The Discovery and Function of Filaggrin
Abstract
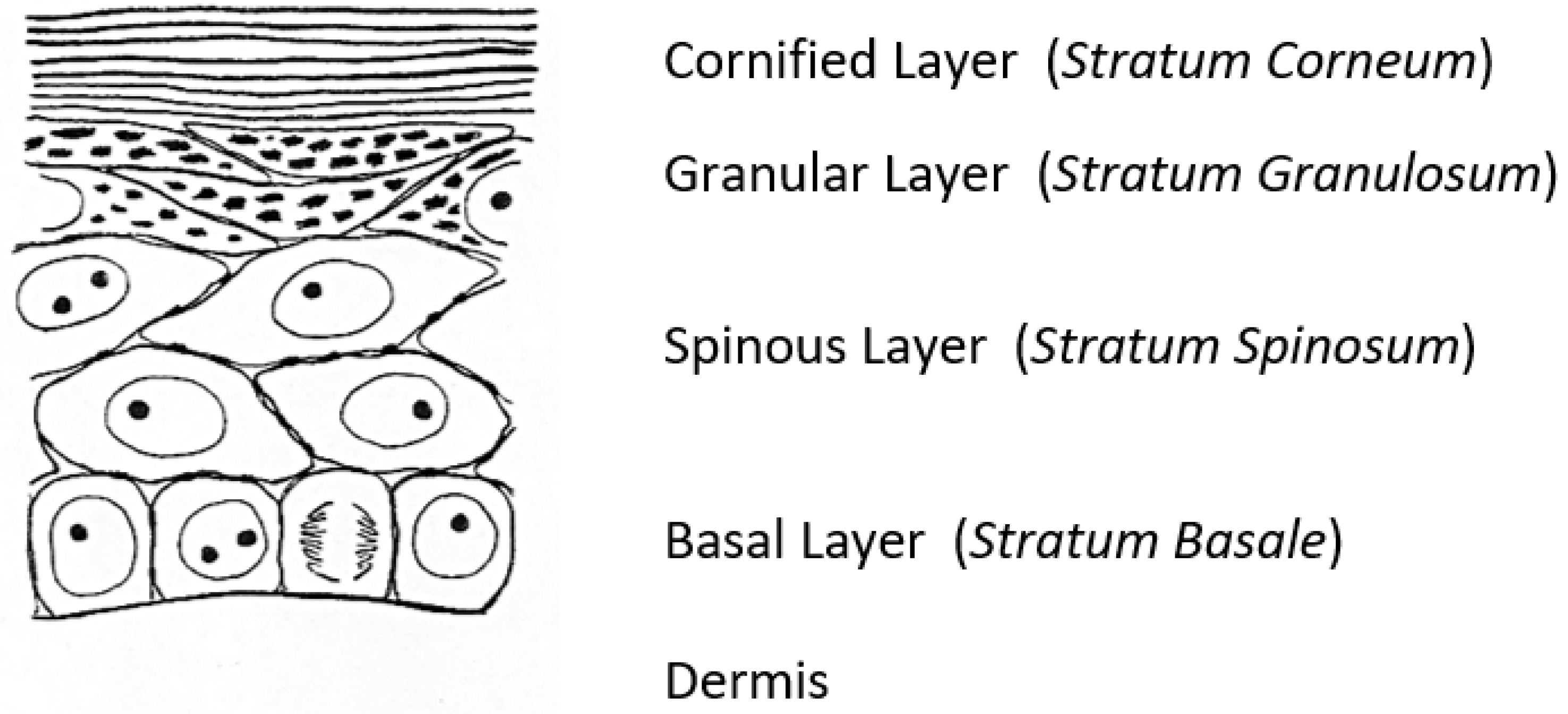
1. Keratohyalin Granules and Histidine
2. Discovery of the ‘Histidine-Rich’ Protein
3. Discovery of the Precursor, Profilaggrin
4. The Short Life of Filaggrin
5. Mutations That Cause Loss of Filaggrin
6. Keratinocyte to Corneocyte Transition
7. Another Role for Transglutaminase?
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Holbrook, K.A. Biologic structure and function: Perspectives on morphologic approaches to the study of the granular layer keratinocyte. J. Investig. Dermatol. 1989, 92, s84–s104. [Google Scholar] [CrossRef]
- Rothman, S. Keratinization in historical perspective. In The Epidermis; Montagna, W., Lobitz, W.C., Jr., Eds.; Academic Press: New York, NY, USA, 1964; pp. 1–14. [Google Scholar]
- Burgdorf, W.H.C.; Bickers, D.R. The scientific legacy of Stephen Rothman. J. Investig. Dermatol. 2015, 135, 954–959. [Google Scholar] [CrossRef] [PubMed]
- Hoober, J.K. The Differential Incorporation of Amino Acids (In Vivo) into Proteins of the Newborn Rat Epidermis. Ph.D. Thesis, University of Michigan, Ann Arbor, MI, USA, 1965. [Google Scholar]
- Reaven, E.P.; Cox, A.J., Jr. The histochemical localization of histidine in the human epidermis and its relationship to zinc binding. J. Histochem. Cytochem. 1963, 11, 782–790. [Google Scholar] [CrossRef]
- Reaven, E.P.; Cox, A.J. Histidine and keratinization. J. Investig. Dermatol. 1965, 45, 422–431. [Google Scholar] [CrossRef] [PubMed]
- Reaven, E.P.; Cox, A.J. Binding of zinc by the transitional layer of the epidermis. J. Investig. Dermatol. 1962, 39, 133–137. [Google Scholar]
- Pauly, H. Über die Konstitution des Histidins. I. Mitteilung. Z. Physiol. Chem. Hoppe-Seyler 1904, 42, 508–518. [Google Scholar] [CrossRef]
- Fukuyama, K.; Bernstein, I. Autoradiographic Studies of the Incorporation of Thymidine-H3into Deoxyribonucleic Acid in the Skin of Young Rats1. J. Investig. Dermatol. 1961, 36, 321–326. [Google Scholar] [CrossRef]
- Fukuyama, K.; Bernstein, I.A. Site of synthesis of ribonucleic acid in mammalian epidermis. J. Investig. Dermatol. 1963, 41, 47–52. [Google Scholar] [CrossRef][Green Version]
- Fukuyama, K.; Nakamura, T.; Bernstein, I.A. Differentially localized incorporation of amino acids in relation to epidermal keratinization in the newborn rat. Anat. Rec. 1965, 152, 525–535. [Google Scholar] [CrossRef]
- Fukuyama, K.; Epstein, W.L. Ultrastructural autoradiographic studies of keratohyalin granule formation. J. Investig. Dermatol. 1967, 49, 595–604. [Google Scholar] [CrossRef]
- Fukuyama, K.; Marshburn, I.; Epstein, W.L. Histidine-rich protein in developing rat epidermis. Dev. Biol. 1981, 81, 201–207. [Google Scholar] [CrossRef]
- Bernstein, I.A.; Chakrabarti, S.G.; Kumaroo, K.K.; Sibrack, L.A. Synthesis of protein in the mammalian epidermis. J. Investig. Dermatol. 1970, 55, 291–302. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hoober, J.K.; Bernstein, I.A. Studies on the mechanism of the localized incorporation of glycine-H3 in newborn rat epidermis. Fed. Proc. 1963, 22, 238. [Google Scholar]
- Hoober, J.K.; Bernstein, I.A. Protein synthesis related to epidermal differentiation. Proc. Natl. Acad. Sci. USA 1966, 56, 594–601. [Google Scholar] [CrossRef]
- Gumucio, J.; Feldkamp, C.; Bernstein, I.A. Studies on localization of “histidine-rich” peptide material present in epidermis of the newborn rat. J. Investig. Dermatol. 1967, 49, 545–551. [Google Scholar] [CrossRef] [PubMed]
- Voorhees, J.J.; Siba, M.D.; Chakrabarti, G.; Bernstein, I.A. The metabolism of “histidine-rich” protein in normal and psoriatic keratinization. J. Investig. Dermatol. 1968, 51, 344–354. [Google Scholar] [CrossRef][Green Version]
