Role of miRNA-145, 148, and 185 and Stem Cells in Prostate Cancer
Abstract
:1. Introduction
1.1. MiR-145
1.1.1. Downregulated miR-145
1.1.2. Upregulated miR-145
1.1.3. MiR-145/mRNA Network in PC
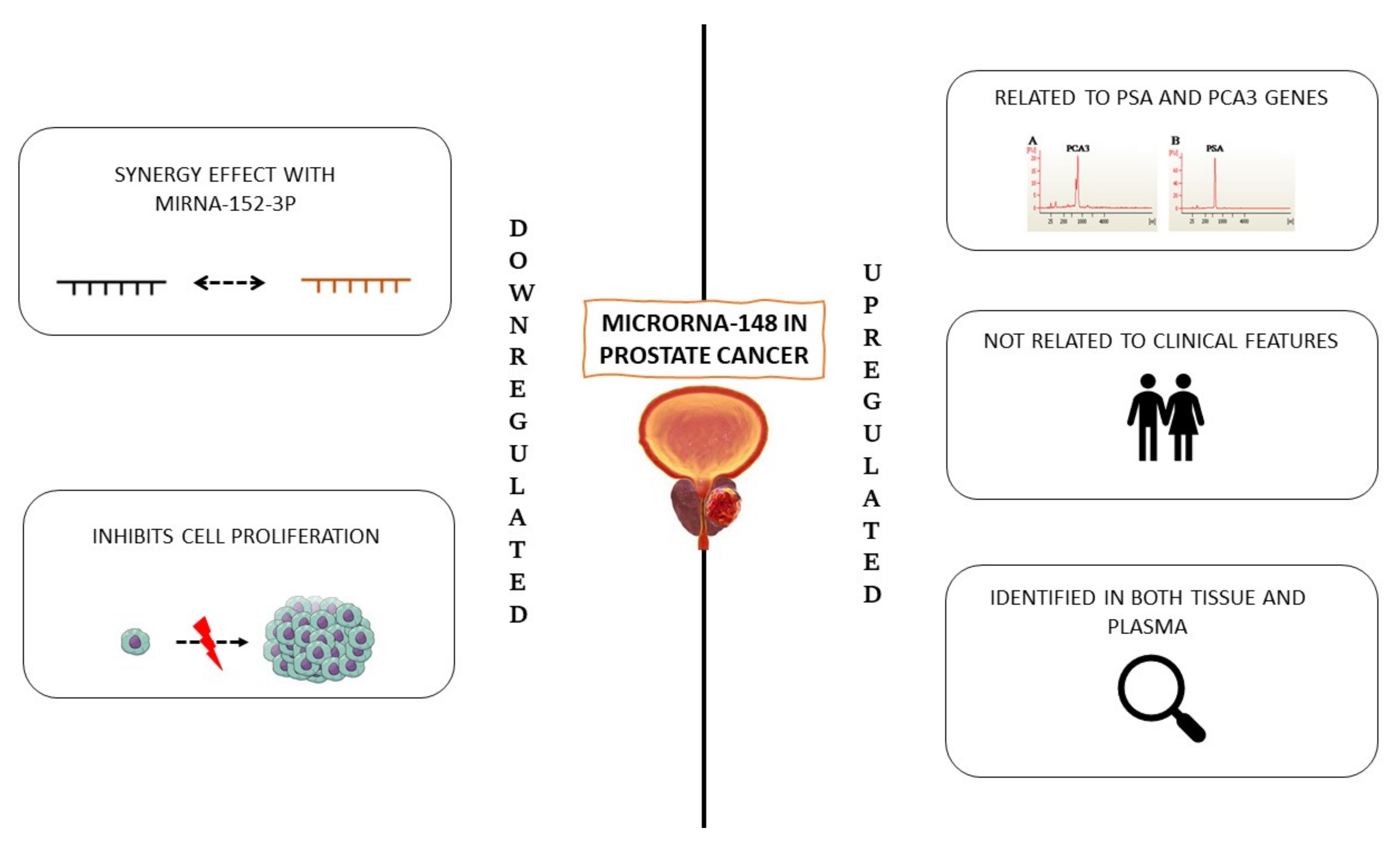
1.2. MiR-148
1.2.1. Downregulated MiR-148
1.2.2. Upregulated miR-148
1.3. MiR-185
1.3.1. Downregulated MiR-185
1.3.2. Upregulated miR-185
1.4. Involvement of Stem Cells in the Development of Prostate Cancer
2. Materials and Methods
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hart, M.; Nolte, E.; Wach, S.; Szczyrba, J.; Taubert, H.; Rau, T.T.; Hartmann, A.; Greasser, F.A.; Wullich, B. Comparative microrna profiling of prostate carcinomas with increasing tumor stage by deep sequencing. Mol. Cancer Res. 2014, 12, 250–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porkka, K.P.; Pfeiffer, M.J.; Waltering, K.K.; Vessella, R.L.; Tammela, T.L.J.; Visakorpi, T. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007, 67, 6130–6135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willard, S.S.; Koochekpour, S. Regulators of gene expression as biomarkers for prostate cancer. Am. J. Cancer Res. 2012, 2, 620–657. [Google Scholar] [PubMed]
- Naeini, M.M.; Ardekani, A.M. Noncoding RNAs and Cancer. Avicenna J. Med. Biotechnol. 2009, 1, 55. [Google Scholar]
- Tong, A.W.; Nemunaitis, J. Modulation of miRNA activity in human cancer: A new paradigm for cancer gene therapy? Cancer Gene Ther. 2008, 15, 341–355. [Google Scholar] [CrossRef]
- Di Leva, G.; Croce, C.M. miRNA profiling of cancer. Curr. Opin. Genet. Dev. 2013, 23, 3–11. [Google Scholar] [CrossRef] [Green Version]
- Brase, J.C.; Johannes, M.; Schlomm, T.; Haese, A.; Steuber, T.; Beissbarth, T.; Kuner, R.; Sültmann, H. Circulating miRNAs are correlated with tumor progression in prostate cancer. Int. J. Cancer 2011, 128, 608–616. [Google Scholar] [CrossRef]
- Selth, L.A.; Townley, S.; Gillis, J.L.; Ochnik, A.M.; Murti, K.; Macfarlane, R.J.; Chi, K.N.; Marshall, V.R.; Tilley, W.D.; Butler, L.M. Discovery of circulating microRNAs associated with human prostate cancer using a mouse model of disease. Int. J. Cancer 2012, 131, 652–661. [Google Scholar] [CrossRef]
- Cheng, H.H.; Yi, H.S.; Kim, Y.; Kroh, E.M.; Chien, J.W.; Eaton, K.D.; Goodman, M.T.; Tait, J.F.; Tewari, M.; Pritchard, C.C. Plasma Processing Conditions Substantially Influence Circulating microRNA Biomarker Levels. PLoS ONE 2013, 8, e64795. [Google Scholar] [CrossRef] [Green Version]
- Corcoran, C.; Rani, S.; O’Driscoll, L. miR-34a is an intracellular and exosomal predictive biomarker for response to docetaxel with clinical relevance to prostate cancer progression. Prostate 2014, 74, 1320–1334. [Google Scholar] [CrossRef] [Green Version]
- Guzel, E.; Karatas, O.F.; Semercioz, A.; Ekici, S.; Aykan, S.; Yentur, S.; Creighton, C.J.; Ittmann, M.; Ozen, M. Identification of microRNAs differentially expressed in prostatic secretions of patients with prostate cancer. Int. J. Cancer 2015, 136, 875–879. [Google Scholar] [CrossRef]
- Song, C.J.; Chen, H.; Chen, L.Z.; Ru, G.M.; Guo, J.J.; Ding, Q.N. The potential of microRNAs as human prostate cancer biomarkers: A meta-analysis of related studies. J. Cell. Biochem. 2018, 119, 2763–2786. [Google Scholar] [CrossRef] [Green Version]
