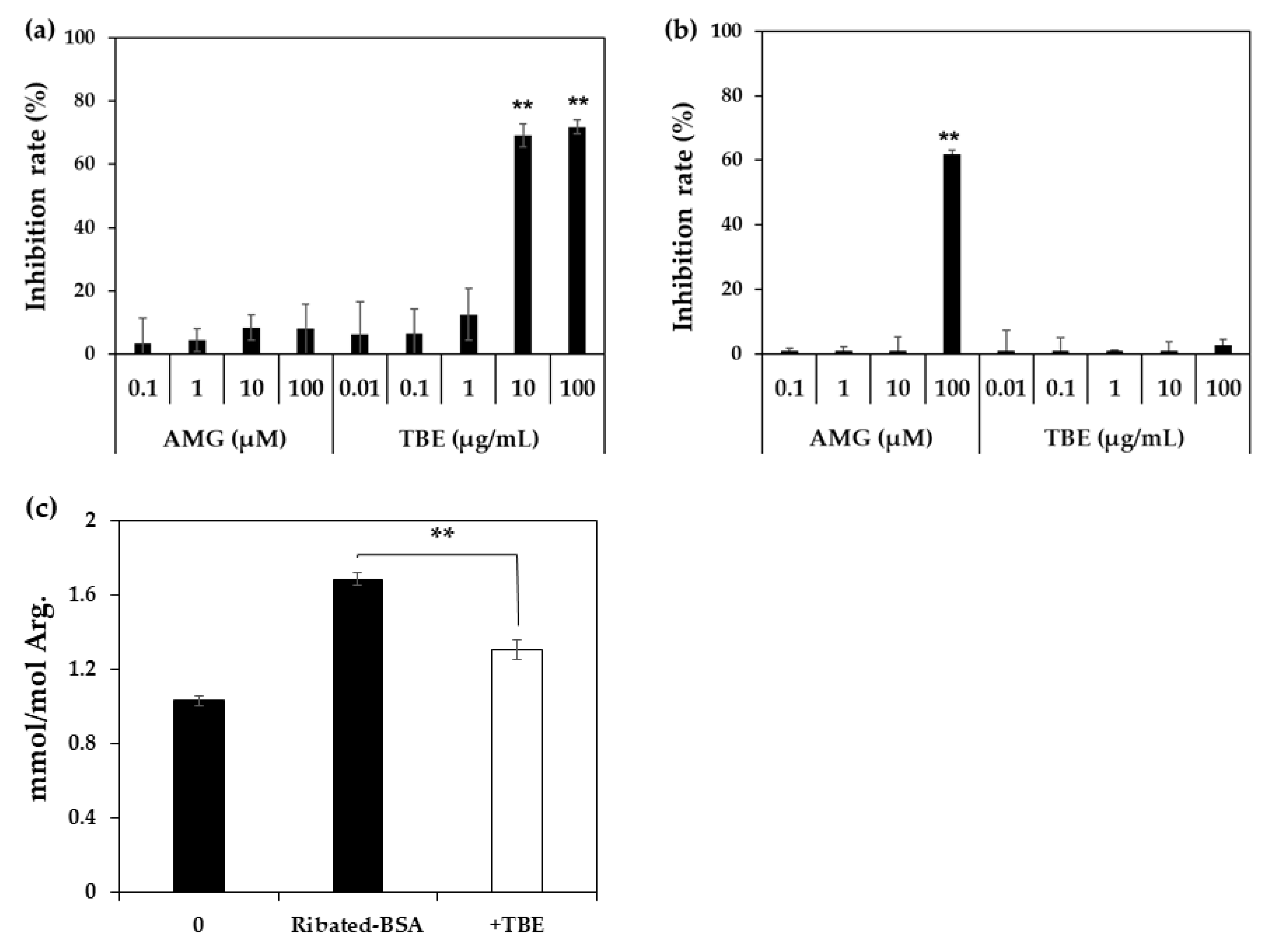
4.5. Inhibitory Effect of TBE on MG-H1 Formation by ELISA
4.6. Inhibitory Effect of TBE on MG-H1 Formation in Ribated-BSA by LC-QTOF
4.7. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Abbreviations
| AEGs | Advanced glycation end-products |
| CML | Nε-(carboxymethyl) lysine |
| MG-H1 | Nδ-(5-hydro-5-methyl-4-imidazolone-2-yl)-ornithine |
| CEL | Nε-(carboxyethyl) lysine |
| CMA | Nω-(carboxymethyl) arginine |
| BSA | Bovine serum albumin |
| NaPB | Sodium phosphate buffer (pH 7.2) |
| Ribated BSA | Amadori-BSA derived from ribose |
| Glycated-BSA | Amadori-BSA derived from glucose |
| AMG | Aminoguanidine |
| DTPA | Diethylenetriaminepentaacetic acid |
| DAN | 2, 3-diaminonaphthalene |
| LC-ESI-QTOF | Liquid chromatography–quadrupole time-of-flight mass spectrometry |
| HPLC | High performance liquid chromatography |
| MG | Methylglyoxal |
| GO | Glyoxal |
| ACN | Acetonitrile |
| FA | Formic acid |
| DAN | 2, 3-diaminonaphthalene |
| ELISA | Enzyme-linked immunosorbent assay |
References
- Lin, C.C.; Li, C.I.; Liu, C.S.; Lin, W.Y.; Fuh, M.M.; Yang, S.Y.; Lee, C.C.; Li, T.C. Impact of lifestyle-related factors on all-cause and cause-specific mortality in patients with type 2 diabetes: The Taichung Diabetes Study. Diabetes Care 2012, 35, 105–112. [Google Scholar] [CrossRef] [Green Version]
- Tabák, A.G.; Herder, C.; Rathmann, W.; Brunner, E.J.; Kivimäki, M. Prediabetes: A high-risk state for diabetes development. Lancet 2012, 379, 2279–2290. [Google Scholar] [CrossRef] [Green Version]
- Vistoli, G.; De Maddis, D.; Cipak, A.; Zarkovic, N.; Carini, M.; Aldini, G. Advanced glycoxidation and lipoxidation endproducts (AGEs and ALEs): An overview of theirmechanisms of formation. Free Radic. Res. 2013, 2013 47, 3–27. [Google Scholar] [CrossRef] [Green Version]