- Ball, R.D.; Walker, G.K.; Bernstein, I.A. Histidine-rich proteins as molecular markers of epidermal differentiation. J. Biol. Chem. 1978, 253, 5861–5868. [Google Scholar] [CrossRef]
- Ugel, A.R. Studies on isolated aggregating oligoribonucleoproteins of the epidermis with histochemical and morphological characteristics of keratohyalin. J. Cell Biol. 1971, 49, 405–422. [Google Scholar] [CrossRef]
- Ugel, A.R.; Idler, W. Further characterization of bovine keratohyalin. J. Cell Biol. 1972, 52, 453–464. [Google Scholar] [CrossRef]
- Sibrack, L.A.; Gray, R.; Bernstein, I. Localization of the Histidine-Rich Protein in Keratohyalin: A Morphologic and Macromolecular Marker in Epidermal Differentiation. J. Investig. Dermatol. 1974, 62, 394–405. [Google Scholar] [CrossRef][Green Version]
- Balmain, A.; Loehren, D.; Alonso, A.; Goerttler, K. Protein synthesis during fetal development of mouse epidermis. II. Biosynthesis of histidine-rich and cystine-rich proteins in vitro and in vivo. Develop. Biol. 1979, 73, 338–344. [Google Scholar] [CrossRef]
- Balmain, A.; Loehren, D.; Fischer, J.; Alonso, A. Protein synthesis during fetal development of mouse epidermis: I. The appearance of “histidine-rich protein”. Dev. Biol. 1977, 60, 442–452. [Google Scholar] [CrossRef]
- Brown, S.J.; McLean, W.I. One Remarkable Molecule: Filaggrin. J. Investig. Dermatol. 2012, 132, 751–762. [Google Scholar] [CrossRef] [PubMed]
- Dale, B.A. Purification and chaaracterization of a basic protein from the stratum corneum of mammalian epidermis. Biochem. Biophys. Acta 1977, 491, 193–204. [Google Scholar] [PubMed]
- Steinert, P.M.; Cantieri, J.S.; Teller, D.C.; Lonsdale-Eccles, J.D.; Dale, B.A. Characterization of a class of cationic proteins that specifically interact with intermediate filaments. Proc. Natl. Acad. Sci. USA 1981, 78, 4097–4101. [Google Scholar] [CrossRef] [PubMed]
- Harding, C.R.; Scott, I.R. Histidine-rich proteins (filaggrins): Structural and functional heterogeneity during epidermal differentiation. J. Mol. Biol. 1983, 170, 651–673. [Google Scholar] [CrossRef]
- Steinert, P.M.; Roop, D.R. Molecular and cellular biology of intermediate filaments. Ann. Rev. Biochem. 1988, 57, 593–625. [Google Scholar] [CrossRef]
- Fuchs, E.; Green, H. Changes in keratin gene expression during terminal differentiation of the keratinocyte. Cell 1980, 19, 1033–1042. [Google Scholar] [CrossRef]
- Meek, R.L.; Lonsdale-Eccles, J.D.; Dale, B.A. Epidermal filaggrin is synthesized on a large messenger ribonucleic acid as a high-molecular-weight precursor. Biochemistry 1983, 22, 4867–4871. [Google Scholar] [CrossRef]
- Lonsdale-Eccles, J.D.; Haugen, J.A.; Dale, B.A. A phosphorylated keratohyalin-derived precursor of epidermal stratum corneum basic protein. J. Biol. Chem. 1980, 255, 2235–2238. [Google Scholar] [CrossRef]
- Fleckman, P.; Dale, B.A.; Holbrook, K.A. Profilaggrin, a High-Molecular-Weight Precursor of Filaggrin in Human Epidermis and Cultured Keratinocytes. J. Investig. Dermatol. 1985, 85, 507–512. [Google Scholar] [CrossRef]
- Haydock, P.V.; Dale, B.A. The repetitive structure of the profilaggrin gene as demonstrated using epidermal profilaggrin cDNA. J. Biol. Chem. 1986, 261, 12520–12525. [Google Scholar] [CrossRef]
- Rothnagel, J.A.; Mehrel, T.; Idler, W.W.; Roop, D.R.; Steinert, P.M. The gene for mouse epidermal filaggrin precursor. J. Biol. Chem. 1987, 262, 15643–15648. [Google Scholar] [CrossRef]
- McKinley-Grant, L.J.; Idler, W.W.; Bernstein, I.A.; Parry, D.A.; Cannizzaro, L.; Croce, C.M.; Huebner, K.; Lessin, S.R.; Steinert, P.M. Characterization of a cDNA clone encoding human filaggrin and localization of the gene to chromosome region 1q21. Proc. Natl. Acad. Sci. USA 1989, 86, 4848–4852. [Google Scholar] [CrossRef]
- Resing, K.A.; Johnson, R.S.; Walsh, K.A. Characterization of protease processing sites during conversion of rat proflaggrin to filaggrin. Biochemistry 1993, 32, 10036–10045. [Google Scholar] [CrossRef]