- Bertoli, G.; Cava, C.; Castiglioni, I. MicroRNAs as Biomarkers for Diagnosis, Prognosis and Theranostics in Prostate Cancer. Int. J. Mol. Sci. 2016, 17, 421. [Google Scholar] [CrossRef] [Green Version]
- Balzano, F.; Campesi, I.; Cruciani, S.; Garroni, G.; Bellu, E.; Giudici, S.D.; Angius, A.; Oggiano, A.; Rallo, V.; Capobianco, G.; et al. Epigenetics, Stem Cells, and Autophagy: Exploring a Path Involving miRNA. Int. J. Mol. Sci. 2019, 20, 5091. [Google Scholar] [CrossRef] [Green Version]
- Ozen, M.; Creighton, C.J.; Ozdemir, M.; Ittmann, M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene 2008, 27, 1788–1793. [Google Scholar] [CrossRef] [Green Version]
- Ambs, S.; Prueitt, R.L.; Yi, M.; Hudson, R.S.; Howe, T.M.; Petrocca, F.; Wallace, T.A.; Liu, C.G.; Volinia, S.; Calin, G.A.; et al. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res. 2008, 68, 6162–6170. [Google Scholar] [CrossRef] [Green Version]
- Leite, K.R.M.; Tomiyama, A.; Reis, S.T.; Sousa-Canavez, J.M.; Sañudo, A.; Camara-Lopes, L.H.; Srougi, M. MicroRNA expression profiles in the progression of prostate cancer—From high-grade prostate intraepithelial neoplasia to metastasis. Urol. Oncol. Semin. Orig. Investig. 2013, 31, 796–801. [Google Scholar] [CrossRef]
- La Rocca, G.; Badin, M.; Shi, B.; Xu, S.Q.; Deangelis, T.; Sepp-Lorenzino, L.; Baserga, R. Mechanism of growth inhibition by MicroRNA 145: The role of the IGF-I receptor signaling pathway. J. Cell. Physiol. 2009, 220, 485–491. [Google Scholar] [CrossRef]
- Pashaei, E.; Guzel, E.; Ozgurses, M.E.; Demirel, G.; Aydin, N.; Ozen, M. A meta-analysis: Identification of common Mir-145 target genes that have similar behavior in different GEO datasets. PLoS ONE 2016, 11, e0161491. [Google Scholar] [CrossRef]
- Karadag, A.; Ozen, A.; Ozkurt, M.; Can, C.; Bozgeyik, I.; Kabadere, S.; Uyar, R. Identification of miRNA signatures and their therapeutic potentials in prostate cancer. Mol. Biol. Rep. 2021, 48, 5531–5539. [Google Scholar] [CrossRef]
- Liu, J.; Li, J.; Ma, Y.; Xu, C.; Wang, Y.; He, Y. MicroRNA miR-145-5p inhibits Phospholipase D 5 (PLD5) to downregulate cell proliferation and metastasis to mitigate prostate cancer. Bioengineered 2021, 12, 3240–3251. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, P.; Kumar, R.K.; Iyer, A.; Boswell, S.; Gerarduzzi, C.; Dadhania, V.P.; Herbert, Z.; Joshi, N.; Luyendyk, J.P.; Humphreys, B.D.; et al. Targeting phospholipase D4 attenuates kidney fibrosis. J. Am. Soc. Nephrol. 2017, 28, 3579–3589. [Google Scholar] [CrossRef] [PubMed]
- Uchida, N.; Okamura, S.I.; Nagamachi, Y.; Yamashita, S. Increased phospholipase D activity in human breast cancer. J. Cancer Res. Clin. Oncol. 1997, 123, 280–285. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Wang, X.; Li, H.; Fan, J.; Qian, X.; Li, H.; Xu, Y. Phospholipase D as a key modulator of cancer progression. Biol. Rev. 2020, 95, 911–935. [Google Scholar] [CrossRef]
- Avgeris, M.; Stravodimos, K.; Fragoulis, E.G.; Scorilas, A. The loss of the tumour-suppressor miR-145 results in the shorter disease-free survival of prostate cancer patients. Br. J. Cancer 2013, 108, 2573–2581. [Google Scholar] [CrossRef] [Green Version]