- Fu, M.X.; Wells-Knecht, K.J.; Blackledge, J.A.; Lyons, T.J.; Thorpe, S.R.; Baynes, J.W. Glycation, glycoxidation, and cross-linking of collagen by glucose. Kinetics, mechanisms, and inhibition of late stages of the Maillard reaction. Diabetes 1994, 43, 676–683. [Google Scholar] [CrossRef] [PubMed]
- Zeng, C.; Li, Y.; Ma, J.; Niu, L.; Tay, F.R. Clinical/Translational Aspects of Advanced Glycation End-Products. Trends Endocrinol. Metab. 2019, 30, 959–973. [Google Scholar] [CrossRef]
- Yamanaka, M.; Matsumura, T.; Ohno, R.; Fujiwara, Y.; Shinagawa, M.; Sugawa, H.; Hatano, K.; Shirakawa, J.; Kinoshita, H.; Ito, K.; et al. Non-invasive measurement of skin autofluorescence to evaluate diabetic complications. J. Clin. Biochem. Nutr. 2016, 58, 135–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, M.X.; Requena, J.R.; Jenkins, A.J.; Lyons, T.J.; Baynes, J.W.; Thorpe, S.R. The advanced glycation end product, Nepsilon-(carboxymethyl)lysine, is a product of both lipid peroxidation and glycoxidation reactions. Biol. Chem. 1996, 271, 9982–9986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rabbani, N.; Ashour, A.; Thornalley, P.J. Mass spectrometric determination of early and advanced glycation in biology. Glycoconj. J. 2016, 33, 553–568. [Google Scholar] [CrossRef] [Green Version]
- Beisswenger, P.J.; Howell, S.K.; Russell, G.B.; Miller, M.E.; Rich, S.S.; Mauer, M. Early progression of diabetic nephropathy correlates with methylglyoxal-derived advanced glycation end products. Diabetes Care 2013, 36, 3234–3239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Yu, L.; Wang, Y.; Wei, Y.; Xu, Y.; He, T.; He, R. d-Ribose contributes to the glycation of serum protein. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 2285–2292. [Google Scholar] [CrossRef]
