Genome-Wide Identification and Characterization of RNA/DNA Differences Associated with Drought Response in Wheat
Abstract
:1. Introduction
2. Results
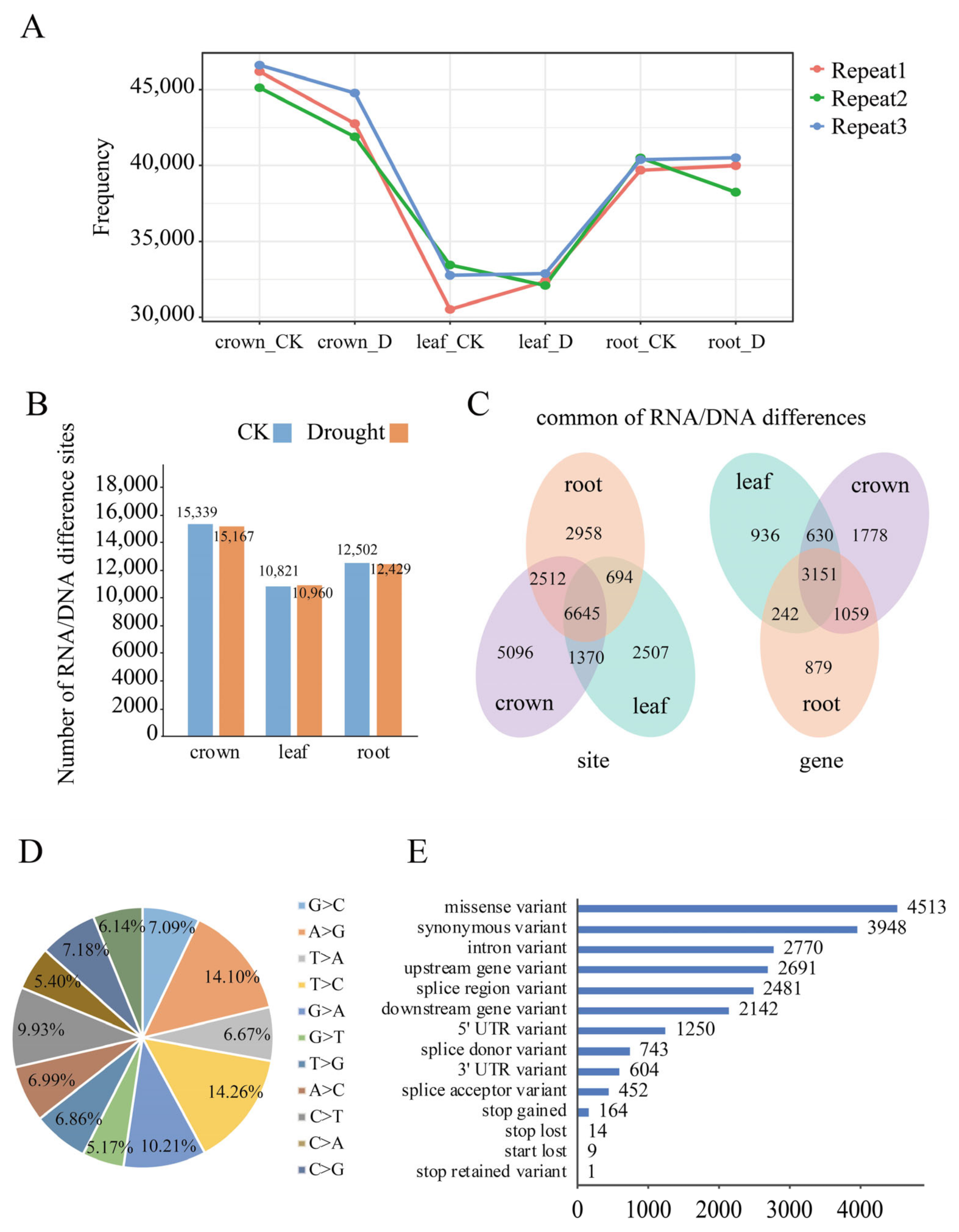
2.1. Identification of RNA/DNA Differences Based on RNA-Seq Data in Wheat
2.2. Identification of RDDs Associated with Drought Response
2.3. Effects of RDDs on Gene Expression
2.4. Effect of RDDs on RNA Secondary Structure and MiRNA Targeting
2.5. Effects of RDDs on Protein Conserved Domain
3. Discussion
4. Materials and Methods
4.1. RNA-Seq Data and Mapping
4.2. Identification of RNA/DNA Difference Sites
- (1)
- GATK VariantFiltration tool was used to correct the systematic errors of the sequencing platform and software, and the initial filtering parameters – Filter “FS > 30.0”, – Filter “QD < 2.0” were selected;
- (2)
- The site was mapped by more than 10 reads with reference read >2 and mutation read >3 retained;
- (3)
- In order to improve the accuracy, only the DNA/RNA difference sites that appeared in all of the three replicates were retained;
- (4)
- the RDDs between control and drought-treated samples were compared to remove the same sites to obtain the potential drought-responsive RDDs;
- (5)
- manually verify each bam file to assure the RDD’s actual occurrence through Integrative Genomics Viewer (IGV) tool.