- Wach, S.; Nolte, E.; Szczyrba, J.; Stöhr, R.; Hartmann, A.; Ørntoft, T.; Dyrskjøt, L.; Eltze, E.; Wieland, W.; Keck, B.; et al. MicroRNA profiles of prostate carcinoma detected by multiplatform microRNA screening. Int. J. Cancer 2012, 130, 611–621. [Google Scholar] [CrossRef]
- Leite, K.R.M.; Tomiyama, A.; Reis, S.T.; Sousa-Canavez, J.M.; Saudo, A.; Dall’Oglio, M.F.; Camara-Lopes, L.H.; Srougi, M. MicroRNA-100 expression is independently related to biochemical recurrence of prostate cancer. J. Urol. 2011, 185, 1118–1122. [Google Scholar] [CrossRef]
- Karatas, O.F.; Guzel, E.; Suer, I.; Ekici, I.D.; Caskurlu, T.; Creighton, C.J.; Ittmann, M.; Ozen, M. miR-1 and miR-133b are differentially expressed in patients with recurrent prostate cancer. PLoS ONE 2014, 9, 98675. [Google Scholar] [CrossRef]
- Shen, J.; Hruby, G.W.; McKiernan, J.M.; Gurvich, I.; Lipsky, M.J.; Benson, M.C.; Santella, R.M. Dysregulation of circulating microRNAs and prediction of aggressive prostate cancer. Prostate 2012, 72, 1469–1477. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Qin, S.; An, T.; Tang, Y.; Huang, Y.; Zheng, L. MiR-145 detection in urinary extracellular vesicles increase diagnostic efficiency of prostate cancer based on hydrostatic filtration dialysis method. Prostate 2017, 77, 1167–1175. [Google Scholar] [CrossRef]
- Larne, O.; Hagman, Z.; Lilja, H.; Bjartell, A.; Edsjö, A.; Ceder, Y. miR-145 suppress the androgen receptor in prostate cancer cells and correlates to prostate cancer prognosis. Carcinogenesis 2015, 36, 858–866. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.D.; Ceniccola, K.; Yang, Q.; Andrawis, R.; Patel, V.; Ji, Y.; Rhim, J.; Olender, J.; Popratiloff, A.; Latham, P.; et al. Identification and functional validation of reciprocal microRNA-mRNA pairings in African American prostate cancer disparities. Clin. Cancer Res. 2015, 21, 4970–4984. [Google Scholar] [CrossRef] [Green Version]
- Audenet, F.; Cancel-Tassin, G.; Bigot, P.; Audouin, M.; Gaffory, C.; Ondet, V.; Thibault, F.; Auribault, K.; Gazut, S.; Benhabiles, N.; et al. Germline genetic variations at 11q13 and 12p11 locus modulate age at onset for renal cell carcinoma. J. Urol. 2014, 191, 487–492. [Google Scholar] [CrossRef]
- Zhu, J.; Wang, S.; Zhang, W.; Qiu, J.; Shan, Y.; Yang, D.; Shen, B. Screening key microRNAs for castration-resistant prostate cancer based on miRNA/mRNA functional synergistic network. Oncotarget 2015, 6, 43819–43830. [Google Scholar] [CrossRef] [PubMed]
- Buttyan, R.; Sawczuk, I.S.; Benson, M.C.; Siegal, J.D.; Olsson, C.A. Enhanced expression of the c-myc protooncogene in high-grade human prostate cancers. Prostate 1987, 11, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Schwartzman, J.; Gibbs, A.; Lisac, R.; Kleinschmidt, R.; Wilmot, B.; Bottomly, D.; Coleman, I.; Nelson, P.; McWeeney, S.; et al. Androgen Receptor Promotes Ligand-Independent Prostate Cancer Progression through c-Myc Upregulation. PLoS ONE 2013, 8, 63563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spizzo, R.; Nicoloso, M.S.; Lupini, L.; Lu, Y.; Fogarty, J.; Rossi, S.; Zagatti, B.; Fabbri, M.; Veronese, A.; Liu, X.; et al. MiR-145 participates with TP53 in a death-promoting regulatory loop and targets estrogen receptor-α in human breast cancer cells. Cell Death Differ. 2010, 17, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Coarfa, C.; Fiskus, W.; Eedunuri, V.K.; Rajapakshe, K.; Foley, C.; Chew, S.A.; Shah, S.S.; Geng, C.; Shou, J.; Mohamed, J.S.; et al. Comprehensive proteomic profiling identifies the androgen receptor axis and other signaling pathways as targets of microRNAs suppressed in metastatic prostate cancer. Oncogene 2016, 35, 2345–2356. [Google Scholar] [CrossRef] [Green Version]