- Gan, S.Q.; McBride, O.W.; Idler, W.W.; Markova, N.; Steinert, P.M. Organization, structure, and polymorphisms of the human profilaggrin gene. Biochemistry 1990, 29, 9432–9440. [Google Scholar] [CrossRef]
- Presland, R.B.; Haydock, P.V.; Fleckman, P.; Nirunsuksiri, W.; Dale, B.A. Characterization of the human epidermal profilaggrin gene. Genomic organization and identification of an S-100-like calcium binding domain at the amino terminus. J. Biol. Chem. 1992, 267, 23772–23781. [Google Scholar] [CrossRef]
- Markova, N.G.; Marekov, L.N.; Chipev, C.C.; Gan, S.-Q.; Idler, W.W.; Steinert, P.M. Profilaggrin is a major epidermal calcium-binding protein. Mol. Cell. Biol. 1993, 13, 613–625. [Google Scholar]
- Rothnagel, J.A.; Steinert, P.M. The structure of the gene for mouse filaggrin and a comparison of the repeating units. J. Biol. Chem. 1990, 265, 1862–1865. [Google Scholar] [CrossRef]
- Haydock, P.V.; Dale, B.A. Filaggrin, an Intermediate Filament-Associated Protein: Structural and Functional Implications from the Sequence of a cDNA from Rat. DNA Cell Biol. 1990, 9, 251–261. [Google Scholar] [CrossRef]
- Bilkle, D.D.; Zie, Z.; Tu, C.-L. Calcium regulation of keratinocyte differentiation. Expert Rev. Endocrinol. Metab. 2012, 7, 461–472. [Google Scholar] [CrossRef]
- Lee, S.E.; Lee, S.H. Skin barrier and calcium. Ann. Dermatol. 2018, 30, 265–275. [Google Scholar] [CrossRef]
- Lee, A.-Y. Molecular Mechanism of Epidermal Barrier Dysfunction as Primary Abnormalities. Int. J. Mol. Sci. 2020, 21, 1194. [Google Scholar] [CrossRef]
- Mauro, T.; Elias, P.; Cullander, C.; Bench, G.; Sidderas-Haddad, E.; Feingold, K. Acute Barrier Perturbation Abolishes the Ca2+ and K+ Gradients in Murine Epidermis: Quantitative Measurement Using PIXE. J. Investig. Dermatol. 1998, 111, 1198–1201. [Google Scholar] [CrossRef]
- Aho, S.; Harding, C.R.; Lee, J.-M.; Meldrum, H.; Bosko, C.A. Regulatory Role for the Profilaggrin N-Terminal Domain in Epidermal Homeostasis. J. Investig. Dermatol. 2012, 132, 2376–2385. [Google Scholar] [CrossRef]
- Sandilands, A.; Sutherland, C.; Irvine, A.D.; McLean, W.H.I. Filaggrin in the frontline: Role in skin barrier function and disease. J. Cell Sci. 2009, 122, 1285–1294. [Google Scholar] [CrossRef]
- Ishida-Yamamoto, A.; Senshu, T.; Eady, R.A.J.; Takahashi, H.; Shimizu, H.; Akiyama, M.; Iizukz, H. Sequential reorganization of cornified cell keratin filaments involving filaggrin-mediated compaction and keratin 1 dimination. J. Investig. Dermatol. 2002, 118, 282–287. [Google Scholar] [CrossRef]
- Presland, R.; Kimball, J.R.; Kautsky, M.B.; Lewis, S.P.; Lo, C.Y.; Dale, B.A. Evidence for Specific Proteolytic Cleavage of the N-Terminal Domain of Human Profilaggrin During Epidermal Differentiation. J. Investig. Dermatol. 1997, 108, 170–178. [Google Scholar] [CrossRef][Green Version]
- Pearton, D.; Nirunsuksiri, W.; Rehemtulla, A.; Lewis, S.P.; Presland, R.; Dale, B.A. Proprotein convertase expression and localization in epidermis: Evidence for multiple roles and substrates. Exp. Dermatol. 2001, 10, 193–203. [Google Scholar] [CrossRef]
- Matsui, T.; Miyamoto, K.; Kubo, A.; Kawasaki, H.; Ebihara, T.; Hata, K.; Tanahashi, S.; Ichinose, S.; Imoto, I.; Inazawa, J.; et al. SASPase regulates stratum corneum hydration through profilaggrin-to-filaggrin processing. EMBO Mol. Med. 2011, 3, 320–333. [Google Scholar] [CrossRef]
- Leyvraz, C.; Charles, R.-P.; Rubera, I.; Guitard, M.; Rolman, S.; Breiden, B.; Sandhoff, K.; Hummler, E. The epidermal barrier function is dependent upon the serine protease CAP1/Prss8. J. Cell Biol. 2005, 170, 487–496. [Google Scholar] [CrossRef]
- List, K.; Szabo, R.; Wertz, P.W.; Segre, J.; Haudenschild, C.C.; Kim, S.-Y.; Bugge, T.H. Loss of proteolytically processed filaggrin caused by epidermal deletion of Matriptase/MT-SP1. J. Cell Biol. 2003, 163, 901–910. [Google Scholar] [CrossRef]