4.3. Validation of RNA/DNA Difference Site Using RT-PCR
4.4. RNA Secondary Structure and MiRna Target Analysis
4.5. Conserved Domain Prediction and Protein Structure Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shiferaw, B.; Smale, M.; Braun, H.J.; Duveiller, E.; Reynolds, M.; Muricho, G. Crops that feed the world 10. Past successes and future challenges to the role played by wheat in global food security. Food Secur. 2013, 5, 291–317. [Google Scholar] [CrossRef] [Green Version]
- FAO Statistics. Available online: http://www.fao.org/unfao/procurement/statistics-from-2010-2020/statistics-2019/en/ (accessed on 4 June 2021).
- Asseng, S.; Ewert, F.; Martre, P.; Rötter, R.P.; Lobell, D.B.; Cammarano, D.; Kimball, B.A.; Ottman, M.J.; Wall, G.; White, J.W.; et al. Rising temperatures reduce global wheat production. Nat. Clim. Chang. 2015, 5, 143–147. [Google Scholar] [CrossRef]
- Zampieri, M.; Ceglar, A.; Dentener, F.; Toreti, A. Wheat yield loss attributable to heat waves, drought and water excess at the global, national and subnational scales. Environ. Res. Lett. 2017, 12, 064008. [Google Scholar] [CrossRef]
- Stratonovitch, P.; Semenov, M.A. Heat tolerance around flowering in wheat identified as a key trait for increased yield potential in Europe under climate change. J. Exp. Bot. 2015, 66, 3599–3609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lipiec, J.; Doussan, C.; Nosalewicz, A.; Kondracka, K. Effect of drought and heat stresses on plant growth and yield: A review. Int. Agrophys. 2013, 27, 463–477. [Google Scholar] [CrossRef]
- Basu, S.; Ramegowda, V.; Kumar, A.; Pereira, A. Plant adaptation to drought stress. F1000Research 2016, 5. [Google Scholar] [CrossRef]
- Fahad, S.; Bajwa, A.A.; Nazir, U.; Anjum, S.A.; Farooq, A.; Zohaib, A.; Sadia, S.; Nasim, W.; Adkins, S.; Saud, S.; et al. Crop production under drought and heat stress: Plant responses and management options. Front. Plant Sci. 2017, 8, 1147. [Google Scholar] [CrossRef] [Green Version]
- Peng, Z.; Cheng, Y.; Tan, B.C.M.; Kang, L.; Tian, Z.; Zhu, Y.; Zhang, W.; Liang, Y.; Hu, X.; Tan, X.; et al. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat. Biotechnol. 2012, 30, 253–260. [Google Scholar] [CrossRef]
- Nishikura, K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 2016, 17, 83–96. [Google Scholar] [CrossRef] [Green Version]
- Gray, M. Diversity and evolution of mitochondrial RNA editing systems. IUBMB Life 2003, 55, 227–233. [Google Scholar] [CrossRef]
- Bian, Z.; Ni, Y.; Xu, J.R.; Liu, H. A-to-I mRNA editing in fungi: Occurrence, function, and evolution. Cell. Mol. Life Sci. 2019, 76, 329–340. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.L.; Li, L.; Wang, B.; Chen, Z.; Dombrovska, O.; Lee, J.; Kent, L.; Li, R.; Jobson, R.W.; Hendry, T.A.; et al. A nonflowering land plant phylogeny inferred from nucleotide sequences of seven chloroplast, mitochondrial, and nuclear genes. Int. J. Plant Sci. 2007, 168, 691–708. [Google Scholar] [CrossRef]
- Takenaka, M.; Verbitskiy, D.; van der Merwe, J.A.; Zehrmann, A.; Brennicke, A. The process of RNA editing in plant mitochondria. Mitochondrion 2008, 8, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Sun, Y.; Song, X.; Wang, Z.; Zhang, Y.; Zhao, Y.; Peng, X.; Zhang, X.; Li, C.; Gao, C.J.; et al. Hepatitis B virus evades immune recognition via RNA adenosine deaminase ADAR1-mediated viral RNA editing in hepatocytes. Cell. Mol. Immunol. 2021, 18, 1871–1882. [Google Scholar] [CrossRef]
