Clinical Breakpoint of Apramycin to Swine Salmonella and Its Effect on Ileum Flora
Abstract
:1. Introduction
2. Results
2.1. MIC Distribution and COWT
2.2. PD Study of APR against Swine Salmonella Strain 10# In Vitro and Ex Vivo
2.3. PK/PD Analysis in the Plasma and Ileum Fluid and Establishment of COPD
2.4. Dosage Determination and Establishment of COCL of APR against Swine Salmonella
2.5. DNA Sequence Data and Bacterial Community Structure
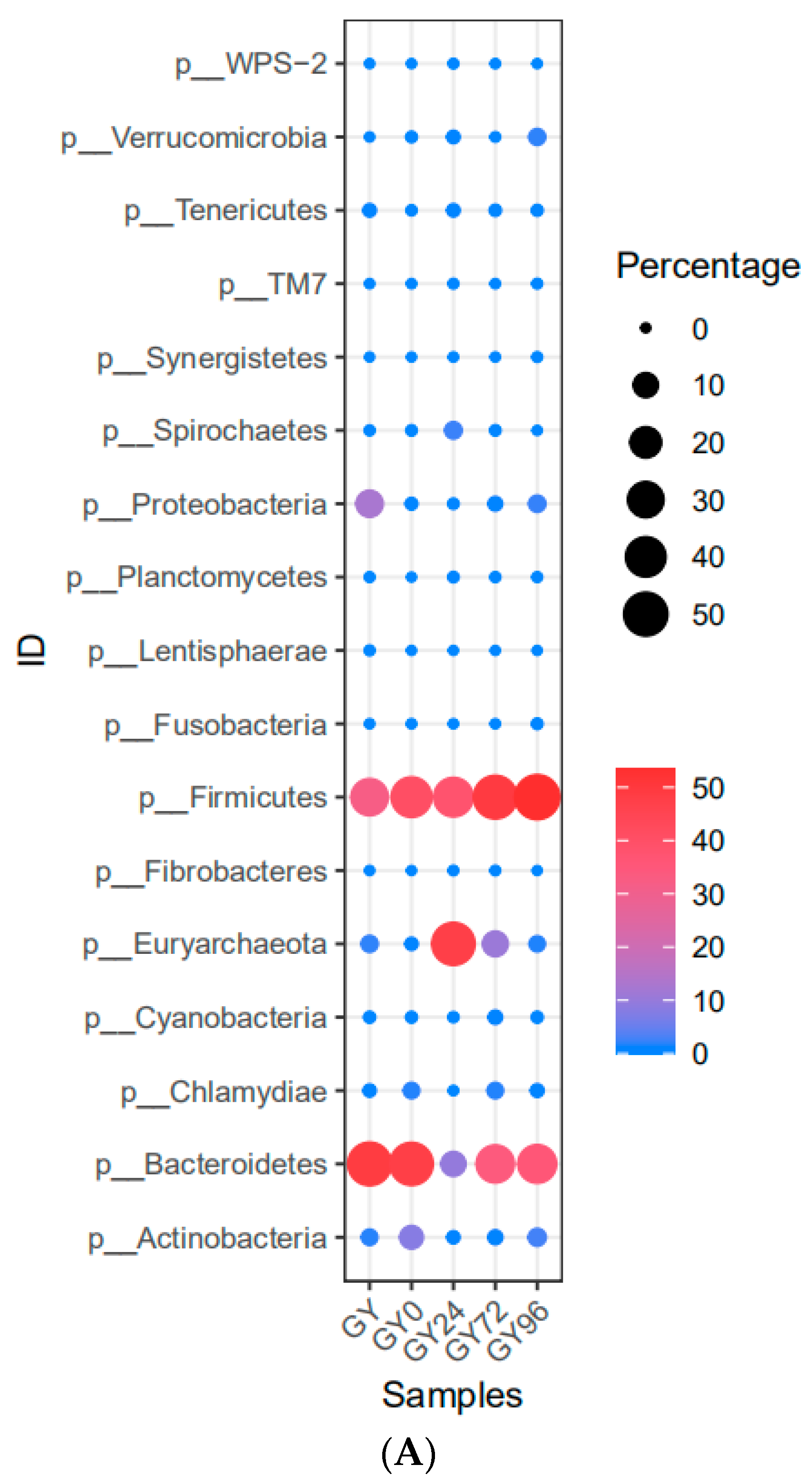
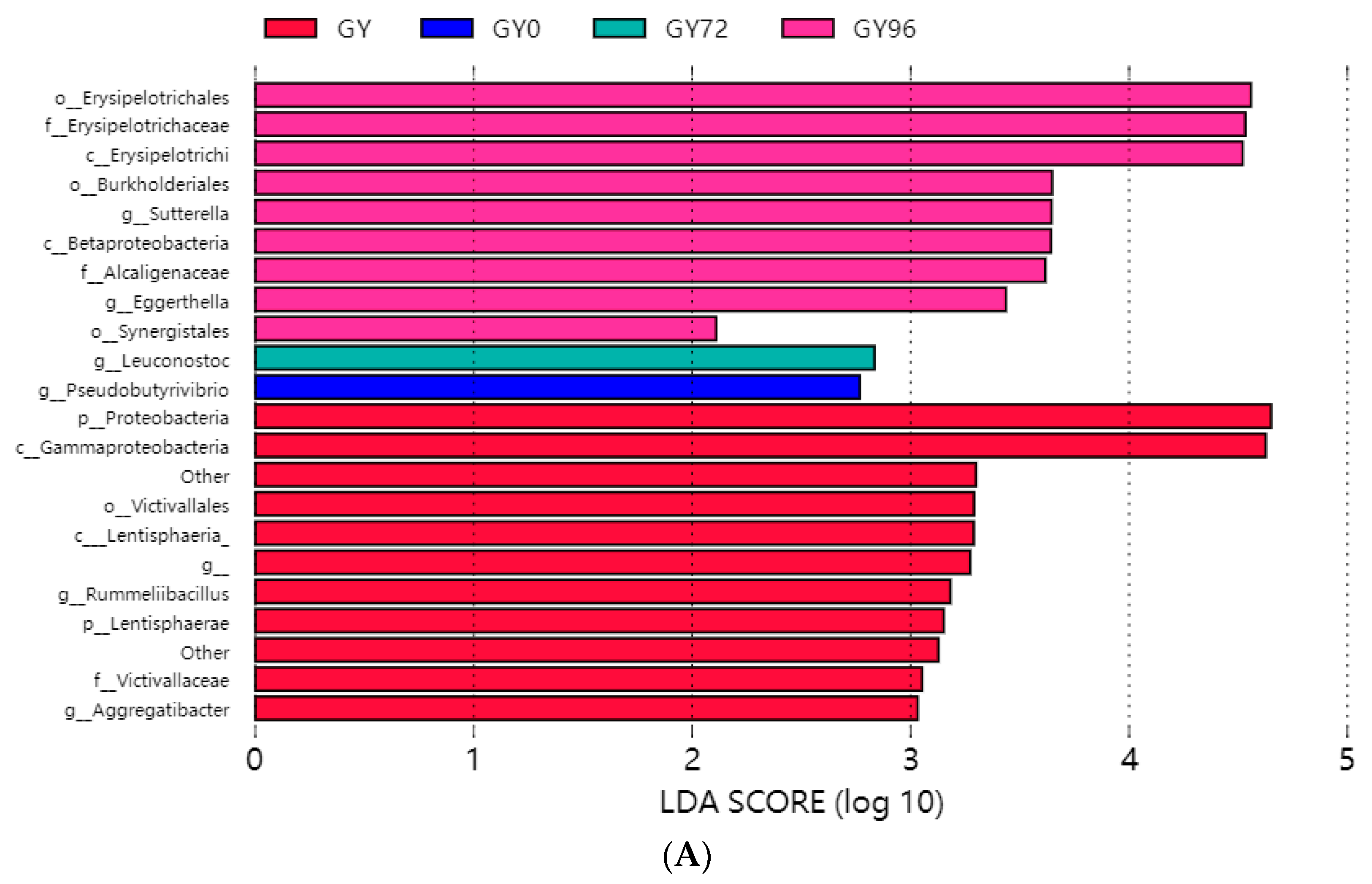
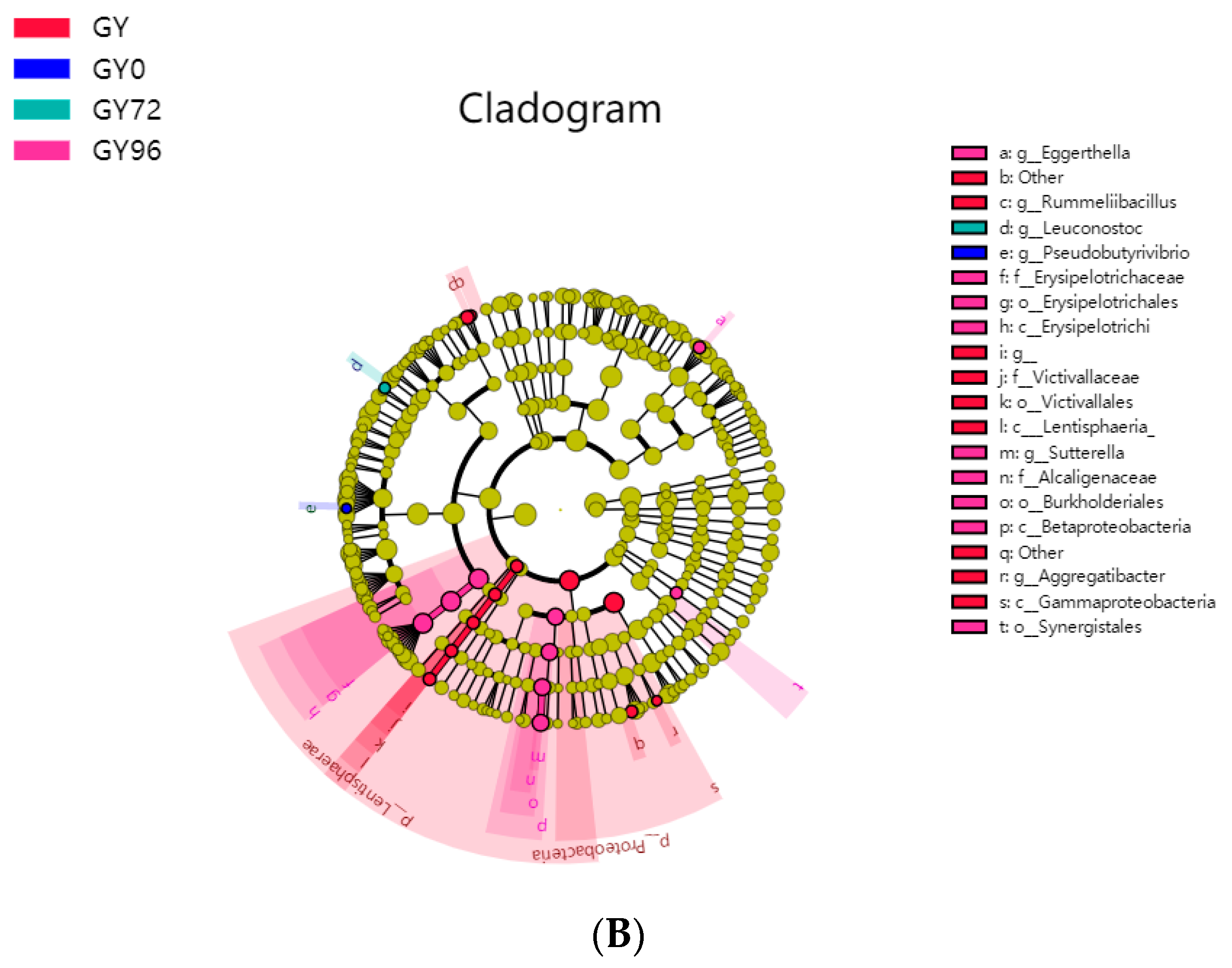
2.6. Analysis of Microbial Flora Composition in Ileum of Piglets under Different Taxonomic Levels
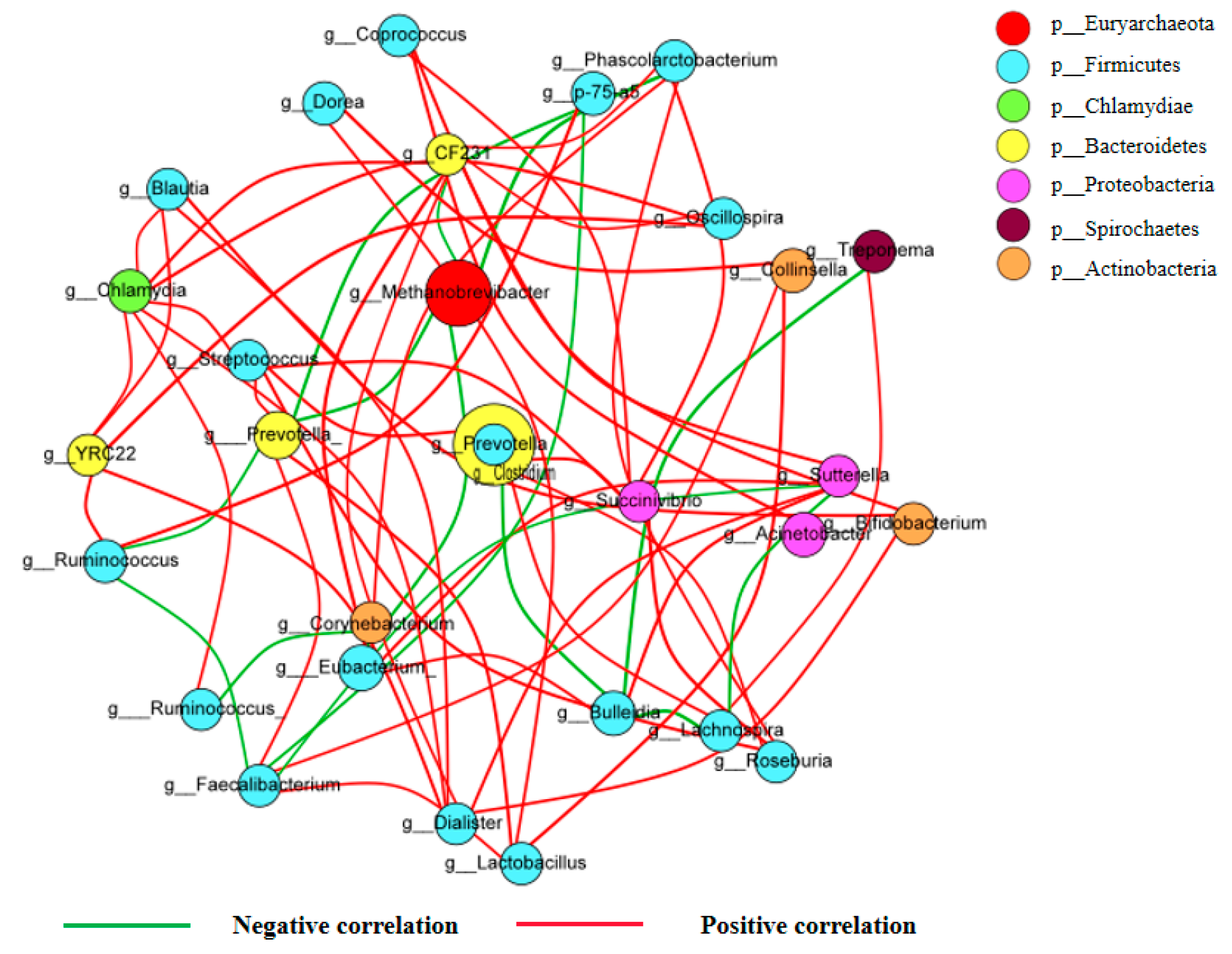
2.7. Correlations of the Different Dominant Bacterial Communities
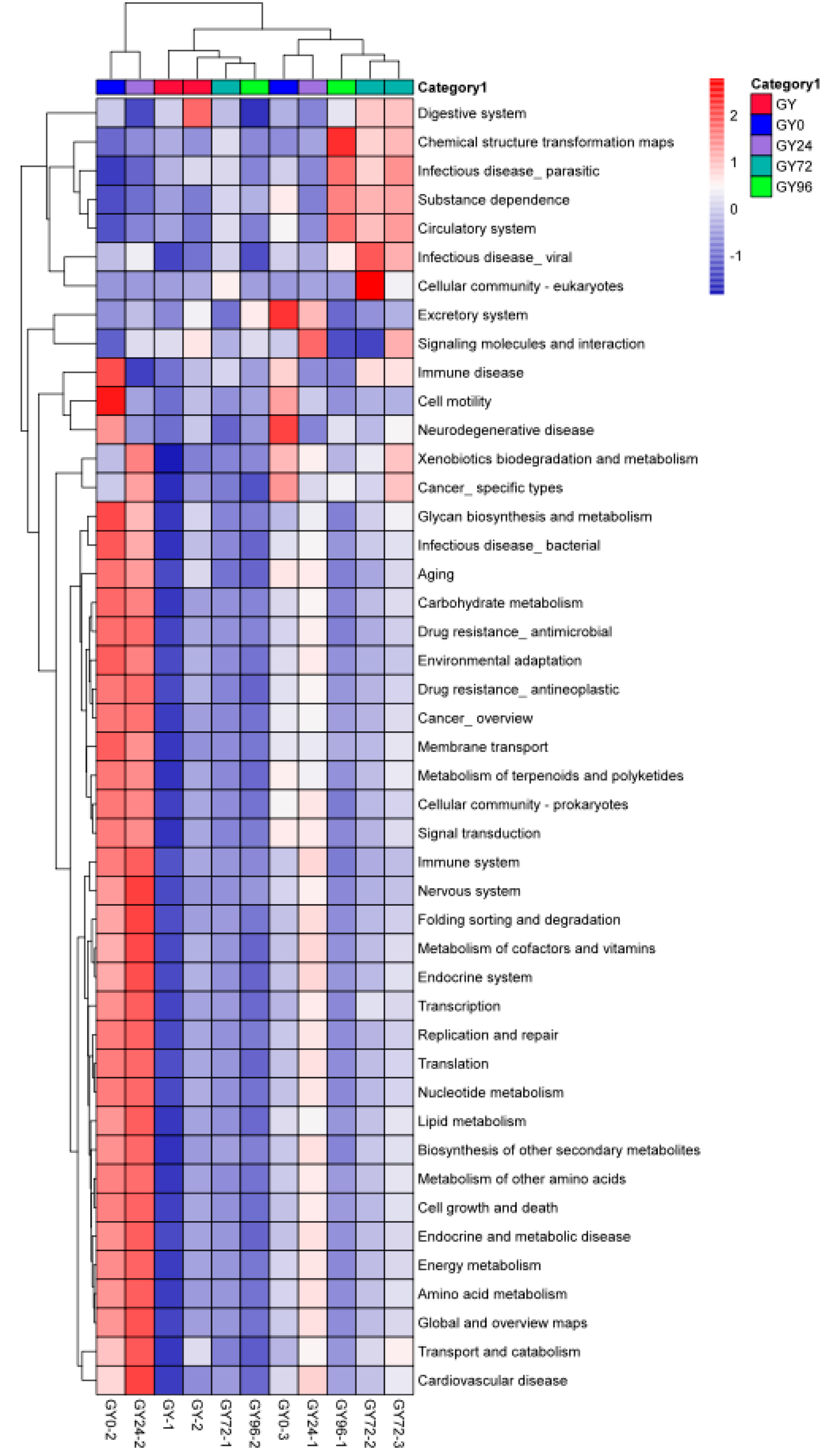
2.8. Predicted Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Salmonella Isolates
4.3. Animals
4.4. Establishment of COWT
4.5. Model of Salmonella Infection
4.6. PD Study In Vitro and Ex Vivo
4.7. Samples Collection for PK Study
4.8. Samples Analysis for PK
4.9. PK and PD Analyses, Dosage Prediction and Establishment of COPD
4.10. Infection Model and Clinical Trials
4.11. Statistical Analysis for Establishment of COCL
4.12. Samples Collection of Gut Flora 16S rRNA Gene Sequencing
4.13. 16S rRNA Gene Sequencing
4.14. Analysis of Microbial Diversity
4.15. Microbial Taxonomic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Garrido, V.; Sánchez, S.; Román, B.S.; Fraile, L.; Migura, L.; Grilló, M.-J. Salmonella Infection in Mesenteric Lymph Nodes of Breeding Sows. Foodborne Pathog. Dis. 2019, 17, 411–417. [Google Scholar] [CrossRef] [PubMed]