- Hsu, C.-Y.; Henry, J.; Raymond, A.-A.; Méchin, M.-C.; Pendaries, V.; Nassar, D.; Simon, M.; Serre, G.; Paul, C.; Takahara, H.; et al. Deimination of human filaggrin-2 promotes its proteolysis by Calpain1. J. Biol. Chem. 2011, 286, 23222–23233. [Google Scholar] [CrossRef]
- Hoste, E.; Kemperman, P.; Devos, M.; Denecker, G.; Kezic, S.; Yau, N.; Gilbert, B.; Lippens, S.; De Groote, P.; Roelandt, R.; et al. Caspase-14 Is Required for Filaggrin Degradation to Natural Moisturizing Factors in the Skin. J. Investig. Dermatol. 2011, 131, 2233–2241. [Google Scholar] [CrossRef]
- Kamata, Y.; Taniguchi, A.; Yamamoto, M.; Nomura, J.; Ishihara, K.; Takahara, H.; Hibino, T.; Takeda, A. Neutral Cysteine Protease Bleomycin Hydrolase Is Essential for the Breakdown of Deiminated Filaggrin into Amino Acids. J. Biol. Chem. 2009, 284, 12829–12836. [Google Scholar] [CrossRef]
- Hanson, K.M.; Simon, J.D. Epidermal trans-urocanic acid and the UV-A-induced photoaging of the skin. Proc. Natl. Acad. Sci. USA 1998, 95, 10576–10578. [Google Scholar] [CrossRef]
- Gibbs, N.K.; Norval, M. Urocanic Acid in the Skin: A Mixed Blessing? J. Investig. Dermatol. 2011, 131, 14–17. [Google Scholar] [CrossRef]
- Rawlings, A.V.; Harding, C.R. Moisturization and skin barrier function. Dermatol. Ther. 2004, 17, 43–48. [Google Scholar] [CrossRef]
- Kezic, S.; Kemperman, P.M.; Koster, E.S.; de Jongh, C.M.; Thio, H.B.; Campbell, L.E.; Irvine, A.; McLean, W.H.I.; Puppels, G.J.; Caspers, P.J. Loss-of-Function Mutations in the Filaggrin Gene Lead to Reduced Level of Natural Moisturizing Factor in the Stratum Corneum. J. Investig. Dermatol. 2008, 128, 2117–2119. [Google Scholar] [CrossRef]
- Kezic, S.; Novak, N.; Jakasa, I.; Jungersted, J.M.; Simon, M.; Brandner, J.M.; Middelkamp-Hup, M.A.; Weidinger, S. Skin barrier in atopic dermatitis. Front. Biosci. 2014, 19, 542–556. [Google Scholar] [CrossRef]
- De Veer, S.J.; Furio, L.; Harris, J.M.; Hovnanian, A. Proteases: Common culprits in human skin disorders. Trends Mol. Med. 2014, 20, 166–178. [Google Scholar] [CrossRef]
- Smith, F.J.D.; Irvine, A.; Terron-Kwiatkowski, A.; Sandilands, A.; Campbell, L.E.; Zhao, Y.; Liao, H.; Evans, A.T.; Goudie, D.R.; Lewis-Jones, S.; et al. Loss-of-function mutations in the gene encoding filaggrin cause ichthyosis vulgaris. Nat. Genet. 2006, 38, 337–342. [Google Scholar] [CrossRef]
- Irvine, A.; McLean, W.H.I.; Leung, D.Y. Filaggrin Mutations Associated with Skin and Allergic Diseases. N. Engl. J. Med. 2011, 365, 1315–1327. [Google Scholar] [CrossRef]
- McLean, W.H.I. Filaggrin failure—From ichthyosis vulgaris to atopic eczema and beyond. Br. J. Dermatol. 2016, 175 (Suppl. S2), 4–7. [Google Scholar] [CrossRef]
- Salama, R.H.; Rasheed, Z.; Ahmed, A.A.; Bin Saif, G.A.; Elkholy, M.M.; El-Moniem, A.E.A.; Salem, T.; Zedan, K.; Al Robaee, A.A.; Alzolibani, A.A. Missense, silent, non-sense and frame-shift mutations in exon 3 of the filaggrin gene in patients with bronchial asthma, atopic dermatitis, allergic rhinitis and mixed atopy. Nucleosides Nucleotides Nucleic Acids 2021, 40, 357–367. [Google Scholar] [CrossRef]
- O’Regan, G.M.; Sandilands, A.; McLean, W.H.I.; Irvine, A.D. Filaggrin in atopic dermatitis. J. Allergy Clin. Immunol. 2008, 122, 689–693. [Google Scholar] [CrossRef]
- Osawa, R.; Akiyama, M.; Shimizu, H. Filaggrin Gene Defects and the Risk of Developing Allergic Disorders. Allergol. Int. 2011, 60, 1–9. [Google Scholar] [CrossRef]
- Rodríguez, E.; Illig, T.; Weidinger, S. Filaggrin loss-of-function mutations and association with allergic diseases. Pharmacogenomics 2008, 9, 399–413. [Google Scholar] [CrossRef]