- Savva, Y.A.; Rieder, L.E.; Reenan, R.A. The ADAR protein family. Genome Biol. 2012, 13, 252. [Google Scholar] [CrossRef]
- Nishikura, K. Editor meets silencer: Crosstalk between RNA editing and RNA interference. Nat. Rev. Mol. Cell Biol. 2006, 7, 919–931. [Google Scholar] [CrossRef] [Green Version]
- Jin, Y.; Zhang, W.; Li, Q. Origins and evolution of ADAR-mediated RNA editing. IUBMB Life 2009, 61, 572–578. [Google Scholar] [CrossRef]
- Salter, J.D.; Bennett, R.P.; Smith, H.C. The APOBEC protein family: United by structure, divergent in function. Trends Biochem. Sci. 2016, 41, 578–594. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Wang, I.X.; Li, Y.; Bruzel, A.; Richards, A.L.; Toung, J.M.; Cheung, V.G. Widespread RNA and DNA sequence differences in the human transcriptome. Science 2011, 333, 53–58. [Google Scholar] [CrossRef] [Green Version]
- Liscovitch-Brauer, N.; Alon, S.; Porath, H.T.; Elstein, B.; Unger, R.; Ziv, T.; Admon, A.; Levanon, E.Y.; Rosenthal, J.J.; Eisenberg, E. Trade-off between transcriptome plasticity and genome evolution in cephalopods. Cell 2017, 169, 191–202. [Google Scholar] [CrossRef] [Green Version]
- Qulsum, U.; Azad, M.T.A.; Tsukahara, T. Analysis of tissue-specific RNA editing events of genes involved in RNA editing in Arabidopsis thaliana. J. Plant Biol. 2019, 62, 351–358. [Google Scholar] [CrossRef]
- Small, I.D.; Schallenberg-Rüdinger, M.; Takenaka, M.; Mireau, H.; Ostersetzer-Biran, O. Plant organellar RNA editing: What 30 years of research has revealed. Plant J. 2020, 101, 1040–1056. [Google Scholar] [CrossRef]
- Wang, X.; An, Y.; Xu, P.; Xiao, J. Functioning of PPR Proteins in organelle RNA metabolism and chloroplast biogenesis. Front. Plant Sci. 2021, 12, 1. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.Y.; Jiang, R.C.; Wang, Y.; Tang, J.J.; Sun, F.; Yang, Y.Z.; Tan, B.C. ZmPPR26, a DYW-type pentatricopeptide repeat protein, is required for C-to-U RNA editing at atpA-1148 in maize chloroplasts. J. Exp. Bot. 2021, 72, 4809–4821. [Google Scholar] [CrossRef]
- Ichinose, M.; Sugita, M. The DYW domains of pentatricopeptide repeat RNA editing factors contribute to discriminate target and non-target editing sites. Plant Cell Physiol. 2018, 59, 1652–1659. [Google Scholar] [CrossRef]
- Malbert, B.; Burger, M.; Lopez-Obando, M.; Baudry, K.; Launay-Avon, A.; Härtel, B.; Verbitskiy, D.; Jörg, A.; Berthomé, R.; Lurin, C.; et al. The Analysis of the Editing Defects in the dyw2 Mutant Provides New Clues for the Prediction of RNA Targets of Arabidopsis E+-Class PPR Proteins. Plants 2020, 9, 280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Meng, Y.; Chen, D.; Jin, Y.; Mao, C.; Wu, P.; Chen, M. RNA editing of nuclear transcripts in Arabidopsis thaliana. BMC Genom. 2010, 11 (Suppl. S4), S12. [Google Scholar] [CrossRef] [Green Version]
- Yang, G.; Pan, Y.; Zhang, R.; Huang, J.; Pan, W.; Cui, L.; Song, W.; Nie, X. Genome-wide identification and characterization of DNA/RNA differences associated with Fusarium graminearum infection in wheat. bioRxiv 2021. [Google Scholar] [CrossRef]