- Chen, X.; Su, T.; Chen, Y.; He, Y.; Liu, Y.; Xu, Y.; Wei, Y.; Li, J.; He, R. d-Ribose as a Contributor to Glycated Haemoglobin. EBioMedicine 2017, 25, 143–153. [Google Scholar] [CrossRef] [Green Version]
- Han, C.; Lu, Y.; Wei, Y.; Liu, Y.; He, R. D-ribose induces cellular protein glycation and impairs mouse spatial cognition. PLoS ONE 2011, 6, e24623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bunn, H.F.; Higgins, P.J. Reaction of monosaccharides with proteins: Possible evolutionary significance. Science 1981, 213, 222–224. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Lu, Y.; Wei, Y.; Wu, B.; Liu, Y.; He, R. D-ribosylation induces cognitive impairment through RAGE-dependent astrocytic inflammation. Cell Death Dis. 2014, 5, e1117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mou, L.; Hu, P.; Cao, X.; Chen, Y.; Xu, Y.; He, T.; Wei, Y.; He, R. Comparison of bovine serum albumin glycation by ribose and fructose in vitro and in vivo. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1868, 166283. [Google Scholar] [CrossRef] [PubMed]
- Sugawa, H.; Ohno, R.; Shirakawa, J.; Nakajima, A.; Kanagawa, A.; Hirata, T.; Ikeda, T.; Moroishi, N.; Nagai, M.; Nagai, R. Eucommia ulmoides extracts prevent the formation of advanced glycation end products. Food Funct. 2016, 7, 2566–2573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kinoshita, S.; Mera, K.; Ichikawa, H.; Shimasaki, S.; Nagai, M.; Taga, Y.; Iijima, K.; Hattori, S.; Fujiwara, Y.; Shirakawa, J.I.; et al. Nω-(Carboxymethyl)arginine Is One of the Dominant Advanced Glycation End Products in Glycated Collagens and Mouse Tissues. Oxid. Med. Cell. Longev. 2019, 2019, 9073451. [Google Scholar]