- Bin, L.; Malley, C.; Taylor, P.; Boorgula, M.P.; Chavan, S.; Daya, M.; Mathias, M.; Shankar, G.; Rafaels, N.; Vergara, C.; et al. Whole genome sequencing identifies novel genetic mutations in patients with eczema herpeticum. Allergy 2021, 76, 2510–2523. [Google Scholar] [CrossRef]
- Esparza-Gordillo, J.; Matanovic, A.; Marenholz, I.; Bauerfeind, A.; Rohde, K.; Nemat, K.; Lee-Kirsch, M.-A.; Nordenskjöld, M.; Winge, M.C.G.; Keil, T.; et al. Maternal Filaggrin Mutations Increase the Risk of Atopic Dermatitis in Children: An Effect Independent of Mutation Inheritance. PLoS Genet. 2015, 11, e1005076. [Google Scholar] [CrossRef]
- Butler, D.C.; Simpson, E.; Guttman-Yassky, E.; Eichenfield, L.F.; Golant, A.K.; Koo, J.Y.M.; Armstrong, A.W.; Alexis, A.F.; Lio, P.A.; Marson, J.W.; et al. The atopic dermatitis spectrum disorder. Recognizing the clinical heterogeneity in patients with atopic related skin conditions in order to improve therapeutic decision-making and outcomes: An expert panel consensus statement. J. Dermatol. Treat. 2021, 33, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Shaw, T.E.; Currie, G.P.; Koudelka, C.W.; Simpson, E.L. Eczema Prevalence in the United States: Data from the 2003 National Survey of Children’s Health. J. Investig. Dermatol. 2011, 131, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Nutten, S. Atopic Dermatitis: Global Epidemiology and Risk Factors. Ann. Nutr. Metab. 2015, 66 (Suppl. S1), 8–16. [Google Scholar] [CrossRef] [PubMed]
- McAleer, M.A.; Jakasa, I.; Raj, N.; O’Donnell, C.P.F.; Lane, M.E.; Rawlings, A.V.; Irvine, A.D.; Voegeli, R.; McLean, W.H.I.; Kezic, S. Early-life regional and temporal variation in filaggrin-derived moisturizing factor, filaggrin-processing enzyme activity, corneocyte phenotypes and plasmin activity: Implications for atopic dermatitis. Br. J. Dermatol. 2018, 179, 431–441. [Google Scholar] [CrossRef]
- Werfel, T.; Allam, J.-P.; Biedermann, T.; Eyerich, K.; Gilles, S.; Guttman-Yassky, E.; Akdis, C.A.; Bieber, T.; Knol, E.; Lauener, R.; et al. Cellular and molecular immunologic mechanisms in patients with atopic dermatitis. J. Allergy Clin. Immunol. 2016, 138, 336–349. [Google Scholar] [CrossRef]
- Sözener, Z.C.; Cevhertas, L.; Nadeau, K.; Akdis, M.; Akdis, C.A. Environmental factors in epithelial barrier dysfunction. J. Allergy Clin. Immunol. 2020, 145, 1517–1528. [Google Scholar] [CrossRef]
- Peng, W.; Novak, N. Pathogenesis of atopic dermatitis. Clin. Exp. Allergy 2015, 45, 566–574. [Google Scholar] [CrossRef]
- Thyssen, J.P.; Kezic, S. Causes of epidermal filaggrin reduction and their role in the pathogenesis of atopic dermatitis. J. Allergy Clin. Immunol. 2014, 134, 792–799. [Google Scholar] [CrossRef]
- Dębińska, A. New Treatments for Atopic Dermatitis Targeting Skin Barrier Repair via the Regulation of FLG Expression. J. Clin. Med. 2021, 10, 2506. [Google Scholar] [CrossRef]
- Jin, H.; He, R.; Oyoshi, M.; Geha, R.S. Animal models of atopic dermatitis. J. Investig. Dermatol. 2009, 129, 31–40. [Google Scholar] [CrossRef]
- Martel, B.C.; Lovato, P.; Bäumer, W.; Olivry, T. Translational Animal Models of Atopic Dermatitis for Preclinical Studies. Yale J. Biol. Med. 2017, 90, 389–402. [Google Scholar] [PubMed]
- Stout, T.E.; McFarland, T.; Mitchell, J.C.; Appukuttan, B.; Stout, J.T. Recombinant Filaggrin Is Internalized and Processed to Correct Filaggrin Deficiency. J. Investig. Dermatol. 2014, 134, 423–429. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, H.; Nagao, K.; Kubo, A.; Hata, T.; Shimizu, A.; Mizuno, H.; Yamada, T.; Amagai, M. Altered stratum corneum barrier and enhanced percutaneous immune responses in filaggrin-null mice. J. Allergy Clin. Immunol. 2012, 129, 1538–1546.e6. [Google Scholar] [CrossRef]
- Van Ruissen, F.; Le, M.; Carroll, J.M.; van der Valk, P.G.; Schalkwijk, J. Differential Effects of Detergents on Keratinocyte Gene Expression. J. Investig. Dermatol. 1998, 110, 358–363. [Google Scholar] [CrossRef] [PubMed]