- Ramos, M.J.; Faísca-Silva, D.; Coito, J.L.; Cunha, J.; Silva, H.G.; Viegas, W.; Costa, M.M.R.; Amâncio, S.; Rocheta, M. RNA editing in inflorescences of wild grapevine unveils association to sex and development. bioRxiv 2020. [Google Scholar] [CrossRef]
- Ichinose, M.; Sugita, M. RNA editing and its molecular mechanism in plant organelles. Genes 2017, 8, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, B.; Yao, H.; Gao, Y.; Wang, R.; Jin, L. Overexpression of chalcone synthase gene improves flavonoid accumulation and drought tolerance in tobacco. Preprints 2019. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.K.; Jung, H.; Jang, G.; Jeong, J.S.; Kim, Y.S.; Ha, S.H.; Choi, Y.D.; Kim, J.K. Overexpression of the OsERF71 Transcription Factor Alters Rice Root Structure and Drought Resistance. Plant Physiol. 2016, 172, 575–588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dethoff, E.A.; Chugh, J.; Mustoe, A.M.; Al-Hashimi, H.M. Functional complexity and regulation through RNA dynamics. Nature 2012, 482, 322–330. [Google Scholar] [CrossRef] [Green Version]
- Andronescu, M.; Fejes, A.P.; Hutter, F.; Hoos, H.H.; Condon, A.J. A New Algorithm for RNA Secondary Structure Design. J. Mol. Biol. 2004, 336, 607–624. [Google Scholar] [CrossRef]
- Yuan, L.; Zhang, S. Finding stable local optimal RNA secondary structures. Bioinformatics 2011, 27, 2994–3001. [Google Scholar]
- Picardi, E.; Horner, D.S.; Chiara, M.; Schiavon, R.; Valle, G.; Pesole, G. Large-scale detection and analysis of RNA editing in grape mtDNA by RNA deep-sequencing. Nucleic Acids Res. 2010, 38, 4755–4767. [Google Scholar] [CrossRef] [Green Version]
- Vij, S.; Giri, J.; Dansana, P.K.; Kapoor, S.; Tyagi, A.K. The receptor-like cytoplasmic kinase (OsRLCK) gene family in rice: Organization, phylogenetic relationship, and expression during development and stress. Mol. Plant 2008, 1, 732–750. [Google Scholar] [CrossRef]
- Gott, J.M. Expanding genome capacity via RNA editing. Comptes Rendus Biol. 2003, 326, 901–908. [Google Scholar] [CrossRef]
- González-Guzmán, M.; Apostolova, N.; Bellés, J.M.; Barrero, J.M.; Piqueras, P.; Ponce, M.R.; Micol, J.L.; Serrano, R.; Rodríguez, P.L. The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell 2002, 14, 1833–1846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Endo, A.; Nelson, K.M.; Thoms, K.; Abrams, S.R.; Nambara, E.; Sato, Y.J. Functional characterization of xanthoxin dehydrogenase in rice. J. Plant Physiol. 2014, 171, 1231–1240. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.; Sharma, M.; Pandey, G.K. Small and large g proteins in biotic and abiotic stress responses in plants. In Elucidation of Abiotic Stress Signaling in Plants: Functional Genomics Perspectives; Pandey, G.K., Ed.; Springer: New York, NY, USA, 2015; pp. 231–270. [Google Scholar]