- Goto, Y.; Kurozumi, A.; Arai, T.; Nohata, N.; Kojima, S.; Okato, A.; Kato, M.; Yamazaki, K.; Ishida, Y.; Naya, Y.; et al. Impact of novel miR-145-3p regulatory networks on survival in patients with castration-resistant prostate cancer. Br. J. Cancer 2017, 117, 409–420. [Google Scholar] [CrossRef]
- Walter, B.A.; Valera, V.A.; Pinto, P.A.; Merino, M.J. Comprehensive microRNA profiling of prostate cancer. J. Cancer 2013, 4, 350–357. [Google Scholar] [CrossRef] [Green Version]
- Feng, F.; Liu, H.; Chen, A.; Xia, Q.; Zhao, Y.; Jin, X.; Huang, J. miR-148-3p and miR-152-3p synergistically regulate prostate cancer progression via repressing KLF4. J. Cell. Biochem. 2019, 120, 17228–17239. [Google Scholar] [CrossRef] [PubMed]
- Arámbula-Meraz, E.; Bergez-Hernández, F.; Leal-León, E.; Romo-Martínez, E.; Picos-Cárdenas, V.; Luque-Ortega, F.; Romero-Quintana, J.; Alvarez-Arrazola, M.; García-Magallanes, N. Expression of miR-148b-3p is correlated with overexpression ofbiomarkers in prostate cancer. Genet. Mol. Biol. 2020, 43. [Google Scholar] [CrossRef]
- Paunescu, I.A.; Bardan, R.; Marcu, A.; Nitusca, D.; Dema, A.; Negru, S.; Balacescu, O.; Balacescu, L.; Cumpanas, A.; Sirbu, I.O.; et al. Biomarker Potential of Plasma MicroRNA-150-5p in Prostate Cancer. Medicina 2019, 55, 564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hellwinkel, O.J.C.; Sellier, C.; Sylvester, Y.M.J.; Brase, J.C.; Isbarn, H.; Erbersdobler, A.; Steuber, T.; Sültmann, H.; Schlomm, T.; Wagner, C. A cancer-indicative microRNA pattern in normal prostate tissue. Int. J. Mol. Sci. 2013, 14, 5239–5249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dakubo, G.D.; Jakupciak, J.P.; Birch-Machin, M.A.; Parr, R.L. Clinical implications and utility of field cancerization. Cancer Cell Int. 2007, 7, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braakhuis, B.J.M.; Leemans, C.R.; Brakenhoff, R.H. A genetic progression model of oral cancer: Current evidence and clinical implications. J. Oral Pathol. Med. 2004, 33, 317–322. [Google Scholar] [CrossRef] [PubMed]
- Kakizoe, T. Development and progression of urothelial carcinoma. Cancer Sci. 2006, 97, 821–828. [Google Scholar] [CrossRef] [PubMed]
- Ostadrahimi, S.; Fayaz, S.; Parvizhamidi, M.; Abedi-Valugerdi, M.; Hassan, M.; Kadivar, M.; Teimoori-Toolabi, L.; Asgari, M.; Shahrokh, H.; Abolhasani, M.; et al. Downregulation of miR-1266-5P, miR-185-5P and miR-30c-2 in prostatic cancer tissue and cell lines. Oncol. Lett. 2018, 15, 8157–8164. [Google Scholar] [CrossRef]
- Kristensen, H.; Thomsen, A.R.; Haldrup, C.; Dyrskjøt, L.; Høyer, S.; Borre, M.; Mouritzen, P.; Ørntoft, T.F.; Sørensen, K.D. Novel diagnostic and prognostic classifiers for prostate cancer identified by genome-wide microRNA profiling. Oncotarget 2016, 7, 30760–30771. [Google Scholar] [CrossRef]
