Identification and Characterization of WRKY41, a Gene Conferring Resistance to Powdery Mildew in Wild Tomato (Solanum habrochaites) LA1777
Abstract
:1. Introduction
2. Results
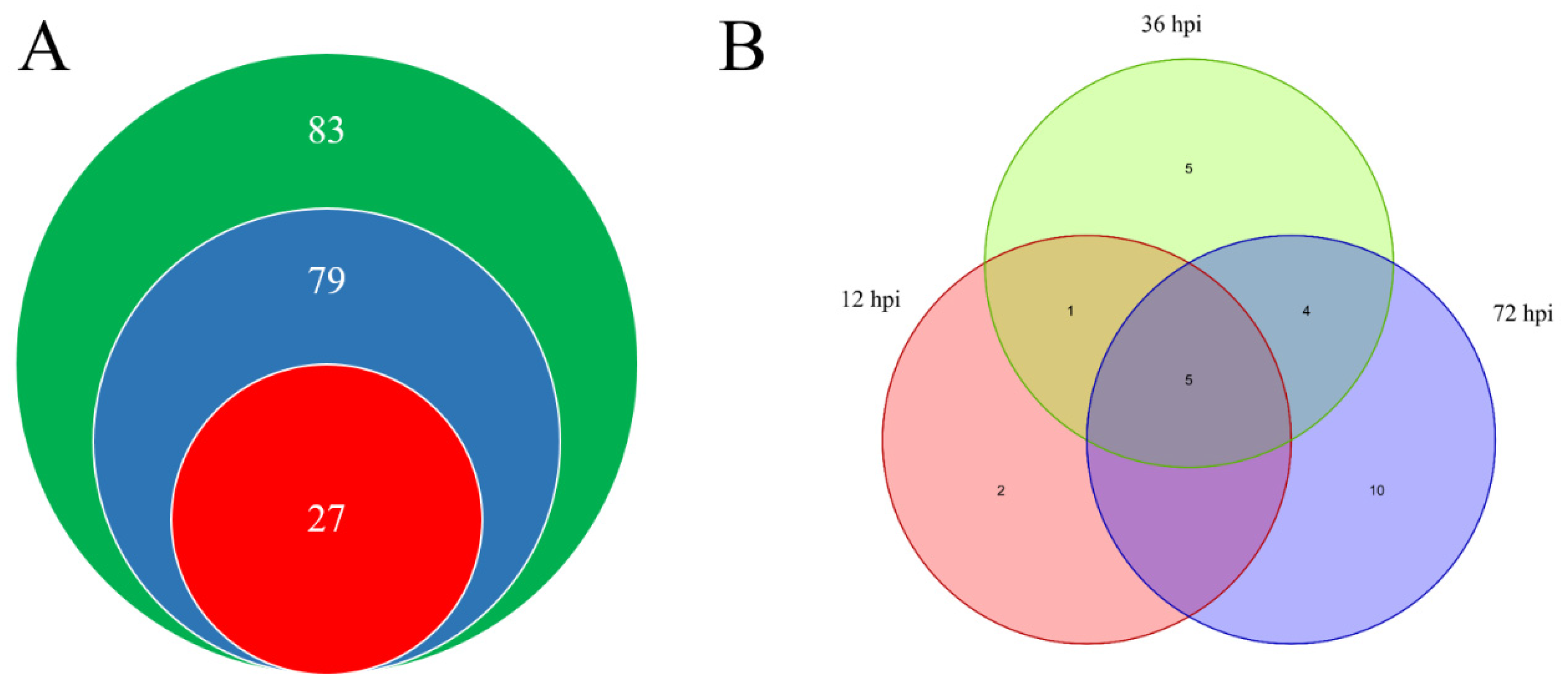
2.1. A Total of 27 WRKYs Are Differentially Expressed in LA1777 under On-lz Infection
2.2. Categorization and Function Analysis of Differentially Expressed WRKYs in Wild Tomato LA1777 under On-lz Infection
2.3. The Molecular Function Analysis of ShWRKY41
2.4. ShWRKY41 Silencing Results in Host Susceptibility to On-lz in Wild Tomato LA1777 Seedlings
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Plant, Pathogen Growth, and Inoculation Experiments
4.2. Sample Collection, Library Preparation and Transcriptome Sequencing
4.3. Bioinformatics Analysis of WRKYs
4.4. The Expression Analysis of Target WRKYs
4.5. The Promoter Analysis of ShWRKY41
4.6. Expression Analysis of ShWRKY41 in Wild Tomato LA1777 under Abiotic Stresses
4.7. TRV Vectors Construction and Plant Transformation
4.8. Subcellular Localization Analysis
4.9. Fungal Biomass Analysis and Quantification of Disease Severity
4.10. The expression Analysis of Plant Hormone Biosynthesis Related Genes
4.11. Data Collection and Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kiss, L. Identification of two powdery mildew fungi, Oidium neolycopersici sp. nov. and an Oidium subgenus Reticuloidium, infecting tomato in different parts of the world. Mycol. Res. 2001, 105, 684–697. [Google Scholar] [CrossRef]
- Whipps, J.M.; Budge, S.P.; Fenlon, J.S. Characteristics and host range of tomato powdery mildew. Plant Pathol. 1998, 47, 36–48. [Google Scholar] [CrossRef]
- Jones, H.; Whipps, J.M.; Gurr, S.J. The tomato powdery mildew fungus Oidium neolycopersici. Mol. Plant Pathol. 2001, 2, 303–309. [Google Scholar] [CrossRef] [Green Version]
- Jankovics, T.; Bai, Y.; Kovács, G.M.; Bardin, M.; Nicot, P.C.; Toyoda, H.; Matsuda, Y.; Niks, R.E.; Kiss, L. Oidium neolycopersici: Intraspecific variability inferred from amplified fragment length polymorphism analysis and relationship with closely related powdery mildew fungi infecting various plant species. Phytopathology 2008, 98, 529–540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindhout, P.; Beek, H.; Pet, G. Wild Lycopersicon Species as Sources for Resistance to Powdery Mildew (Oidium lycopersicum): Mapping of the Resistance Gene OL-1 on Chromosome 6 of L. hirsutum; International Society for Horticultural Science (ISHS): Leuven, Belgium, 1994; pp. 387–394. [Google Scholar]