- Besser, J.M. Salmonella epidemiology: A whirlwind of change. Food Microbiol. 2018, 71, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Forge, A.; Schacht, J. Aminoglycoside antibiotics. Audiol. Neurotol. 2004, 5, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Shi, K.; Caldwell, S.J.; Fong, D.H.; Berghuis, A.M. Prospects for circumventing aminoglycoside kinase mediated antibiotic resistance. Front. Cell. Infect. Microbiol. 2013, 3, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Connor, S.; Lam, L.K.T.; Jones, N.D.; Chaney, M.O. Apramycin, a unique aminocyclitol antibiotic. J. Org. Chem. 1976, 41, 2087–2092. [Google Scholar] [CrossRef]
- Ramirez, M.S.; Tolmasky, M.E. Aminoglycoside modifying enzymes. Drug Resist. Updat. 2010, 13, 151–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antunes, E.D.A.B.; Lourenço, F.R.; Pinto, T.D.J.A. Determination of apramycin in oral soluble powder by a HPLC method using pre-column derivatization with o-phthalaldehyde and UV detection. Braz. J. Pharm. Sci. 2011, 47, 261–268. [Google Scholar] [CrossRef] [Green Version]
- Bischoff, K.; Edrington, T.; Callaway, T.; Genovese, K.; Nisbet, D. Characterization of antimicrobial resistant Salmonella Kinshasa from dairy calves in Texas. Lett. Appl. Microbiol. 2004, 38, 140–145. [Google Scholar] [CrossRef]
- Choi, M.-J.; Lim, S.-K.; Nam, H.-M.; Kim, A.-R.; Jung, S.-C.; Kim, M.-N. Apramycin and gentamicin resistances in indicator and clinical escherichia coli isolates from farm animals in Korea. Foodborne Pathog. Dis. 2011, 8, 119–123. [Google Scholar] [CrossRef]
- Yates, C.M.; Pearce, M.C.; Woolhouse, M.E.J.; Amyes, S.G.B. High frequency transfer and horizontal spread of apramycin resistance in calf faecal Escherichia coli. J. Antimicrob. Chemother. 2004, 54, 534–537. [Google Scholar] [CrossRef] [Green Version]