- Jinno, M.; Nagai, R.; Takeuchi, M.; Watanabe, A.; Teruya, K.; Sugawa, H.; Hatakeyama, N.; Jinno, Y. Trapa bispinosa Roxb. extract lowers advanced glycation end-products and increases live births in older patients with assisted reproductive technology: A randomized controlled trial. Reprod. Biol. Endocrinol. 2021, 19, 149. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, H.; Nagai, M.; Sugawa, H.; Yasuda, H.; Nagai, R. Development of a conventional immunochemical detection system for determination of N δ-(5-hydro-5-methyl-4-imidazolone-2-yl)-ornithine in methylglyoxal-modified proteins. Glycoconj. J. 2021, 38, 293–301. [Google Scholar] [CrossRef] [PubMed]
- Phillips, S.A.; Thornalley, P.J. The formation of methylglyoxal from triose phosphates. Investigation using a specific assay for methylglyoxal. Eur. J. Biochem. 1993, 212, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Thornalley, P.J.; Langborg, A.; Minhas, H.S. Formation of glyoxal, methylglyoxal and 3-deoxyglucosone in the glycation of proteins by glucose. Biochem. J. 1999, 344, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Nagai, R.; Matsumoto, K.; Ling, X.; Suzuki, H.; Araki, T.; Horiuchi, S. Glycolaldehyde, a reactive intermediate for advanced glycation end products, plays an important role in the generation of an active ligand for the macrophage scavenger receptor. Diabetes 2000, 49, 1714–1723. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, M.; Thorpe, S.R.; Baynes, J.W. Identification of N epsilon-carboxymethyllysine as a degradation product of fructoselysine in glycated protein. J. Biol. Chem. 1986, 261, 4889–4894. [Google Scholar] [CrossRef]
- Kinoshita, S.; Sugawa, H.; Nanri, T.; Ohno, R.; Shirakawa, J.; Sato, H.; Katsuta, N.; Sakake, S.; Nagai, R. Trapa bispinosa Roxb. and lutein ameliorate cataract in type 1 diabetic rats. J. Clin. Biochem. Nutr. 2020, 66, 8–14. [Google Scholar]