- Bai, Y.; Huang, C.C.; van der Hulst, R.; Meijer-Dekens, F.; Bonnema, G.; Lindhout, P. QTLs for tomato powdery mildew resistance (Oidium lycopersici) in Lycopersicon parviflorum G1.1601 co-localize with two qualitative powdery mildew resistance genes. Mol. Plant-Microbe Interact. 2003, 16, 169–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, Y.; van der Hulst, R.; Bonnema, G.; Marcel, T.C.; Meijer-Dekens, F.; Niks, R.E.; Lindhout, P. Tomato defense to Oidium neolycopersici: Dominant Ol genes confer isolate-dependent resistance via a different mechanism than recessive ol-2. Mol. Plant-Microbe Interact. 2005, 18, 354–362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, C.C.; Cui, Y.Y.; Weng, C.R.; Zabel, P.; Lindhout, P. Development of diagnostic PCR markers closely linked to the tomato powdery mildew resistance gene Ol-1 on chromosome 6 of tomato. Theor. Appl. Genet. 2000, 101, 918–924. [Google Scholar] [CrossRef]
- Sun, G.; Feng, C.; Guo, J.; Zhang, A.; Xu, Y.; Wang, Y.; Day, B.; Ma, Q. The tomato Arp2/3 complex is required for resistance to the powdery mildew fungus Oidium neolycopersici. Plant Cell Environ. 2019, 42, 2664–2680. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, K.; Pei, D.; Yu, D.; Zhang, J.; Li, X.; Chen, G.; Yang, H.; Zhou, W.; Li, C. ShORR-1, a novel tomato gene, confers enhanced host resistance to Oidium neolycopersici. Front. Plant Sci. 2019, 10, 1400. [Google Scholar] [CrossRef] [Green Version]
- Ulker, B.; Somssich, I.E. WRKY transcription factors: From DNA binding towards biological function. Curr. Opin. Plant Biol. 2004, 7, 491–498. [Google Scholar] [CrossRef] [Green Version]
- Poll, A.A.; Lee, J.; Sanderson, R.A.; Byrne, E.; Gatehouse, J.A.; Sadanandom, A.; Gatehouse, A.M.R.; Edwards, M.G. Septoria leaf blotch and reduced nitrogen availability alter WRKY transcription factor expression in a codependent manner. Int. J. Mol. Sci. 2020, 21, 4165. [Google Scholar] [CrossRef] [PubMed]
- Rosado, D.; Ackermann, A.; Spassibojko, O.; Rossi, M.; Pedmale, U.V. WRKY transcription factors and ethylene signaling modify root growth during the shade avoidance response. Plant Physiol. 2021, 2021, kiab493. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zheng, W.; Li, J.; Liu, P.; Zhong, K.; Jin, P.; Xu, M.; Yang, J.; Chen, J. NbWRKY40 positively regulates the response of Nicotiana benthamiana to Tomato Mosaic Virus via salicylic acid signaling. Front. Plant Sci. 2020, 11, 603518. [Google Scholar] [CrossRef] [PubMed]
- Ahammed, G.J.; Li, X.; Mao, Q.; Wan, H.; Zhou, G.; Cheng, Y. The SlWRKY81 transcription factor inhibits stomatal closure by attenuating nitric oxide accumulation in the guard cells of tomato under drought. Physiol. Plant 2021, 172, 885–895. [Google Scholar] [CrossRef]
- Shu, P.; Zhang, S.; Li, Y.; Wang, X.; Yao, L.; Sheng, J.; Shen, L. Over-expression of SlWRKY46 in tomato plants increases susceptibility to Botrytis cinerea by modulating ROS homeostasis and SA and JA signaling pathways. Plant Physiol. Biochem. 2021, 166, 1–9. [Google Scholar] [CrossRef]
- Ishiguro, S.; Nakamura, K. Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and β-amylase from sweet potato. Mol. Gen. Genet. 1994, 244, 563–571. [Google Scholar] [CrossRef]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Huang, Y.; Li, M.-Y.; Wu, P.; Xu, Z.-S.; Que, F.; Wang, F.; Xiong, A.-S. Members of WRKY Group III transcription factors are important in TYLCV defense signaling pathway in tomato (Solanum lycopersicum). BMC Genom. 2016, 17, 788. [Google Scholar] [CrossRef] [Green Version]