- Jensen, V.F.; Jakobsen, L.; Emborg, H.-D.; Seyfarth, A.M.; Hammerum, A.M. Correlation between apramycin and gentamicin use in pigs and an increasing reservoir of gentamicin-resistant Escherichia coli. J. Antimicrob. Chemother. 2006, 58, 101–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toutain, P.-L.; Bousquet-Melou, A.; Damborg, P.; Ferran, A.A.; Mevius, D.; Pelligand, L.; Veldman, K.T.; Lees, P. En route towards European clinical breakpoints for veterinary antimicrobial susceptibility testing: A position paper explaining the vetcast approach. Front. Microbiol. 2017, 8, 2344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuetz, A.N. Role of bacterial and fungal epidemiological cutoff values for antimicrobial susceptibility testing in the microbiology laboratory. Clin. Microbiol. Newsl. 2020, 42, 9–17. [Google Scholar] [CrossRef]
- Turnidge, J.D.; Martinez, M.N. Proposed method for estimating clinical cut-off (CO CL ) values: An attempt to address challenges encountered when setting clinical breakpoints for veterinary antimicrobial agents. Vet. J. 2017, 228, 33–37. [Google Scholar] [CrossRef]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef]
- Ashida, H.; Ogawa, M.; Kim, M.; Mimuro, H.; Sasakawa, C. Bacteria and host interactions in the gut epithelial barrier. Nat. Chem. Biol. 2012, 8, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Bauer, E.; Williams, B.A.; Smidt, H.; Verstegen, M.W.; Mosenthin, R. Influence of the gastrointestinal microbiota on development of the immune system in young animals. Curr. Issues Intest. Microbiol. 2006, 7, 35–42. [Google Scholar]