- Östling, P.; Leivonen, S.K.; Aakula, A.; Kohonen, P.; Mäkelä, R.; Hagman, Z.; Edsjö, A.; Kangaspeska, S.; Edgren, H.; Nicorici, D.; et al. Systematic analysis of microRNAs targeting the androgen receptor in prostate cancer cells. Cancer Res. 2011, 71, 1956–1967. [Google Scholar] [CrossRef] [Green Version]
- Qu, F.; Cui, X.; Hong, Y.; Wang, J.; Li, Y.; Chen, L.; Liu, Y.; Gao, Y.; Xu, D.; Wang, Q. MicroRNA-185 suppresses proliferation, invasion, migration, and tumorigenicity of human prostate cancer cells through targeting androgen receptor. Mol. Cell. Biochem. 2013, 377, 121–130. [Google Scholar] [CrossRef]
- Li, X.; Chen, Y.T.; Josson, S.; Mukhopadhyay, N.K.; Kim, J.; Freeman, M.R.; Huang, W.C. MicroRNA-185 and 342 Inhibit Tumorigenicity and Induce Apoptosis through Blockade of the SREBP Metabolic Pathway in Prostate Cancer Cells. PLoS ONE 2013, 8, e70987. [Google Scholar] [CrossRef]
- Di Leva, G.; Garofalo, M.; Croce, C.M. MicroRNAs in cancer. Annu. Rev. Pathol. Mech. Dis. 2014, 9, 287–314. [Google Scholar] [CrossRef] [Green Version]
- McDonald, A.C.; Vira, M.; Walter, V.; Shen, J.; Raman, J.D.; Sanda, M.G.; Patil, D.; Taioli, E. Circulating microRNAs in plasma among men with low-grade and high-grade prostate cancer at prostate biopsy. Prostate 2019, 79, 961–968. [Google Scholar] [CrossRef]
- Gurbuz, V.; Kiliccioglu, I.; Dikmen, A.U.; Bilen, C.Y.; Sozen, S.; Konac, E. Comparative analysis of epi-miRNA expression levels in local/locally advanced and metastatic prostate cancer patients. Gene 2020, 758, 144963. [Google Scholar] [CrossRef]
- Phi, L.T.H.; Sari, I.N.; Yang, Y.G.; Lee, S.H.; Jun, N.; Kim, K.S.; Lee, Y.K.; Kwon, H.Y. Cancer Stem Cells (CSCs) in Drug Resistance and their Therapeutic Implications in Cancer Treatment. Stem Cells Int. 2018, 2018, 5416923. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Steed, A.; Co, M.; Chen, X. Cancer stem cells, epithelial-mesenchymal transition, ATP and their roles in drug resistance in cancer. Cancer Drug Resist. 2021, 4, 684–709. [Google Scholar] [CrossRef]
- Khan, A.; Ahmed, E.; Elareer, N.; Junejo, K.; Steinhoff, M.; Uddin, S. Role of miRNA-Regulated Cancer Stem Cells in the Pathogenesis of Human Malignancies. Cells 2019, 8, 840. [Google Scholar] [CrossRef] [Green Version]
- Ullah, I.; Subbarao, R.B.; Rho, G.J. Human mesenchymal stem cells—Current trends and future prospective. Biosci. Rep. 2015, 35, e00191. [Google Scholar] [CrossRef]
- Toivanen, R.; Shen, M.M. Prostate organogenesis: Tissue induction, hormonal regulation and cell type specification. Development 2017, 144, 1382. [Google Scholar] [CrossRef] [Green Version]
- Ferraro, F.; Lo Celso, C.; Scadden, D. Adult stem cels and their niches. Adv. Exp. Med. Biol. 2010, 695, 155–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lang, S.H.; Frame, F.M.; Collins, A.T. Prostate cancer stem cells. J. Pathol. 2009, 217, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Prajapati, A.; Gupta, S.; Mistry, B. Prostate stem cells in the development of benign prostate hyperplasia and prostate cancer: Emerging role and concepts. Biomed Res. Int. 2013, 2013, 107954. [Google Scholar] [CrossRef] [Green Version]