- Karkute, S.G.; Gujjar, R.S.; Rai, A.; Akhtar, M.; Singh, M.; Singh, B. Genome wide expression analysis of WRKY genes in tomato (Solanum lycopersicum) under drought stress. Plant Gene 2018, 13, 8–17. [Google Scholar] [CrossRef]
- Abdullah-Zawawi, M.R.; Ahmad-Nizammuddin, N.F.; Govender, N.; Harun, S.; Mohd-Assaad, N.; Mohamed-Hussein, Z.A. Comparative genome-wide analysis of WRKY, MADS-box and MYB transcription factor families in Arabidopsis and rice. Sci. Rep. 2021, 11, 19678. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Qiao, L.; Guo, H.; Guo, L.; Ren, F.; Bai, J.; Wang, Y. Genome-wide identification of wheat WRKY gene family reveals that TaWRKY75-A is referred to drought and salt resistances. Front. Plant Sci. 2021, 12, 663118. [Google Scholar] [CrossRef] [PubMed]
- Ramos, R.N.; Martin, G.B.; Pombo, M.A.; Rosli, H.G. WRKY22 and WRKY25 transcription factors are positive regulators of defense responses in Nicotiana benthamiana. Plant Mol. Biol. 2021, 105, 65–82. [Google Scholar] [CrossRef] [PubMed]
- Rinerson, C.I.; Rabara, R.C.; Tripathi, P.; Shen, Q.J.; Rushton, P.J. The evolution of WRKY transcription factors. BMC Plant Biol. 2015, 15, 66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eulgem, T.; Somssich, I.E. Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 2007, 10, 366–371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bakshi, M.; Oelmüller, R. WRKY transcription factors: Jack of many trades in plants. Plant Signal. Behav. 2014, 9, e27700. [Google Scholar] [CrossRef] [Green Version]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.M.; Zhang, Y. Plant immunity: Danger perception and signaling. Cell 2020, 181, 978–989. [Google Scholar] [CrossRef]
- Xu, X.; Chen, C.; Fan, B.; Chen, Z. Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 2006, 18, 1310–1326. [Google Scholar] [CrossRef] [Green Version]
- Shen, Q.H.; Saijo, Y.; Mauch, S.; Biskup, C.; Bieri, S.; Keller, B.; Seki, H.; Ulker, B.; Somssich, I.E.; Schulze-Lefert, P. Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 2007, 315, 1098–1103. [Google Scholar] [CrossRef] [Green Version]
- Peng, Y.; Bartley, L.E.; Chen, X.; Dardick, C.; Chern, M.; Ruan, R.; Canlas, P.E.; Ronald, P.C. OsWRKY62 is a negative regulator of basal and Xa21-mediated defense against Xanthomonas oryzae pv. oryzae in rice. Mol. Plant 2008, 1, 446–458. [Google Scholar] [CrossRef] [Green Version]
- Qiu, D.; Xiao, J.; Ding, X.; Xiong, M.; Cai, M.; Cao, Y.; Li, X.; Xu, C.; Wang, S. OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. Mol. Plant-Microbe Interact. 2007, 20, 492–499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimono, M.; Sugano, S.; Nakayama, A.; Jiang, C.J.; Ono, K.; Toki, S.; Takatsuji, H. Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell 2007, 19, 2064–2076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chujo, T.; Takai, R.; Akimoto-Tomiyama, C.; Ando, S.; Minami, E.; Nagamura, Y.; Kaku, H.; Shibuya, N.; Yasuda, M.; Nakashita, H.; et al. Involvement of the elicitor-induced gene OsWRKY53 in the expression of defense-related genes in rice. Biochim. Biophys. Acta 2007, 1769, 497–505. [Google Scholar] [CrossRef]