- Sun, J.; Du, L.; Li, X.; Zhong, H.; Ding, Y.; Liu, Z.; Ge, L. Identification of the core bacteria in rectums of diarrheic and non-diarrheic piglets. Sci. Rep. 2019, 9, 18675. [Google Scholar] [CrossRef] [PubMed]
- Kang, A.D.; Smith, K.P.; Eliopoulos, G.M.; Berg, A.H.; McCoy, C.; Kirby, J.E. In vitro apramycin activity against multidrug-resistant Acinetobacter baumannii and Pseudomonas aeruginosa. Diagn. Microbiol. Infect. Dis. 2017, 88, 188–191. [Google Scholar] [CrossRef]
- Randall, L.; Ridley, A.; Lemma, F.; Hale, C.; Davies, R. In vitro investigations into the use of antimicrobials in combination to maintain efficacy of fluoroquinolones in poultry. Res. Vet. Sci. 2016, 108, 47–53. [Google Scholar] [CrossRef]
- Tian, E.; Muhammad, I.; Hu, W.; Wu, Z.; Li, R.; Lu, X.; Chen, C.; Li, J. Tentative epidemiologic cut-off value and resistant characteristic detection of apramycin against Escherichia coli from chickens. FEMS Microbiol. Lett. 2019, 16, 16. [Google Scholar] [CrossRef] [PubMed]
- Theuretzbacher, U. Tissue penetration of antibacterial agents: How should this be incorporated into pharmacodynamic analyses? Curr. Opin. Pharmacol. 2007, 7, 498–504. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Lepak, A.J.; Andes, D.R. Animal models in the pharmacokinetic/pharmacodynamic evaluation of antimicrobial agents. Bioorg. Med. Chem. 2016, 24, 6390–6400. [Google Scholar] [CrossRef] [PubMed]
- Dai, C.; Zhao, T.; Yang, X.; Xiao, X.; Velkov, T.; Tang, S. Pharmacokinetics and relative bioavailability of an oral amoxicillin-apramycin combination in pigs. PLoS ONE 2017, 12, e0176149. [Google Scholar] [CrossRef]