- Tominaga, Y.; Sugawa, H.; Hirabayashi, K.; Ikeda, T.; Hoshi, Y.; Nagai, R. Drosera tokaiensis extract containing multiple phenolic compounds inhibits the formation of advanced glycation end-products. Arch. Biochem. Biophys. 2020, 693, 108586. [Google Scholar] [CrossRef] [PubMed]
- Klöpfer, A.; Spanneberg, R.; Glomb, M.A. Formation of arginine modifications in a model system of Nα-tert-butoxycarbonyl (Boc)-arginine with methylglyoxal. J. Agric. Food Chem. 2011, 59, 394–401. [Google Scholar] [CrossRef]
- Nagai, R.; Unno, Y.; Hayashi, M.C.; Masuda, S.; Hayase, F.; Kinae, N.; Horiuchi, S. Peroxynitrite induces formation of N( epsilon )-(carboxymethyl) lysine by the cleavage of Amadori product and generation of glucosone and glyoxal from glucose: Novel pathways for protein modification by peroxynitrite. Diabetes 2002, 51, 2833–2839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohno, R.; Moroishi, N.; Sugawa, H.; Maejima, K.; Saigusa, M.; Yamanaka, M.; Nagai, M.; Yoshimura, M.; Amakura, Y.; Nagai, R. Mangosteen pericarp extract inhibits the formation of pentosidine and ameliorates skin elasticity. J. Clin. Biochem. 2015, 57, 27–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagai, R.; Nagai, M.; Shimasaki, S.; Baynes, J.W.; Fujiwara, Y. Citric acid inhibits development of cataracts, proteinuria and ketosis in streptozotocin (type 1) diabetic rats. Biochem. Biophys. Res. Commun. 2010, 393, 118–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clark, P.M.; Flores, G.; Evdokimov, N.M.; McCracken, M.N.; Chai, T.; Nair-Gill, E.; O’Mahony, F.; Beaven, S.W.; Faull, K.F.; Phelps, M.E.; et al. Positron emission tomography probe demonstrates a striking concentration of ribose salvage in the liver. Proc. Natl. Acad. Sci. USA 2014, 111, E2866–E2874. [Google Scholar] [CrossRef] [Green Version]