- Liu, Q.; Li, X.; Yan, S.; Yu, T.; Yang, J.; Dong, J.; Zhang, S.; Zhao, J.; Yang, T.; Mao, X.; et al. OsWRKY67 positively regulates blast and bacteria blight resistance by direct activation of PR genes in rice. BMC Plant Biol. 2018, 18, 257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, B.; Hong, Y.B.; Zhang, Y.F.; Li, X.H.; Huang, L.; Zhang, H.J.; Li, D.Y.; Song, F.M. Tomato WRKY transcriptional factor SlDRW1 is required for disease resistance against Botrytis cinerea and tolerance to oxidative stress. Plant Sci. Int. J. Exp. Plant Biol. 2014, 227, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wang, J.; Zheng, Z.; Fan, B.; Yu, J.Q.; Chen, Z. Characterization of the promoter and extended C-terminal domain of Arabidopsis WRKY33 and functional analysis of tomato WRKY33 homologues in plant stress responses. J. Exp. Bot. 2015, 66, 4567–4583. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.; Gao, Y.; Liu, J.; Peng, X.; Niu, X.; Fei, Z.; Cao, S.; Liu, Y. Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol. Genet. Genom. 2012, 287, 495–513. [Google Scholar] [CrossRef] [PubMed]
- Chinnapandi, B.; Bucki, P.; Braun Miyara, S. SlWRKY45, nematode-responsive tomato WRKY gene, enhances susceptibility to the root knot nematode M. javanica infection. Plant Signal. Behav. 2017, 12, e1356530. [Google Scholar] [CrossRef] [Green Version]
- Akbudak, M.A.; Filiz, E. Genome-wide investigation of proline transporter (ProT) gene family in tomato: Bioinformatics and expression analyses in response to drought stress. Plant Physiol. Biochem. 2020, 157, 13–22. [Google Scholar] [CrossRef]
- Finiti, I.; Leyva, M.d.l.O.; Vicedo, B.; Gómez-Pastor, R.; López-Cruz, J.; García-Agustín, P.; Real, M.D.; González-Bosch, C. Hexanoic acid protects tomato plants against Botrytis cinerea by priming defence responses and reducing oxidative stress. Mol. Plant Pathol. 2014, 15, 550–562. [Google Scholar] [CrossRef]
- López-Galiano, M.J.; González-Hernández, A.I.; Crespo-Salvador, O.; Rausell, C.; Real, M.D.; Escamilla, M.; Camañes, G.; García-Agustín, P.; González-Bosch, C.; García-Robles, I. Epigenetic regulation of the expression of WRKY75 transcription factor in response to biotic and abiotic stresses in Solanaceae plants. Plant Cell Rep. 2018, 37, 167–176. [Google Scholar] [CrossRef]
- Hichri, I.; Muhovski, Y.; Žižková, E.; Dobrev, P.I.; Gharbi, E.; Franco-Zorrilla, J.M.; Lopez-Vidriero, I.; Solano, R.; Clippe, A.; Errachid, A.; et al. The Solanum lycopersicum WRKY3 Transcription Factor SlWRKY3 is involved in salt stress tolerance in tomato. Front. Plant Sci. 2017, 8, 1343. [Google Scholar] [CrossRef]