- Shang, R.F.; Hu, Z.Y.; Zhang, X.G.; Li, S.H.; Shi, Y.B.; Xu, Z.Z.; Song, Z.S. Pharmacokinetics and bioavailability of Apramycin sulfate by oral administration in pigs. Chin. J. Vet. Med. 2004, 40, 3–5. [Google Scholar]
- Afifi, N.; Ramadan, A. Kinetic disposition, systemic bioavailability and tissue distribution of apramycin in broiler chickens. Res. Vet. Sci. 1997, 62, 249–252. [Google Scholar] [CrossRef]
- Dorey, L.; Pelligand, L.; Lees, P. Prediction of marbofloxacin dosage for the pig pneumonia pathogens Actinobacillus pleuropneumoniae and Pasteurella multocida by pharmacokinetic/pharmacodynamic modelling. BMC Vet. Res. 2017, 13, 209. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Huang, A.; Luo, X.; Zhang, P.; Huang, L.; Wang, X.; Mi, K.; Fang, S.; Huang, X.; Li, J.; et al. Exploration of Clinical Breakpoint of Danofloxacin for Glaesserella parasuis in Plasma and in PELF. Antibiotics 2021, 10, 808. [Google Scholar] [CrossRef]
- Looft, T.; Johnson, T.A.; Allen, H.K.; Bayles, D.O.; Alt, D.P.; Stedtfeld, R.D.; Sul, W.J.; Stedtfeld, T.M.; Chai, B.; Cole, J.R.; et al. In-feed antibiotic effects on the swine intestinal microbiome. Proc. Natl. Acad. Sci. USA 2012, 109, 1691–1696. [Google Scholar] [CrossRef] [Green Version]
- Shin, N.-R.; Whon, T.W.; Bae, J.-W. Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef]
- Ferrer, M.; Méndez-García, C.; Rojo, D.; Barbas, C.; Moya, A. Antibiotic use and microbiome function. Biochem. Pharmacol. 2016, 134, 114–126. [Google Scholar] [CrossRef]