- Hong, J.; Wang, X.; Zhang, N.; Fu, H.; Li, W. D-ribose induces nephropathy through RAGE-dependent NF-κB inflammation. Arch. Pharm. Res. 2018, 41, 838–847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martens, R.J.H.; Broers, N.J.H.; Canaud, B.; Christiaans, M.H.L.; Cornelis, T.; Gauly, A.; Hermans, M.M.H.; Konings, C.J.A.M.; van der Sande, F.M.; Scheijen, J.L.J.M.; et al. Advanced glycation endproducts and dicarbonyls in end-stage renal disease: Associations with uraemia and courses following renal replacement therapy. Clin. Kidney J. 2019, 13, 855–866. [Google Scholar] [CrossRef]
- Ohno, R.; Ichimaru, K.; Tanaka, S.; Sugawa, H.; Katsuta, N.; Sakake, S.; Tominaga, Y.; Ban, I.; Shirakawa, J.; Yamaguchi, Y.; et al. Glucoselysine is derived from fructose and accumulates in the eye lens of diabetic rats. J. Biol. Chem. 2019, 294, 17326–17338. [Google Scholar] [CrossRef] [PubMed]
- Nakano, M.; Kubota, M.; Owada, S.; Nagai, R. The pentosidine concentration in human blood specimens is affected by heating. Amino Acids 2013, 44, 1451–1456. [Google Scholar] [CrossRef] [PubMed]
- Miki Hayashi, C.; Nagai, R.; Miyazaki, K.; Hayase, F.; Araki, T.; Ono, T.; Horiuchi, S. Conversion of Amadori products of the Maillard reaction to N(epsilon)-(carboxymethyl)lysine by short-term heating: Possible detection of artifacts by immunohistochemistry. Lab. Investig. 2002, 82, 795–808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugawa, H.; Yachi, A.; Fujimoto, Y.; Nagai, R. Accumulation of Nε-(carboxyethyl) lysine in Caenorhabditis elegans is correlated with the formation of ketone body. J. Biochem. 2021, 170, 587–592. [Google Scholar] [CrossRef] [PubMed]