- Barclay, W.W.; Axanova, L.S.; Chen, W.; Romero, L.; Maund, S.L.; Soker, S.; Lees, C.J.; Cramer, S.D. Characterization of adult prostatic progenitor/stem cells exhibiting self-renewal and multilineage differentiation. Stem Cells 2008, 26, 600–610. [Google Scholar] [CrossRef] [Green Version]
- Gutschner, T.; Diederichs, S. The hallmarks of cancer. RNA Biol. 2012, 9, 703–719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gérard, C.; Goldbeter, A. The balance between cell cycle arrest and cell proliferation: Control by the extracellular matrix and by contact inhibition. Interface Focus 2014, 4, 20130075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balzano, F.; Garroni, G.; Cruciani, S.; Bellu, E.; Dei Giudici, S.; Oggiano, A.; Capobianco, G.; Dessole, S.; Ventura, C.; Maioli, M. Behavioral Changes in Stem-Cell Potency by HepG2-Exhausted Medium. Cells 2020, 9, 1890. [Google Scholar] [CrossRef] [PubMed]
- Garroni, G.; Balzano, F.; Cruciani, S.; Pala, R.; Coradduzza, D.; Azara, E.; Bellu, E.; Cossu, M.L.; Ginesu, G.C.; Carru, C.; et al. Adipo-derived stem cell features and mcf-7. Cells 2021, 10, 1754. [Google Scholar] [CrossRef]
- Barclay, W.W.; Woodruff, R.D.; Hall, M.C.; Cramer, S.D. A System for Studying Epithelial-Stromal Interactions Reveals Distinct Inductive Abilities of Stromal Cells from Benign Prostatic Hyperplasia and Prostate Cancer. Endocrinology 2005, 146, 13–18. [Google Scholar] [CrossRef] [Green Version]
- Hata, A.; Kashima, R. Dysregulation of microRNA biogenesis machinery in cancer. Crit. Rev. Biochem. Mol. Biol. 2015, 51, 121–134. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, J.; Ruohola-Baker, H. Regulation of Stem Cell Populations by microRNAs. Adv. Exp. Med. Biol. 2013, 786, 329–351. [Google Scholar] [CrossRef] [Green Version]
- Fang, Y.X.; Chang, Y.L.; Gao, W.Q. MicroRNAs targeting prostate cancer stem cells. Exp. Biol. Med. 2015, 240, 1071–1078. [Google Scholar] [CrossRef] [Green Version]
- Xu, N.; Papagiannakopoulos, T.; Pan, G.; Thomson, J.A.; Kosik, K.S. MicroRNA-145 Regulates OCT4, SOX2, and KLF4 and Represses Pluripotency in Human Embryonic Stem Cells. Cell 2009, 137, 647–658. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.; Guo, W.; Tang, Y.; Ren, D.; Zou, X.; Peng, X. miR-143 and miR-145 inhibit stem cell characteristics of PC-3 prostate cancer cells. Oncol. Rep. 2012, 28, 1831–1837. [Google Scholar] [CrossRef] [Green Version]
- Gurbuz, V.; Sozen, S.; Bilen, C.Y.; Konac, E. miR-148a, miR-152 and miR-200b promote prostate cancer metastasis by targeting DNMT1 and PTEN expression. Oncol. Lett. 2021, 22, 805. [Google Scholar] [CrossRef]
- Murata, T.; Takayama, K.; Katayama, S.; Urano, T.; Horie-Inoue, K.; Ikeda, K.; Takahashi, S.; Kawazu, C.; Hasegawa, A.; Ouchi, Y.; et al. miR-148a is an androgen-responsive microRNA that promotes LNCaP prostate cell growth by repressing its target CAND1 expression. Prostate Cancer Prostatic Dis. 2010, 13, 356–361. [Google Scholar] [CrossRef]
- Tan, Y.; Lu, X.; Cheng, Z.; Pan, G.; Liu, S.; Apiziaji, P.; Wang, H.; Zhang, J.; Abulimiti, Y. miR-148a Regulates the Stem Cell-Like Side Populations Distribution by Affecting the Expression of ACVR1 in Esophageal Squamous Cell Carcinoma. Onco. Targets. Ther. 2020, 13, 8079–8094. [Google Scholar] [CrossRef]
- Li, Y.; Zhong, W.; Zhu, M.; Li, M.; Yang, Z. MiR-185 inhibits prostate cancer angiogenesis induced by the nodal/ALK4 pathway. BMC Urol. 2020, 20, 1–11. [Google Scholar] [CrossRef]