- Li, Y.; Zhu, Y.; Wei, H.; Chen, Y.; Shang, H. Study on the diversity and function of gut microbiota in pigs following long-term antibiotic and antibiotic-free breeding. Curr. Microbiol. 2020, 77, 4114–4128. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Huang, S.; Jiang, L.; Dai, Z.; Li, T.; Han, D.; Wang, J. Characterization of the early life microbiota development and predominant lactobacillus species at distinct gut segments of low- and normal-birth-weight piglets. Front. Microbiol. 2019, 10, 797. [Google Scholar] [CrossRef]
- Rodríguez-Hernández, R.; Bernal, J.; Cifuentes, J.; Fandiño, L.; Herrera-Sánchez, M.; Rondón-Barragán, I.; Garcia, N.V. Prevalence and molecular characterization of Salmonella isolated from broiler farms at the Tolima Region—Colombia. Animals 2021, 11, 970. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing: 28th Informational Supplement; CLSI document M100-S28; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Yang, Y.; Zhang, Y.; Li, J.; Cheng, P.; Xiao, T.; Muhammad, I.; Yu, H.; Liu, R.; Zhang, X. Susceptibility breakpoint for Danofloxacin against swine Escherichia coli. BMC Vet. Res. 2019, 15, 51. [Google Scholar] [CrossRef] [PubMed]
- Turnidge, J.; Kahlmeter, G.; Kronvall, G. Statistical characterisation of bacterial wild-type MIC value distributions and the determination of epidemiological cut-off values. Clin. Microbiol. Infect. 2010, 12, 418–425. [Google Scholar] [CrossRef]
- Wang, F.; Mi, K.; Ahmad, I.; Xie, S.; Hussain, H.I.; Yuan, Z.; Dai, M.; Huang, L. Antibacterial activity of cyadox against Clostridium perfringens in broilers and a dosage regimen design based on pharmacokinetic-pharmacodynamic modeling. Microb. Pathog. 2020, 141, 103981. [Google Scholar] [CrossRef]