- Törma, H.; Lindberg, M.; Berne, B. Skin barrier disruption by sodium lauryl sulfate-exposure alters the expression of involucrin, transglutamnase 1, profilaggrin, and kallikreins during the repair phase in human skin in vivo. J. Investig. Dermatol. 2008, 128, 1212–1219. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.-C.; Chang, T.-M. Ceramide 1 and ceramide 3 act synergistically on skin hydration and the transepidermal water loss of sodium lauryl sulfate-irritated skin. Int. J. Dermatol. 2008, 47, 812–819. [Google Scholar] [CrossRef] [PubMed]
- Kasparek, P.; Ileninova, Z.; Zbodakova, O.; Kanchev, I.; Benada, O.; Chalupsky, K.; Sedlacek, R.; Brattsand, M.; Beck, I.M. KLK5 and KLK7 ablation fully rescues letahlity of Netherton syndrome-like phenotype. PLoS Genet. 2017, 13, e1006566. [Google Scholar] [CrossRef]
- Furio, L.; Pampalakis, G.; Michael, I.; Nagy, A.; Sotiropoulou, G.; Hovnanian, A. KLK5 Inactivation Reverses Cutaneous Hallmarks of Netherton Syndrome. PLoS Genet. 2015, 11, e1005389. [Google Scholar] [CrossRef]
- Miyai, M.; Matsumoto, Y.; Yamanishi, H.; Yamamoto-Tanaka, M.; Tsuboi, R.; Hibino, T. Keratinocyte-specific mesotrypsin contributes to the desquamation process via kallikrein activation and LEKT1 degradation. J. Investig. Dermatol. 2014, 134, 1665–1674. [Google Scholar] [CrossRef]
- Briot, A.; Deraison, C.; Lacroix, M.; Bonnart, C.; Robin, A.; Besson, C.; Hovnanian, A.; Dubus, P. Kalikrein 5 induces atopic dermatitis-like lesions through PAR2-mediated thymic stromal lymphopoietin expression in Netherton syndrome. J. Exp. Med. 2009, 206, 1135–1147. [Google Scholar] [CrossRef]
- Zhao, Y.; Terron-Kwiatkowski, A.; Liao, H.; Lee, S.P.; Allen, M.H.; Hull, P.R.; Campbell, L.E.; Trembath, R.C.; Capon, F.; Griffiths, C.E.; et al. Filaggrin Null Alleles Are Not Associated with Psoriasis. J. Investig. Dermatol. 2007, 127, 1878–1882. [Google Scholar] [CrossRef] [PubMed]
- Hüffmeier, U.; Traupe, H.; Oji, V.; Lascorz, J.; Ständer, M.; Lohmann, J.; Reis, A.; Wendler, J.; Burkhardt, H. Loss-of-function variants of the filaggrin gene are not major suseptibility factors for psoriasis vulgaris or psoriatic arthritis in German patients. J. Investig. Dermatol. 2007, 127, 1367–1370. [Google Scholar] [CrossRef] [PubMed]
- Kalinin, A.; Marekov, L.N.; Steinert, P.M. Assembly of the epidermal cornified cell envelope. J. Cell Sci. 2001, 114, 3069–3070. [Google Scholar] [CrossRef] [PubMed]
- Candi, E.; Schmidt, R.; Melino, G. The cornified envelope: A model of cell death in the skin. Nat. Rev. Mol. Cell Biol. 2005, 6, 328–340. [Google Scholar] [CrossRef] [PubMed]
- Eckert, R.L.; Green, H. Structure and evolution of the human involucrin gene. Cell 1986, 46, 583–589. [Google Scholar] [CrossRef]
- Eckert, R.L.; Sturniolo, M.T.; Broome, A.-M.; Ruse, M.; Rorke, E.A. Transglutaminase Function in Epidermis. J. Investig. Dermatol. 2005, 124, 481–492. [Google Scholar] [CrossRef]
- Simon, M.; Green, H. The glutamine residues reactive in transglutaminase-catalyzed cross-linking of involucrin. J. Biol. Chem. 1988, 263, 18093–18098. [Google Scholar] [CrossRef]
- Ishitsuka, Y.; Roop, D.R. Loricrin: Past, present, and future. Int. J. Mol. Sci. 2020, 21, 2271. [Google Scholar] [CrossRef]
- Steinert, P.M.; Marekov, L.N. The Proteins Elafin, Filaggrin, Keratin Intermediate Filaments, Loricrin, and Small Proline-rich Proteins 1 and 2 Are Isodipeptide Cross-linked Components of the Human Epidermal Cornified Cell Envelope. J. Biol. Chem. 1995, 270, 17702–17711. [Google Scholar] [CrossRef]
- Matsuki, M.; Yamashita, F.; Ishida-Yamamoto, A.; Yamada, K.; Kinoshita, C.; Fushiki, S.; Yamanishi, K.; Ueda, E.; Tabata, K.; Okabe, M.; et al. Defective stratum corneum and early neonatal death in mice lacking the gene for transaminase 1 (keratinocyte transglutamase). Proc. Natl. Acad. Sci. USA 1998, 95, 1044–1049. [Google Scholar] [CrossRef]