- Cui, J.; Xu, P.; Meng, J.; Li, J.; Jiang, N.; Luan, Y. Transcriptome signatures of tomato leaf induced by Phytophthora infestans and functional identification of transcription factor SpWRKY3. Theor. Appl. Genet. 2018, 131, 787–800. [Google Scholar] [CrossRef]
- Li, J.-B.; Luan, Y.-S.; Liu, Z. SpWRKY1 mediates resistance to Phytophthora infestans and tolerance to salt and drought stress by modulating reactive oxygen species homeostasis and expression of defense-related genes in tomato. Plant Cell Tissue Organ Cult. (PCTOC) 2015, 123, 67–81. [Google Scholar] [CrossRef]
- Cui, J.; Jiang, N.; Meng, J.; Yang, G.; Liu, W.; Zhou, X.; Ma, N.; Hou, X.; Luan, Y. LncRNA33732-respiratory burst oxidase module associated with WRKY1 in tomato-Phytophthora infestans interactions. Plant J. Cell Mol. Biol. 2019, 97, 933–946. [Google Scholar] [CrossRef]
- Van Eck, L.; Davidson, R.M.; Wu, S.; Zhao, B.Y.; Botha, A.M.; Leach, J.E.; Lapitan, N.L. The transcriptional network of WRKY53 in cereals links oxidative responses to biotic and abiotic stress inputs. Funct. Integr. Genom. 2014, 14, 351–362. [Google Scholar] [CrossRef] [Green Version]
- Yoo, S.J.; Kim, S.H.; Kim, M.J.; Ryu, C.M.; Kim, Y.C.; Cho, B.H.; Yang, K.Y. Involvement of the OsMKK4-OsMPK1 cascade and its downstream transcription factor OsWRKY53 in the wounding response in Rice. Plant Pathol. J. 2014, 30, 168–177. [Google Scholar] [CrossRef] [Green Version]
- Chujo, T.; Miyamoto, K.; Ogawa, S.; Masuda, Y.; Shimizu, T.; Kishi-Kaboshi, M.; Takahashi, A.; Nishizawa, Y.; Minami, E.; Nojiri, H.; et al. Overexpression of phosphomimic mutated OsWRKY53 leads to enhanced blast resistance in rice. PLoS ONE 2014, 9, e98737. [Google Scholar] [CrossRef] [Green Version]
- Hu, L.; Ye, M.; Li, R.; Zhang, T.; Zhou, G.; Wang, Q.; Lu, J.; Lou, Y. The rice transcription factor WRKY53 suppresses herbivore-induced defenses by acting as a negative feedback modulator of mitogen-activated protein kinase activity. Plant Physiol. 2015, 169, 2907–2921. [Google Scholar]
- Tian, X.; Li, X.; Zhou, W.; Ren, Y.; Wang, Z.; Liu, Z.; Tang, J.; Tong, H.; Fang, J.; Bu, Q. Transcription factor OsWRKY53 positively regulates brassinosteroid signaling and plant architecture. Plant Physiol. 2017, 175, 1337–1349. [Google Scholar] [CrossRef]
- Chen, L.; Song, Y.; Li, S.; Zhang, L.; Zou, C.; Yu, D. The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta 2012, 1819, 120–128. [Google Scholar] [CrossRef]
- Dang, F.; Wang, Y.; She, J.; Lei, Y.; Liu, Z.; Eulgem, T.; Lai, Y.; Lin, J.; Yu, L.; Lei, D.; et al. Overexpression of CaWRKY27, a subgroup IIe WRKY transcription factor of Capsicum annuum, positively regulates tobacco resistance to Ralstonia solanacearum infection. Physiol. Plant 2014, 150, 397–411. [Google Scholar] [CrossRef]
- Tak, H.; Negi, S.; Ganapathi, T.R. The 5′-upstream region of WRKY18 transcription factor from banana is a stress-inducible promoter with strong expression in guard cells. Physiol. Plant 2021, 173, 1335–1350. [Google Scholar] [CrossRef]