- Liu, C.; Cai, L.; Li, H. MiR-185 regulates the growth of osteosarcoma cells via targeting Hexokinase 2. Mol. Med. Rep. 2019, 20, 2774–2782. [Google Scholar] [CrossRef]


| MiRNA | Expression | Sample Types, Author, Year | ||
|---|---|---|---|---|
| Tissue | Serum/Plasma | Urine | ||
| miR-21 | Szczyrba, 2018; Wach, 2012 | NA | NA | |
| Up | Zedan, 2018; Kumar, 2018 | Porzycki, 2018; Kotb, 2014; Egidi, 2013; Zedan, 2018; Agaoglu, 2011; Mello Grand, 2018; Gao, 2016; Endzelinš, 2017—EVs | Ghorbanmehr, 2019; Foj, 2017 | |
| miR-125b | Down | Zedan, 2018; Schaefer, 2010 | NA | Fredsoe, 2018 |
| Up | Walter, 2013; Song, 2015 | Mitchell, 2008; Zedan, 2018 | NA | |
| miR-141 | Down | NA | Kachakova, 2014 | Fredsoe, 2018 |
| Up | Szczyrba, 2010; Kumar, 2018; Nguyen, 2013; Brase, 2014; Kelly, 2015 | Mitchell, 2008; Porzycki, 2018; Cheng, 2013; Guo, 2018; Hao, 2016—EVs Bryant, 2012—EVs | Ghorbanmehr, 2019; Foj, 2017 | |
| miR-143 | Down | Szczyrba, 2010; Wach, 2012; Zedan, 2018; Kumar, 2018; Martens Uzunova, 2012 | NA | Stuopelyte, 2016 Rodriguez, 2017—EV |
| Up | NA | Nitchel, 2008; Zedan, 2018 | NA | |
| miR-145 | Down | Szczyrba, 2010; Wach, 2012; Porkka, 2007; Ozen, 2008; Kang, 2012; Zedan, 2018; Kurul, 2019; Schaefer, 2010; Larne, 2015, Kelly, 2015; Martens-Uzunova, 2012; Yfantis, 2008; Avgeris, 2013; Larne, 2013; Karatas, 2014 Wang, 2015, Zhu, 2015 Goto, 2017, Coarfa, 2016 Karadag, 2021, Liu, 2021 Leite, 2011 | NA | NA |
| Up | NA | Shen, 2012 | Xu, 2017 | |
| miR-148 | Down | Feng, 2019, Walter, 2013 Arámbula-Meraz, 2020 | NA | Stuopelyte, 2016 |
| Up | Szczyrba, 2010; Martens-Uzunova, 2012; Stuopelyte, 2016; Lichner, 2015; Hart, 2013 | Dybos, 2018; Al-Qatati, 2017; Paunescu, 2019 | NA | |
| miR-182 | Down | NA | NA | NA |
| Up | Wach, 2012; Schaefer, 2010; Yfantis, 2008; Tsuchiyama, 2013; Costa-Pinheiro, 2015; Casanova-Salas, 2014 | NA | NA | |
| miR185 | Down | Hellwinkel, 2013, Ostadrahimi, 2018 | Mc Donald, 2019, Gurbuz, 2020 | NA |
| Up | Kristensen, 2016 | NA | NA | |
| miR-200c | Down | Szczyrba, 2010; Wach, 2012; Yfantis, 2008 | NA | Fredsoe, 2018 |
| Up | NA | Cheng, 2013 De Souza, 2017 Endzelinš, 2017—EVs | NA | |
| miR-205 | Down | Scahefer, 2010; Martens-Uzunova, 2012; Yfantis, 2008; Tsuchiyama, 2013; Verdoodt, 2013; Kalogirou, 2013 | Guo, 2018 Osipov, 2016 | Fredsoe, 2018 |
| Up | Walter, 2013 | NA | NA | |
| miR-221 | Down | Szczyrba, 2010; Wach, 2012; Zedan, 2018; Kurul, 2019; Schaefer, 2010; Yfantis, 2008; Porkka, 2007; Tsuchiyama, 2013; Casanova-Salas, 2014; Kneitzm, 2014 | NA | Fredsoe, 2018 |
| Up | Song, 2015 | Kotb, 2014 Agaoglu, 2011 | NA | |
| miR-222 | Down | Wach, 2012; Schaefer, 2010; Martens-Uzunova, 2012; Porkka 2007; Tsuchiyama, 2013 | NA | Fredsoe, 2018 |
| Up | NA | NA | NA | |
| miR-375 | Down | Szczyrba, 2010; Wach, 2012; Schaefer, 2010; Nguyen, 2013; Brase, 2011; Stuopelyte, 2016; Yfantis, 2008; Costa-Pinheiro, 2015; Haldrup, 2014; Nam, 2018 | Kachakova, 2014 | NA |
| Up | NA | Porzycki, 2018; Nguyen, 2013; Brase, 2011; Cheng, 2013; Haldrup, 2014; Wach, 2015; Zedan, 2018; Gao, 2016; Endzelinš, 2017; McDonald, 2018; Huang, 2015—EVs Bryant, 2012—EVs | Foj, 2017; Stuopelyte, 2016 | |