- Liedén, A.; Winge, M.C.G.; Sääf, A.; Kockum, I.; Ekelund, E.; Rodriguez, E.; Fölster-Holst, R.; Franke, A.; Illig, T.; Tengvall-Linder, M.; et al. Genetic Variation in the Epidermal Transglutaminase Genes Is Not Associated with Atopic Dermatitis. PLoS ONE 2012, 7, e49694. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Luo, Y.; Sun, J.; Liu, X.; Ling, S.; Xu, B.; Zhang, Y.; Liu, J.; Li, W.; Wang, B.; et al. Transglutaminase 3 Promotes Skin Inflammation in Atopic Dermatitis by Activating Monocyte-Derived Dendritic Cells via DC-SIGN. J. Investig. Dermatol. 2020, 140, 370–379. [Google Scholar] [CrossRef] [PubMed]
- Griffin, M.; Casadio, R.; Bergamini, C.M. Transglutaminases: Nature’s biological glues. Biochem. J. 2002, 368, 377–396. [Google Scholar] [CrossRef]
- Chou, C.-Y.; Streets, A.J.; Watson, P.F.; Huang, L.; Verderio, E.A.; Johnson, T.S. A Crucial Sequence for Transglutaminase Type 2 Extracellular Trafficking in Renal Tubular Epithelial Cells Lies in Its N-terminal β-Sandwich Domain. J. Biol. Chem. 2011, 286, 27825–27835. [Google Scholar] [CrossRef] [PubMed]
- Zemskov, E.A.; Mikhailenko, I.; Hsia, R.-C.; Zaritskaya, L.; Belkin, A.M. Unconventional Secretion of Tissue Transglutaminase Involves Phospholipid-Dependent Delivery into Recycling Endosomes. PLoS ONE 2011, 6, e19414. [Google Scholar] [CrossRef]
- Nurminskaya, M.V.; Belkin, A.M. Cellular Functions of Tissue Transglutaminase. Int. Rev. Cell. Mol. Biol. 2012, 294, 1–97. [Google Scholar] [CrossRef]
- Telei, D.; Griffin, M. Tissue transaminase (TG2)—A wound response enzyme. Front. Biosci. 2006, 11, 867–882. [Google Scholar]
- Stephens, P.; Grenard, P.; Aeschlimann, P.; Langley, M.; Blain, E.; Errington, R.; Kipling, D.; Thomas, D.; Aeschlimann, D. Crosslinking and G-protein functions of transglutaminase 2 contribute differentially to fibroblast wound healing responses. J. Cell Sci. 2004, 117, 3389–3403. [Google Scholar] [CrossRef]
- Haroon, Z.A.; Hettasch, J.M.; Lai, T.-S.; Dewhirst, M.W.; Greenberg, C.S. Tissue transglutaminase is expressed, active, and directly involved in rat dermal wound healing and angiogenesis. FASEB J. 1999, 13, 1787–1795. [Google Scholar] [CrossRef]
- Verderio, E.A.M.; Johnson, T.; Griffin, M. Tissue transglutaminase in normal and abnormal wound healing: Review article. Amino Acids 2004, 26, 387–404. [Google Scholar] [CrossRef]
- Johnson, T.S.; Scholfield, C.I.; Parry, J.; Griffin, M. Induction of tissue transglutaminase by dexamethasone: Its correlation to receptor number and transglutaminase-mediated cell death in a series of malignant hamster fibrosarcomas. Biochem. J. 1998, 331, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Pinkas, D.; Strop, P.; Brunger, A.; Khosla, C. Transglutaminase 2 Undergoes a Large Conformational Change upon Activation. PLoS ONE 2007, 5, e327. [Google Scholar] [CrossRef]
- Mearns, B.; Nanda, N.; Michalieck, J.; Iismaa, S.; Graham, R. Impaired wound healing and altered fibroblast cytoskeletal dynamics in Gh knockout mice. Minerva Biotecnol. 2002, 14, 218. [Google Scholar]
- Elias, P.M. Epidermal lipids, barrier function, and desquamation. J. Investig. Dermatol. 1983, 80, 544–549. [Google Scholar] [CrossRef] [PubMed]
- Nemes, Z.; Steinert, P.M. Bricks and mortar of the epidermal barrier. Exp. Mol. Med. 1999, 31, 5–19. [Google Scholar] [CrossRef] [PubMed]
- Grubauer, G.; Elias, P.M.; Feingold, K.R. Transepidermal water loss: The signal for recovery of barrier structure and function. J. Lipid Res. 1989, 30, 323–333. [Google Scholar] [CrossRef]
- Elias, P.M.; Wakefield, J.S. Mechanisms of abnormal lamellar body secretion and the dysfunctional skin barrier in patients with atopic dermatitis. J. Allergy Clin. Immunol. 2014, 134, 781–791.e1. [Google Scholar] [CrossRef]