- Negi, N.; Khurana, P. A salicylic acid inducible mulberry WRKY transcription factor, MiWRKY53 is involved in plant defence response. Plant Cell Rep. 2021, 40, 2151–2171. [Google Scholar] [CrossRef]
- Zhao, Z.X.; Feng, Q.; Liu, P.Q.; He, X.R.; Zhao, J.H.; Xu, Y.J.; Zhang, L.L.; Huang, Y.Y.; Zhao, J.Q.; Fan, J.; et al. RPW8.1 enhances the ethylene-signaling pathway to feedback-attenuate its mediated cell death and disease resistance in Arabidopsis. New Phytol. 2021, 229, 516–531. [Google Scholar] [CrossRef]
- Zheng, H.; Dong, L.; Han, X.; Jin, H.; Yin, C.; Han, Y.; Li, B.; Qin, H.; Zhang, J.; Shen, Q.; et al. The TuMYB46L-TuACO3 module regulates ethylene biosynthesis in einkorn wheat defense to powdery mildew. New Phytol. 2020, 225, 2526–2541. [Google Scholar] [CrossRef]
- Ge, X.-M.; Cai, H.-L.; Lei, X.; Zhou, X.; Yue, M.; He, J.-M. Heterotrimeric G protein mediates ethylene-induced stomatal closure via hydrogen peroxide synthesis in Arabidopsis. Plant J. 2015, 82, 138–150. [Google Scholar] [CrossRef]
- Pan, R.; Han, H.; Medison, M.B.; Abou-Elwafa, S.F.; Liu, Y.; Yang, X.; Zhang, W. Aerenchyma formation in the root of leaf-vegetable sweet potato: Programmed cell death initiated by ethylene-mediated H2O2 accumulation. Physiol. Plant 2021, 173, 2361–2375. [Google Scholar] [CrossRef]
- Lian, Q.; Meng, Y.; Zhao, X.; Xu, Y.; Wang, Y.; Day, B.; Ma, Q. ShNPSN11, a vesicle-transport-related gene, confers disease resistance in tomato to Oidium neolycopersici. Biochem. J. 2020, 477, 3851–3866. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Y.; Liu, X.; Xu, Y.; Ma, Q. Microtubule polymerization functions in hypersensitive response and accumulation of h2o2 in wheat induced by the stripe rust. Biomed. Res. Int. 2016, 2016, 7830768. [Google Scholar]
- Zheng, Z.; Appiano, M.; Pavan, S.; Bracuto, V.; Ricciardi, L.; Visser, R.G.F.; Wolters, A.M.A.; Bai, Y. Genome-wide study of the tomato SlMLO gene family and its functional characterization in response to the powdery mildew fungus Oidium neolycopersici. Front. Plant. Sci. 2016, 7, 380. [Google Scholar] [CrossRef] [Green Version]
- Wingett, S.W.; Andrews, S. FastQ Screen: A tool for multi-genome mapping and quality control. F1000Research 2018, 7, 1338. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Luo, X.; Qian, J.; Pang, X.; Song, J.; Qian, G.; Chen, J.; Chen, S. FastUniq: A fast de novo duplicates removal tool for paired short reads. PLoS ONE 2012, 7, e52249. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Iwase, A.; Kondo, Y.; Laohavisit, A.; Takebayashi, A.; Ikeuchi, M.; Matsuoka, K.; Asahina, M.; Mitsuda, N.; Shirasu, K.; Fukuda, H.; et al. WIND transcription factors orchestrate wound-induced callus formation, vascular reconnection and defense response in Arabidopsis. New Phytol. 2021, 232, 734–752. [Google Scholar] [CrossRef]