| let-7a | Down | Szczyrba, 2010; Wach, 2012; Kelly, 2015; Porkka, 2007; Tian, 2015 | Endzelinš, 2017—EV | Fredsoe, 2018 |
| Up | Haldrup, 2014 | Mello-Grand, 2018 | NA | |
| Author, Year, Place | Expression | Prostate Cancer Patients (n) | Negative Control (n) | Samples Type | Reference |
|---|---|---|---|---|---|
| Leite, 2011, Brazil | Downregulation | 49 | 10 | Tissue | [27] |
| Shen, 2012, USA | Upregulation | 82 | Plasma | [29] | |
| Wach, 2012, Germany | Downregulation | 76 | Tissue | [26] | |
| Avgeris, 2013, Greece | Downregulation | 73 | 64 | Tissue | [25] |
| Leite, 2013, Brazil | Downregulation | 63 | Tissue | [17] | |
| Hart, 2013, Germany Karatas, 2014, Turkey, USA | Downregulation | 40 | 40 | Tissue | [1] |
| Downregulation | 82 | Tissue | [28] | ||
| Larne, 2015, Sweden | Downregulation | 49 | 25 | Tissue | [31] |
| Wang, 2015, USA | Downregulation | 35 | Tissue | [32] | |
| Zhu, 2015, China | Downregulation | 5 | Cell lines | [34] | |
| Coarfa, 2016, USA | Downregulation | 113 | 28 | Tissue | [38] |
| Goto, 2017, Japan | Downregulation | 34 | 19 | Tissue | [39] |
| Xu, 2017, China | Upregulation | 60 | 24 | Urine | [30] |
| Karadag, 2021, Turkey | Downregulation | 18 | 18 | Cell culture | [20] |
| Liu, 2021, China | Downregulation | 18 | 18 | Cell culture | [21] |
| Author, Year, Place | Expression | Prostate Cancer Patients (n) | Negative Control (n) | Samples Type | Reference |
|---|---|---|---|---|---|
| Walter, 2013, USA | Downregulation | 40 | 20 | Tissue | [40] |
| Hart, 2014, Germany | Upregulation | 40 | 40 | Tissue | [1] |
| Feng, 2019, China | Downregulation | 42 | 42 | Tissue | [41] |
| Paunescu, 2019, Romania | Upregulation | 14 | 15 | Plasma | [43] |
| Arámbula-Meraz, 2020, Mexico | Downregulation | 12 | 12 | Tissue | [42] |
| Author, Year, Place | Expression | Prostate Cancer Patients (n) | Negative Control (n) | Samples Type | Reference |
|---|---|---|---|---|---|
| Hellwinkel, 2013, Germany | Downregulation | 31 | 31 | Tissue | [44] |
| Kristensen, 2016, Denmark | Upregulation | 385 | 60 | Tissue | [49] |
| Ostadrahimi, 2018, Iran | Downregulation | 30 | 30 | Tissue and cell lines | [48] |
| Mc Donald, 2019, USA | Upregulation | 66 | / | Plasma | [54] |
| Gurbuz, 2020, Turkey | Upregulation | 75 | 25 | Blood | [55] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coradduzza, D.; Cruciani, S.; Arru, C.; Garroni, G.; Pashchenko, A.; Jedea, M.; Zappavigna, S.; Caraglia, M.; Amler, E.; Carru, C.; et al. Role of miRNA-145, 148, and 185 and Stem Cells in Prostate Cancer. Int. J. Mol. Sci. 2022, 23, 1626. https://doi.org/10.3390/ijms23031626
Coradduzza D, Cruciani S, Arru C, Garroni G, Pashchenko A, Jedea M, Zappavigna S, Caraglia M, Amler E, Carru C, et al. Role of miRNA-145, 148, and 185 and Stem Cells in Prostate Cancer. International Journal of Molecular Sciences. 2022; 23(3):1626. https://doi.org/10.3390/ijms23031626
Chicago/Turabian StyleCoradduzza, Donatella, Sara Cruciani, Caterina Arru, Giuseppe Garroni, Aleksei Pashchenko, Mosab Jedea, Silvia Zappavigna, Michele Caraglia, Evzen Amler, Ciriaco Carru, and et al. 2022. "Role of miRNA-145, 148, and 185 and Stem Cells in Prostate Cancer" International Journal of Molecular Sciences 23, no. 3: 1626. https://doi.org/10.3390/ijms23031626