- Nagai, R.; Miki Hayashi, C.; Xia, L.; Takeya, M.; Horiuchi, S. Identification in human atherosclerotic lesions of GA-pyridine, a novel structure derived from glycolaldehyde-modified proteins. J. Bio. Chem. 2002, 277, 48905–48912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanda, Y. Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transplant. 2013, 48, 452–458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
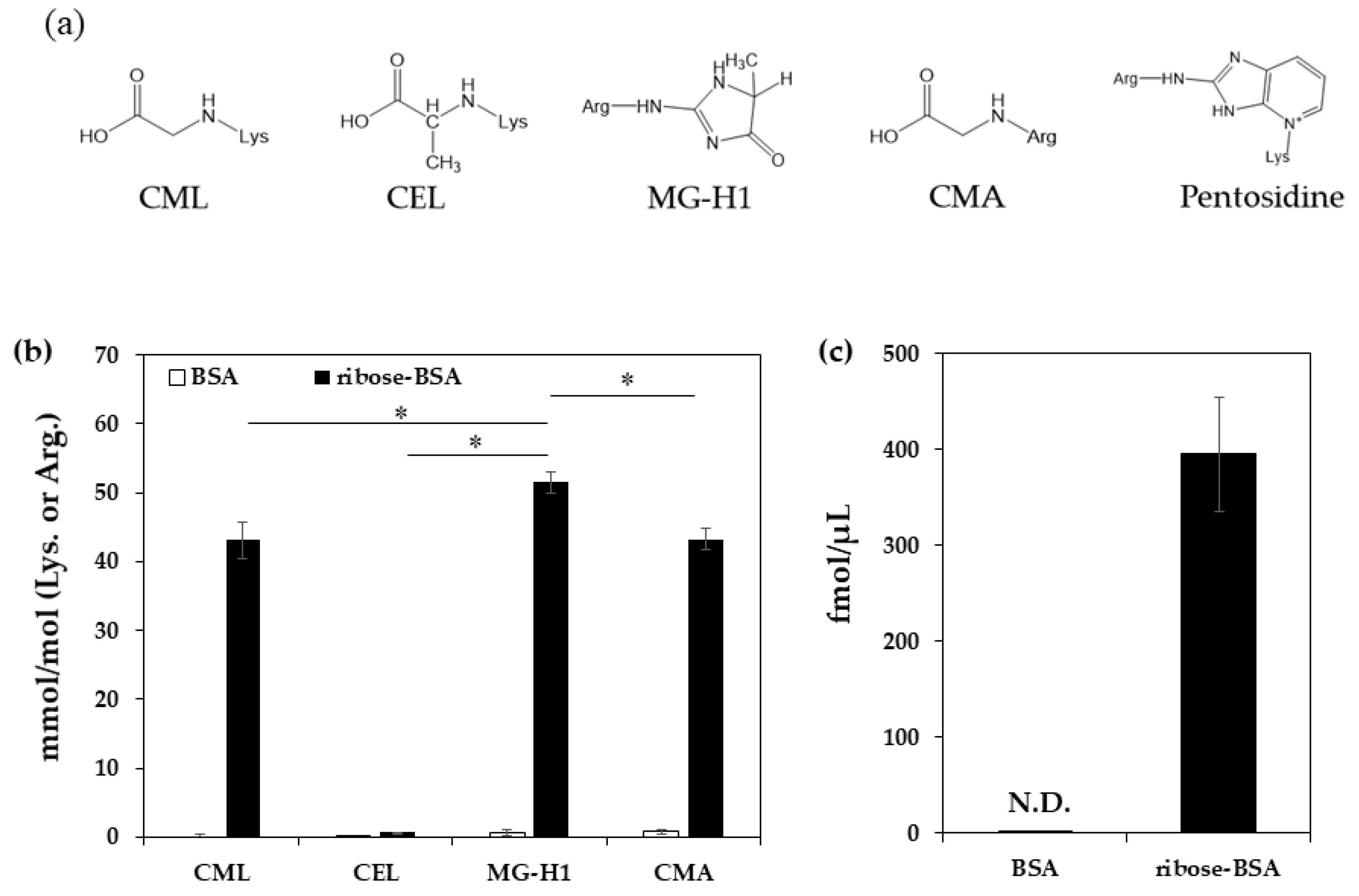
| pmol/μg Protein | BSA | Ribose-BSA |
|---|---|---|
| CML | 0.00 ± 0.31 | 21.20 ± 0.76 |
| CEL | 0.04 ± 0.00 | 0.33 ± 0.02 |
| MG-H1 | 0.20 ± 0.13 | 13.06 ± 0.73 |
| CMA | 0.24 ± 0.11 | 10.95 ± 0.46 |
| pentosidine | N.D. | 0. 40 ± 0.06 |
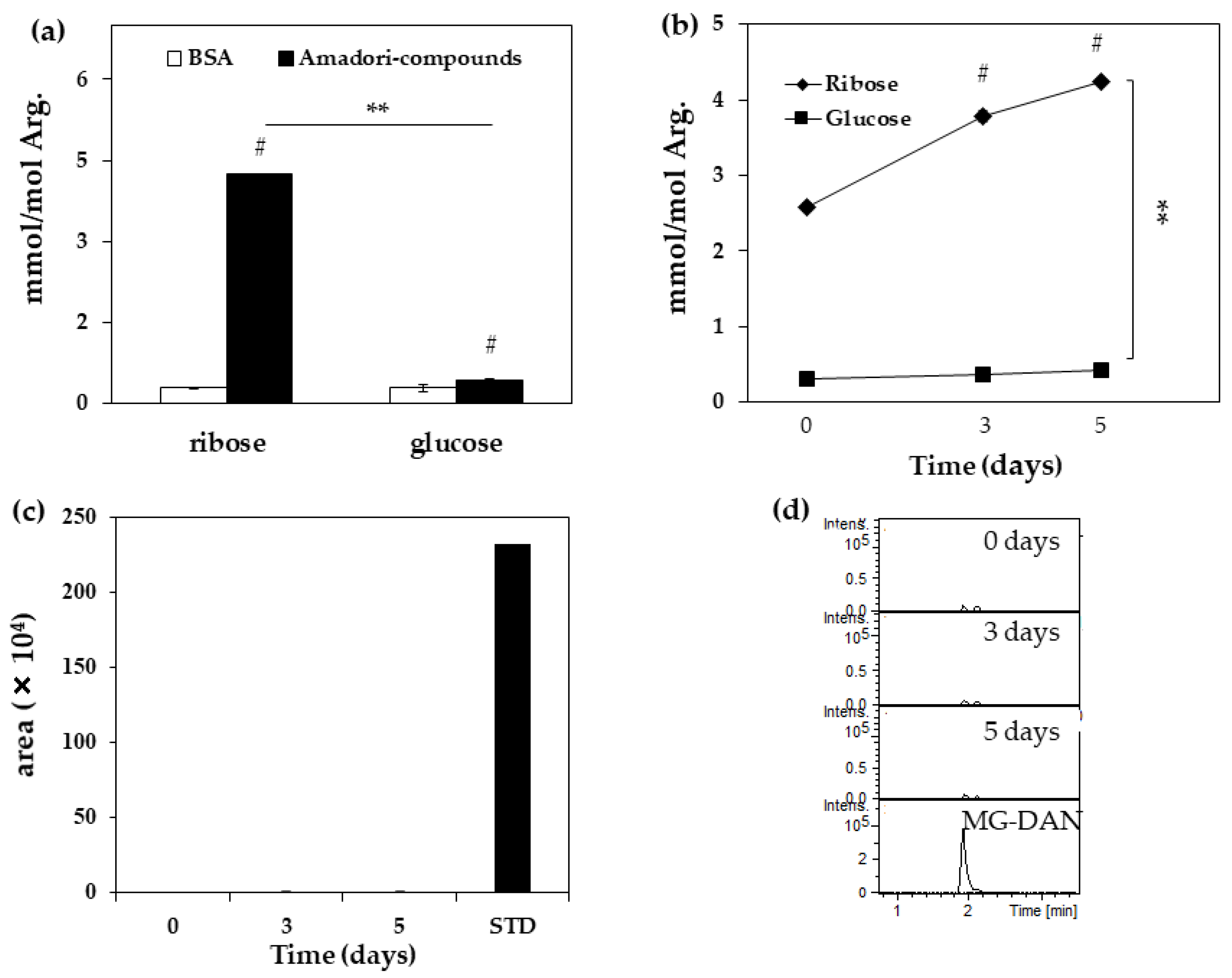
| Arg (nmol) | Change of Arg (%) | |
|---|---|---|
| BSA | 27.6 | 0 |
| Ribated-BSA | ||
| Day 0 | 20.0 | 27.8 |
| Day 3 | 19.9 | 28.0 |
| Day 5 | 19.6 | 29.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ban, I.; Sugawa, H.; Nagai, R. Protein Modification with Ribose Generates Nδ-(5-hydro-5-methyl-4-imidazolone-2-yl)-ornithine. Int. J. Mol. Sci. 2022, 23, 1224. https://doi.org/10.3390/ijms23031224
Ban I, Sugawa H, Nagai R. Protein Modification with Ribose Generates Nδ-(5-hydro-5-methyl-4-imidazolone-2-yl)-ornithine. International Journal of Molecular Sciences. 2022; 23(3):1224. https://doi.org/10.3390/ijms23031224
Chicago/Turabian StyleBan, Ikuho, Hikari Sugawa, and Ryoji Nagai. 2022. "Protein Modification with Ribose Generates Nδ-(5-hydro-5-methyl-4-imidazolone-2-yl)-ornithine" International Journal of Molecular Sciences 23, no. 3: 1224. https://doi.org/10.3390/ijms23031224
APA StyleBan, I., Sugawa, H., & Nagai, R. (2022). Protein Modification with Ribose Generates Nδ-(5-hydro-5-methyl-4-imidazolone-2-yl)-ornithine. International Journal of Molecular Sciences, 23(3), 1224. https://doi.org/10.3390/ijms23031224