- Lian, Q.; Zhang, J.; Gan, L.; Ma, Q.; Zong, Z.; Wang, Y. The biocontrol efficacy of Streptomyces pratensis LMM15 on Botrytis cinerea in tomato. Biomed. Res. Int. 2017, 2017, 9486794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Prot. 2008, 6, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xie, X.; Yao, W.; Wang, J.; Ma, F.; Wang, C.; Yang, Y.; Tong, W.; Zhang, J.; Xu, Y.; et al. RING-H2-type E3 gene VpRH2 from Vitis pseudoreticulata improves resistance to powdery mildew by interacting with VpGRP2A. J. Exp. Bot. 2017, 68, 1669–1687. [Google Scholar] [CrossRef]
- Wakeel, A.; Ali, I.; Wu, M.; Liu, B.; Gan, Y. Dichromate-induced ethylene biosynthesis, perception, and signaling regulate the variance in root growth inhibition among Shaheen basmati and basmati-385 rice varieties. Environ. Sci. Pollut. Res. Int. 2021, 28, 38016–38025. [Google Scholar] [CrossRef]
- Jefferson, R.A. Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol. Biol. Report. 1987, 5, 387–405. [Google Scholar] [CrossRef]
- Fernandez-Pozo, N.; Rosli, H.G.; Martin, G.B.; Mueller, L.A. The SGN VIGS tool: User-friendly software to design virus-induced gene silencing (VIGS) constructs for functional genomics. Mol. Plant 2015, 8, 486–488. [Google Scholar] [CrossRef] [Green Version]
- Senthil-Kumar, M.; Hema, R.; Anand, A.; Kang, L.; Udayakumar, M.; Mysore, K. A systematic study to determine the extent of gene silencing in Nicotiana benthamiana and other Solanaceae species when heterologous gene sequences are used for virus-induced gene silencing. New Phytol. 2007, 176, 782–791. [Google Scholar] [CrossRef]
- Weigel, D.; Glazebrook, J. Transformation of Agrobacterium using the freeze-thaw method. CSH Protoc 2006, 2006. [Google Scholar] [CrossRef]
- Correll, J.C.; Gordon, T.R.; Elliot, V.J. Powdery mildew of tomato: The effect of planting date and triadimefon on disease onset, progress, incidence, and severity. Phytopathology 1988, 78, 512–519. [Google Scholar] [CrossRef]
- Wang, J.; Hai, Z.; Yan, H.; Feng, C.; Yang, W.; Ma, Q. Evaluation of actin cytoskeleton in non-host resistance of pepper to Puccinia striiformis f. sp. tritici stress. Phyiol. Mol. Plant Pathol. 2015, 92, 112–118. [Google Scholar] [CrossRef]
- Rekhter, D.; Lüdke, D.; Ding, Y.; Feussner, K.; Zienkiewicz, K.; Lipka, V.; Wiermer, M.; Zhang, Y.; Feussner, I. Isochorismate-derived biosynthesis of the plant stress hormone salicylic acid. Science 2019, 365, 498–502. [Google Scholar] [CrossRef] [PubMed]
- Torrens-Spence, M.P.; Bobokalonova, A.; Carballo, V.; Glinkerman, C.M.; Pluskal, T.; Shen, A.; Weng, J.K. PBS3 and EPS1 complete salicylic acid biosynthesis from isochorismate in Arabidopsis. Mol. Plant 2019, 12, 1577–1586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, D.O.; Yang, S.F. Ethylene biosynthesis: Identification of 1-aminocyclopropane-1-carboxylic acid as an intermediate in the conversion of methionine to ethylene. Proc. Natl. Acad. Sci. USA 1979, 76, 170–174. [Google Scholar] [CrossRef] [Green Version]