- Caubet, C.; Jonca, N.; Brattsand, M.; Guerrin, M.; Bernard, D.; Schmidt, R.; Egelrud, T.; Simon, M.; Serre, G. Degradation of Corneodesmosome Proteins by Two Serine Proteases of the Kallikrein Family, SCTE/KLK5/hK5 and SCCE/KLK7/hK7. J. Investig. Dermatol. 2004, 122, 1235–1244. [Google Scholar] [CrossRef]
- Igawa, S.; Kishibe, M.; Honma, M.; Murakami, M.; Mizuno, Y.; Suga, Y.; Seishima, M.; Ohguchi, Y.; Akiyama, M.; Hirose, K.; et al. Aberrant distribution patterns of corneodesmosomal components of tape-stripped corneocytes in atopic dermatitis and related skin conditions (ichthyosis vulgaris, Netherton syndrome and peeling skin syndrome type B). J. Dermatol. Sci. 2013, 72, 54–60. [Google Scholar] [CrossRef]
- Simon, M.; Montézin, M.; Guerrin, M.; Durieux, J.-J.; Serre, G. Characterization and Purification of Human Corneodesmosin, an Epidermal Basic Glycoprotein Associated with Corneocyte-specific Modified Desmosomes. J. Biol. Chem. 1997, 272, 31770–31776. [Google Scholar] [CrossRef]
- Guerrin, M.; Simon, M.; Montézin, M.; Haftek, M.; Vincent, C.; Serre, G. Expression Cloning of Human Corneodesmosin Proves Its Identity with the Product of the S Gene and Allows Improved Characterization of Its Processing during Keratinocyte Differentiation. J. Biol. Chem. 1998, 273, 22640–22647. [Google Scholar] [CrossRef] [PubMed]
- Riethmuller, C.; McAleer, M.A.; Koppes, S.A.; Abdayem, R.; Franz, J.; Haftek, M.; Campbell, L.E.; MacCallum, S.F.; McLean, W.I.; Irvine, A.D.; et al. Filaggrin breakdown products determine corneocyte conformation in patients with atopic dermatitis. J. Allergy Clin. Immunol. 2015, 136, 1573–1580.e2. [Google Scholar] [CrossRef] [PubMed]
- Towell, A.M.; Feuillie, C.; Vitry, P.; Da Costa, T.M.; Mathelié-Guiniet, M.; Kezic, S.; Geoghegan, J.A.; Irvine, D.A.; Fleury, M.O.; McAleer, M.A.; et al. Staphylococcus aureus binds to the N-terminal region of corneodesmosin to adhere to the stratum corneum in atopic dermatitis. Proc. Natl. Acad. Sci. USA 2021, 118, e2014444118. [Google Scholar] [CrossRef] [PubMed]
- Vader, L.W.; De Ru, A.; Van Der Wal, Y.; Kooy, Y.M.; Benckhuijsen, W.; Mearin, M.L.; Drijfhout, J.W.; van Veelen, P.; Koning, F. Specificity of Tissue Transglutaminase Explains Cereal Toxicity in Celiac Disease. J. Exp. Med. 2002, 195, 643–649. [Google Scholar] [CrossRef]
- Fleckenstein, B.; Molberg, Ø.; Qiao, S.-W.; Schmid, D.G.; von der Mu¨lbe, F.; Elgstøen, K.; Jung, G.; Sollid, L.M. Gliadin T Cell Epitope Selection by Tissue Transglutaminase in Celiac Disease. J. Biol. Chem. 2002, 277, 34109–34116. [Google Scholar] [CrossRef]
- Eggink, L.L.; Roby, K.F.; Cote, R.; Hoober, J.K. An innovative immunotherapeutic strategy for ovarian cancer: CLEC10A and glycomimetic peptides. J. Immunother. Cancer 2018, 6, 28. [Google Scholar] [CrossRef]
- Teshima, H.; Kato, M.; Tatsukawa, H.; Hitomi, K. Analysis of the expression of transglutaminases in the reconstructed human epidermis using a three-dimensional cell culture. Anal. Biochem. 2020, 603, 113606. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hoober, J.K.; Eggink, L.L. The Discovery and Function of Filaggrin. Int. J. Mol. Sci. 2022, 23, 1455. https://doi.org/10.3390/ijms23031455
Hoober JK, Eggink LL. The Discovery and Function of Filaggrin. International Journal of Molecular Sciences. 2022; 23(3):1455. https://doi.org/10.3390/ijms23031455
Chicago/Turabian StyleHoober, J. Kenneth, and Laura L. Eggink. 2022. "The Discovery and Function of Filaggrin" International Journal of Molecular Sciences 23, no. 3: 1455. https://doi.org/10.3390/ijms23031455
APA StyleHoober, J. K., & Eggink, L. L. (2022). The Discovery and Function of Filaggrin. International Journal of Molecular Sciences, 23(3), 1455. https://doi.org/10.3390/ijms23031455