- Zhou, Y.-F.; Bu, M.-X.; Liu, P.; Sun, J.; Liu, Y.-H.; Liao, X.-P. Epidemiological and PK/PD cutoff values determination and PK/PD-based dose assessment of gamithromycin against Haemophilus parasuis in piglets. BMC Vet. Res. 2020, 16, 81. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Liu, Y.; Liu, J.; Wang, H.; Guo, Y.; Du, M.; Cai, C.; Zhao, Y.; Lu, C.; Guo, X.; et al. Composition of the fecal microbiota of piglets at various growth stages. Front. Vet. Sci. 2021, 8, 661–671. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Toth, F.; Frank, N.; Elliott, S.B.; Geor, R.; Boston, R.C. Effects of an intravenous endotoxin challenge on glucose and insulin dynamics in horses. Am. J. Vet. Res. 2004, 69, 82–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, P.; Shi, Y.; Liu, P.; Yang, Y.; Zhou, C.; Li, G.; Luo, J.; Zhang, C.; Cao, H.; Hu, G.; et al. 16S rRNA gene sequencing reveals an altered composition of the gut microbiota in chickens infected with a nephropathogenic infectious bronchitis virus. Sci. Rep. 2020, 10, 3556. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [Green Version]
- Deng, Y.; Jiang, Y.-H.; Yang, Y.; He, Z.; Luo, F.; Zhou, J. Molecular ecological network analyses. BMC Bioinform. 2012, 13, 113. [Google Scholar] [CrossRef] [Green Version]
- Parks, D.H.; Beiko, R.G. Identifying biologically relevant differences between metagenomic communities. Bioinformatics 2010, 26, 715–721. [Google Scholar] [CrossRef] [Green Version]

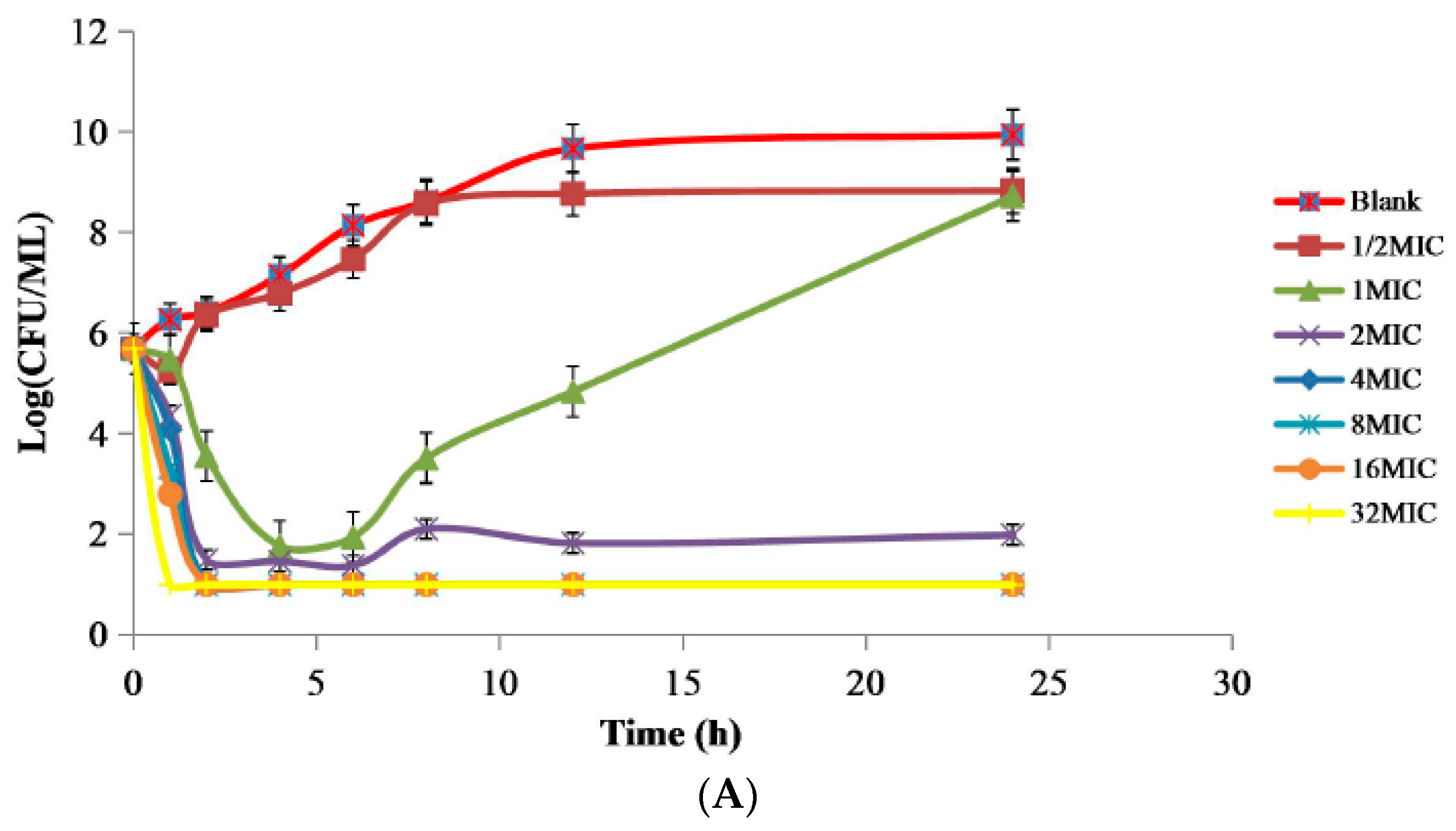

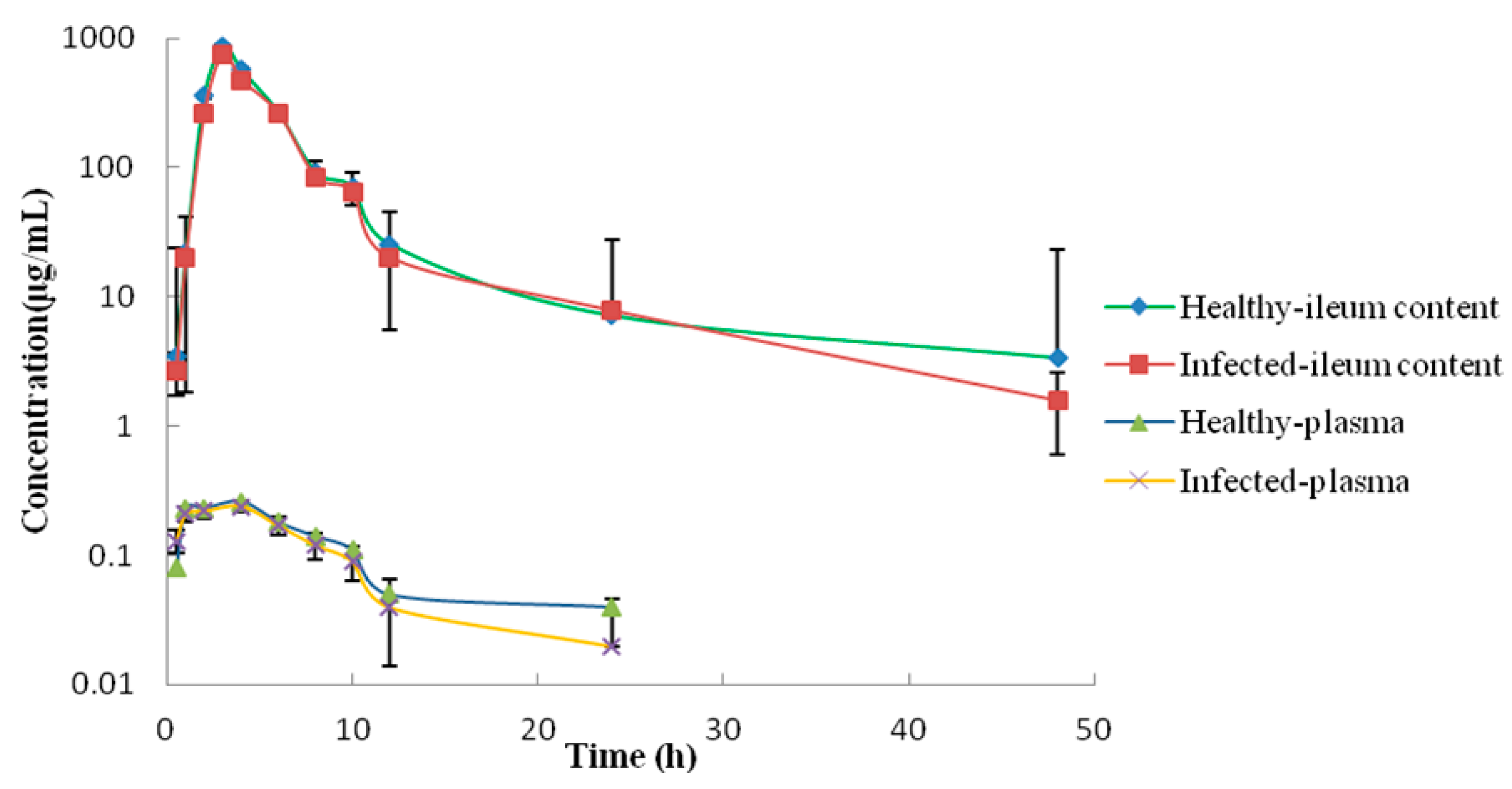

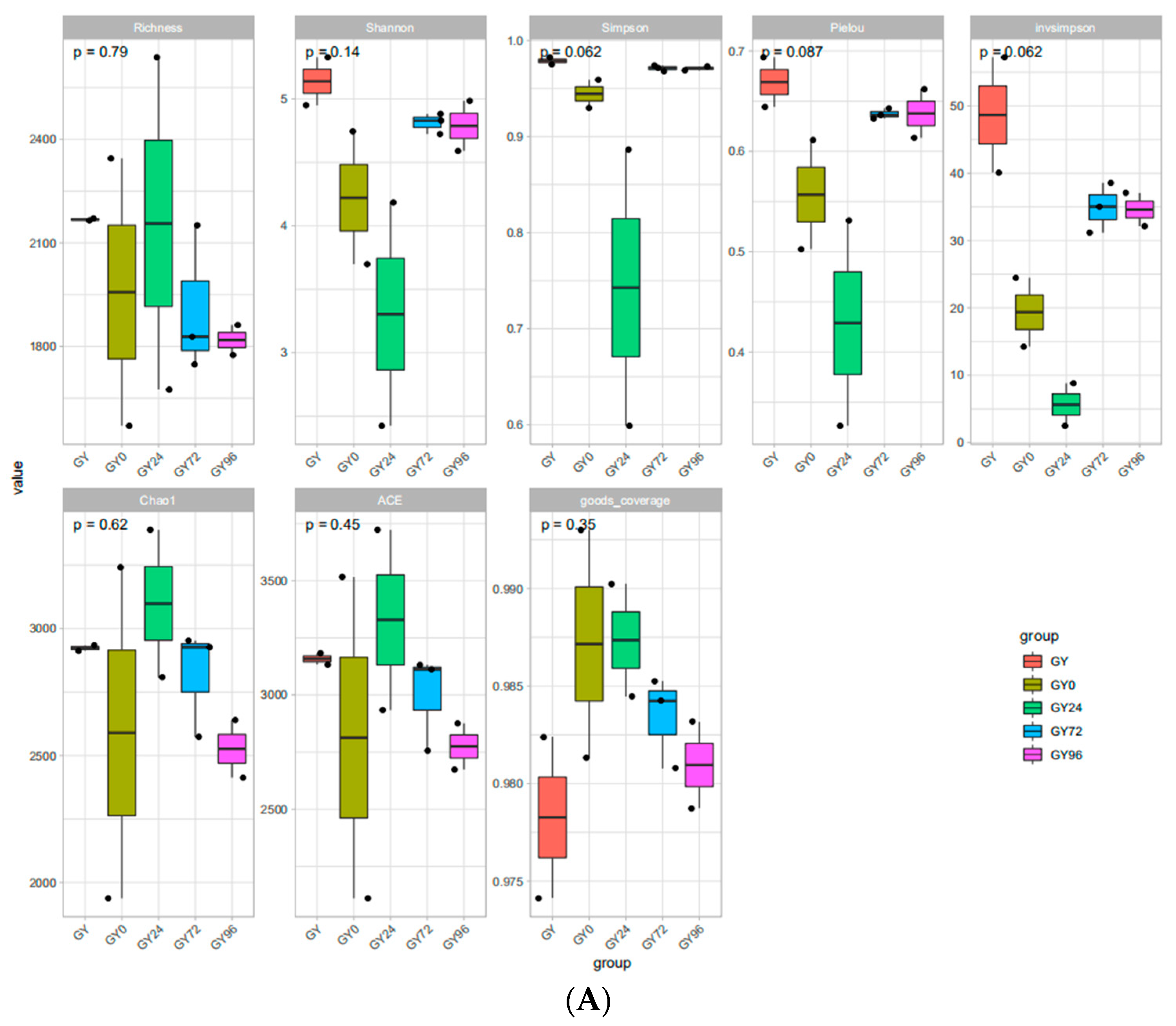
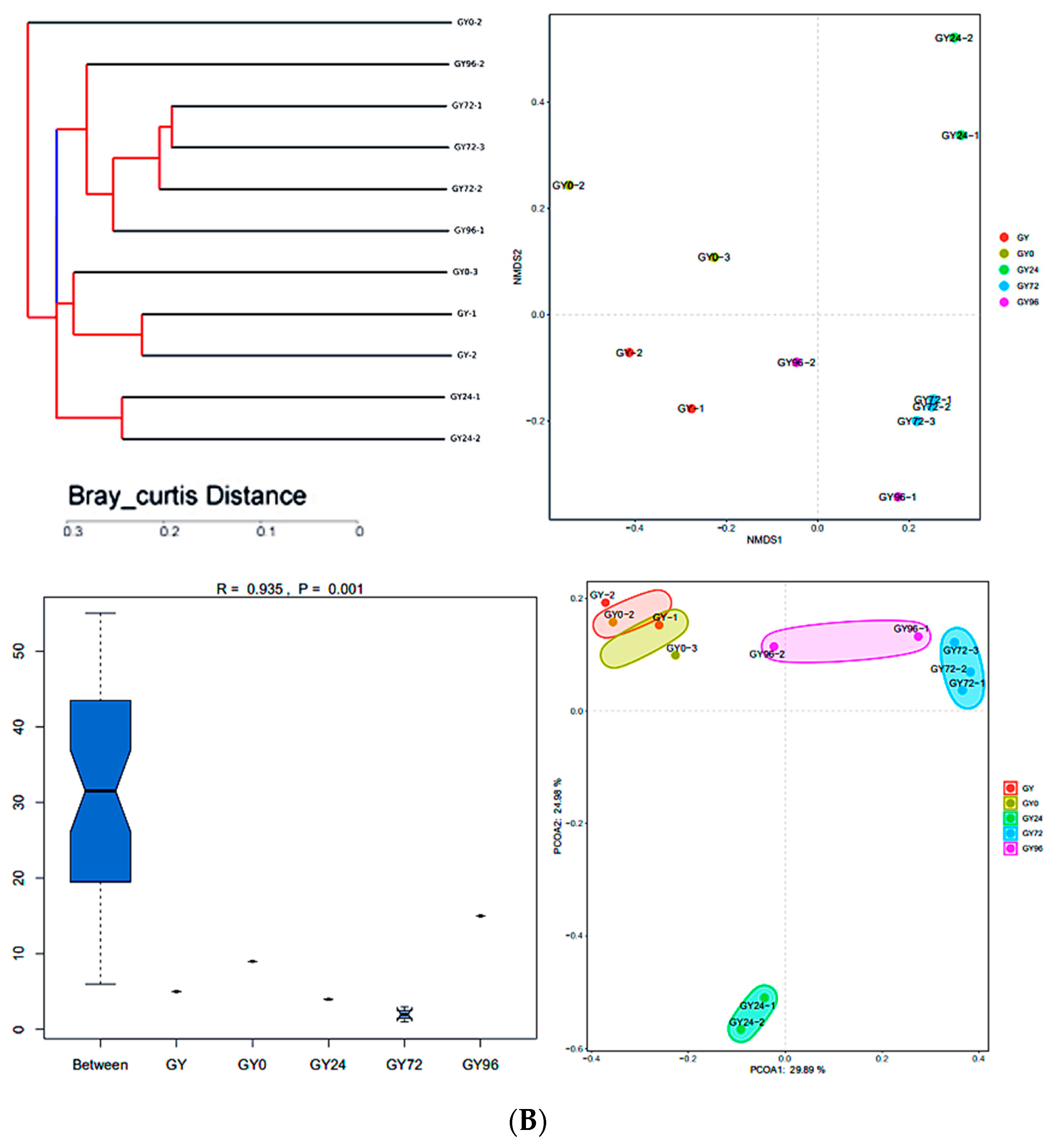

| Parameter | Units | Plasma | Ileum Contents | ||
|---|---|---|---|---|---|
| Healthy Group | Infected Group | Healthy Group | Infected Group | ||
| Cmax | μg/mL | 0.26 ± 0.01 | 0.24 ± 0.01 | 863.4 ± 18.2 | 750.5 ± 24.82 |