- Chang, C. Q&A: How do plants respond to ethylene and what is its importance? BMC Biol. 2016, 14, 7. [Google Scholar]
- Frankowski, K.; Kesy, J.; Kopcewicz, J. Regulation of ethylene biosynthesis in plants. Postepy Biochem. 2007, 53, 66–73. [Google Scholar]








| Gene_Description | Group | log2FoldChange | padj | |
|---|---|---|---|---|
| 12 hpi | ShWRKY44 | I | 4.99 | 0.000017 |
| ShWRKY1 | I | 2.58 | 0.000000 | |
| ShWRKY40 | II-a | 2.57 | 0.000442 | |
| ShWRKY51 | II-c | −1.83 | 0.000173 | |
| ShWRKY IId-1 | II-d | 1.46 | 0.010183 | |
| ShWRKY22-like | II-e | 1.90 | 0.021111 | |
| ShWRKY29 | II-e | 1.64 | 0.000160 | |
| ShWRKY41 | III | 2.37 | 0.000000 | |
| 36 hpi | ShWRKY44 | I | 5.73 | 0.007329 |
| ShWRKY33B | I | 2.71 | 0.000026 | |
| ShWRKY1 | I | 1.66 | 0.003228 | |
| ShWRKY3 | I | −1.33 | 0.047367 | |
| ShWRKY45 | II-a | 4.45 | 0.011781 | |
| ShWRKY40 | II-a | 2.76 | 0.000469 | |
| ShWRKY9 | II-b | 5.18 | 0.029976 | |
| ShWRKY30 | II-c | 2.72 | 0.009789 | |
| ShWRKY30-like | II-c | 1.48 | 0.029717 | |
| ShWRKY IId-1 | II-d | 1.51 | 0.013714 | |
| ShWRKY22-like | II-e | 2.06 | 0.004722 | |
| ShWRKY26 | II-e | 1.75 | 0.029760 | |
| ShWRKY53 | III | 2.49 | 0.000276 | |
| ShWRKY53 | III | 2.26 | 0.000432 | |
| ShWRKY41 | III | 2.10 | 0.000002 | |
| 72 hpi | ShWRKY33B | I | 3.13 | 0.000000 |
| ShWRKY1 | I | 3.04 | 0.000000 | |
| ShWRKY31 | I | 1.76 | 0.000000 | |
| ShWRKY14 | I | −2.69 | 0.038904 | |
| ShWRKY40 | II-a | 4.54 | 0.000000 | |
| ShWRKY48 | II-c | 2.28 | 0.000019 | |
| ShWRKY23 | II-c | 1.68 | 0.001020 | |
| ShWRKY56 | II-c | 5.45 | 0.009433 | |
| ShWRKY75 | II-c | −1.57 | 0.048565 | |
| ShWRKY IId-1 | II-d | 2.28 | 0.000000 | |
| ShWRKY7 | II-d | 1.44 | 0.000000 | |
| ShWRKYIId-4 | II-d | 1.20 | 0.003540 | |
| ShWRKY21 | II-d | −1.14 | 0.025213 | |
| ShWRKY22-like | II-e | 3.26 | 0.000000 | |
| ShWRKY26 | II-e | 3.12 | 0.000000 | |
| ShWRKY70 | II-e | 1.55 | 0.000000 | |
| ShWRKY41 | III | 3.06 | 0.000000 | |
| ShWRKY53 | III | 3.00 | 0.000000 | |
| ShWRKY53 | III | 2.11 | 0.000000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lian, Q.; He, X.; Zhang, B.; Wang, Y.; Ma, Q. Identification and Characterization of WRKY41, a Gene Conferring Resistance to Powdery Mildew in Wild Tomato (Solanum habrochaites) LA1777. Int. J. Mol. Sci. 2022, 23, 1267. https://doi.org/10.3390/ijms23031267
Lian Q, He X, Zhang B, Wang Y, Ma Q. Identification and Characterization of WRKY41, a Gene Conferring Resistance to Powdery Mildew in Wild Tomato (Solanum habrochaites) LA1777. International Journal of Molecular Sciences. 2022; 23(3):1267. https://doi.org/10.3390/ijms23031267
Chicago/Turabian StyleLian, Qinggui, Xinyi He, Bingbing Zhang, Yang Wang, and Qing Ma. 2022. "Identification and Characterization of WRKY41, a Gene Conferring Resistance to Powdery Mildew in Wild Tomato (Solanum habrochaites) LA1777" International Journal of Molecular Sciences 23, no. 3: 1267. https://doi.org/10.3390/ijms23031267