- Pandey, S.; Assmann, S.M. The Arabidopsis putative G protein–coupled receptor GCR1 interacts with the G protein α subunit GPA1 and regulates abscisic acid signaling. Plant Cell 2004, 16, 1616–1632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Qin, J.; Tian, X.; Xu, S.; Wang, Y.; Li, H.; Wang, X.; Peng, H.; Yao, Y.; Hu, Z.; et al. Global profiling of alternative splicing landscape responsive to drought, heat and their combination in wheat (Triticum aestivum L.). Plant Biotechnol. J. 2018, 16, 714–726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thatcher, S.R.; Danilevskaya, O.N.; Meng, X.; Beatty, M.; Zastrow-Hayes, G.; Harris, C.; Van Allen, B.; Habben, J.; Li, B. Genome-wide analysis of alternative splicing during development and drought stress in maize. Plant Physiol. 2016, 170, 586–599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, H.; Lou, Q.; Xu, K.; Yan, M.; Xia, H.; Ma, X.; Yu, X.; Luo, L. Alternative splicing complexity contributes to genetic improvement of drought resistance in the rice maintainer HuHan2B. Sci. Rep. 2017, 7, 11686. [Google Scholar] [CrossRef]
- Ding, Y.; Wang, Y.; Qiu, C.; Qian, W.; Xie, H.; Ding, Z. Alternative splicing in tea plants was extensively triggered by drought, heat and their combined stresses. PeerJ 2020, 8, e8258. [Google Scholar] [CrossRef]
- Zhu, T.; Wang, L.; Rimbert, H.; Rodriguez, J.C.; Deal, K.R.; De Oliveira, R.; Choulet, F.; Keeble-Gagnère, G.; Tibbits, J.; Rogers, J.; et al. Optical maps refine the bread wheat Triticum aestivum cv. Chinese Spring genome assembly. Plant J. 2021, 107, 303–314. [Google Scholar] [CrossRef]
- Borchert, G.M.; Gilmore, B.L.; Spengler, R.M.; Xing, Y.; Lanier, W.; Bhattacharya, D.; Davidson, B.L. Adenosine deamination in human transcripts generates novel microRNA binding sites. Hum. Mol. Genet. 2009, 18, 4801–4807. [Google Scholar] [CrossRef]
- Available online: https://www.ncbi.nlm.nih.gov/sra/?term=SRP098756 (accessed on 10 January 2021).
- Available online: http://picard.sourceforge.net/ (accessed on 15 March 2021).
- Available online: http://plants.ensem-bl.org/index.html (accessed on 25 March 2021).
- Available online: http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi (accessed on 13 May 2021).
- Dai, X.; Zhuang, Z.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef] [Green Version]
- Available online: https://swissmodel.expasy.org/ (accessed on 27 May 2021).





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, Y.; Li, M.; Huang, J.; Pan, W.; Shi, T.; Guo, Q.; Yang, G.; Nie, X. Genome-Wide Identification and Characterization of RNA/DNA Differences Associated with Drought Response in Wheat. Int. J. Mol. Sci. 2022, 23, 1405. https://doi.org/10.3390/ijms23031405
Pan Y, Li M, Huang J, Pan W, Shi T, Guo Q, Yang G, Nie X. Genome-Wide Identification and Characterization of RNA/DNA Differences Associated with Drought Response in Wheat. International Journal of Molecular Sciences. 2022; 23(3):1405. https://doi.org/10.3390/ijms23031405
Chicago/Turabian StylePan, Yan, Mengqi Li, Jiaqian Huang, Wenqiu Pan, Tingrui Shi, Qifan Guo, Guang Yang, and Xiaojun Nie. 2022. "Genome-Wide Identification and Characterization of RNA/DNA Differences Associated with Drought Response in Wheat" International Journal of Molecular Sciences 23, no. 3: 1405. https://doi.org/10.3390/ijms23031405
APA StylePan, Y., Li, M., Huang, J., Pan, W., Shi, T., Guo, Q., Yang, G., & Nie, X. (2022). Genome-Wide Identification and Characterization of RNA/DNA Differences Associated with Drought Response in Wheat. International Journal of Molecular Sciences, 23(3), 1405. https://doi.org/10.3390/ijms23031405