| AUC0–24h | h·μg/mL | 4.12 ± 0.76 | 3.41 ± 0.48 | 3287.25 ± 23.41 | 2855.83 ± 19.81 |
| AUMC0–24h | h2·μg/mL | 63.68 ± 2.12 | 45.13 ± 1.71 | 19,965.83 ± 151.2 | 17,421.25 ± 132.5 |
| Tmax | H | 4.0 ± 0.0 | 4.0 ± 0.0 | 3.0 ± 0.0 | 3.0 ± 0.0 |
| T1/2 | H | 20.00 ± 1.02 | 15.77 ± 1.93 | 8.74 ± 1.01 | 7.83 ± 0.78 |
| MRT0–24h | H | 15.47 ± 0.92 | 13.22 ± 1.02 | 6.07 ± 0.24 | 6.10 ± 0.42 |
| CL/F | mL/h/kg | 5.69 ± 0.01 | 7.76 ± 0.02 | 9.0 ± 0.02 | 10.04 ± 0.01 |
| Parameter | Unit | Value | |
|---|---|---|---|
| Healthy Group | Infected Group | ||
| Emax | Log10 CFU/mL | 4.25 | 3.48 |
| E0 | h | −5.69 | −5.33 |
| EC50 | Log10 CFU/mL | 47.16 | 49.37 |
| N | - | 27.25 | 6.10 |
| Emax-E0 | Log10 CFU/mL | 9.94 | 8.81 |
| AUC0–24h/MIC (E = 0) | h | 46.65 | 46 |
| AUC0–24h/MIC (E = −3) | h | 48.90 | 58.38 |
| AUC0–24h/MIC (E = −4) | h | 49.98 | 65.53 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, X.; Gu, Y.; Guo, J.; Huang, L.; Cheng, G.; Peng, D.; Hao, H. Clinical Breakpoint of Apramycin to Swine Salmonella and Its Effect on Ileum Flora. Int. J. Mol. Sci. 2022, 23, 1424. https://doi.org/10.3390/ijms23031424
Dai X, Gu Y, Guo J, Huang L, Cheng G, Peng D, Hao H. Clinical Breakpoint of Apramycin to Swine Salmonella and Its Effect on Ileum Flora. International Journal of Molecular Sciences. 2022; 23(3):1424. https://doi.org/10.3390/ijms23031424
Chicago/Turabian StyleDai, Xinyu, Yufeng Gu, Jinli Guo, Lingli Huang, Guyue Cheng, Dapeng Peng, and Haihong Hao. 2022. "Clinical Breakpoint of Apramycin to Swine Salmonella and Its Effect on Ileum Flora" International Journal of Molecular Sciences 23, no. 3: 1424. https://doi.org/10.3390/ijms23031424





