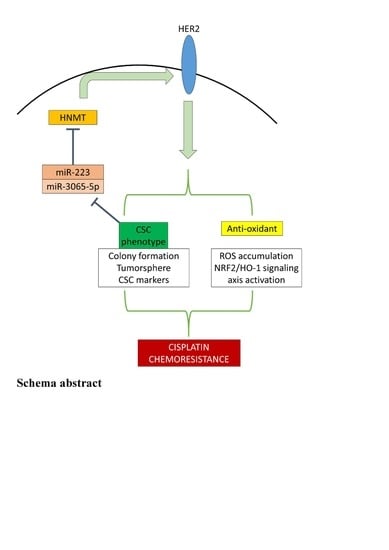
HNMT Upregulation Induces Cancer Stem Cell Formation and Confers Protection against Oxidative Stress through Interaction with HER2 in Non-Small-Cell Lung Cancer
Abstract
:1. Introduction
2. Results
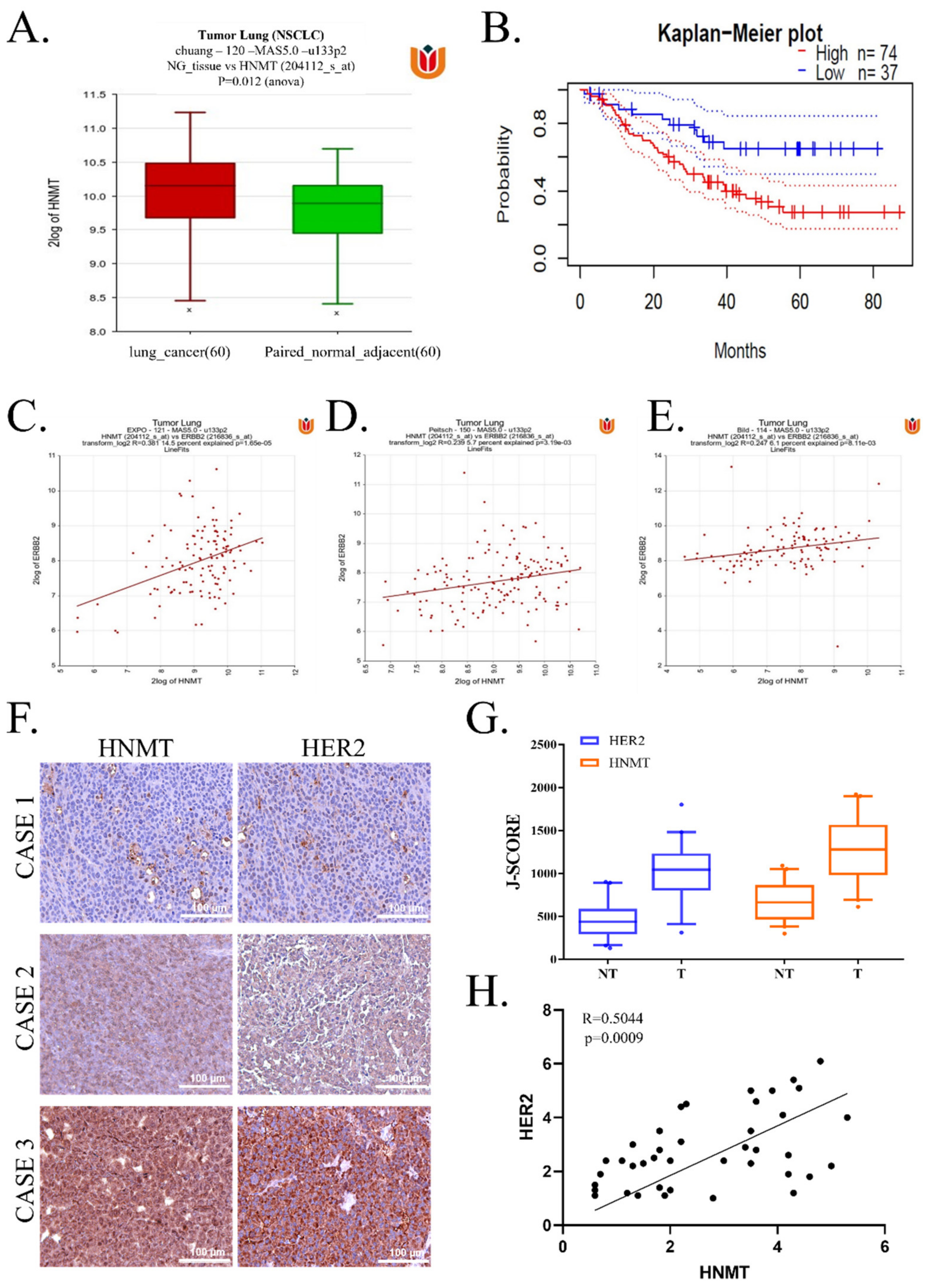
2.1. HNMT Upregulation in NSCLC Tissues Is Related to Worsened Prognosis and Significantly Coexpressed with HER2
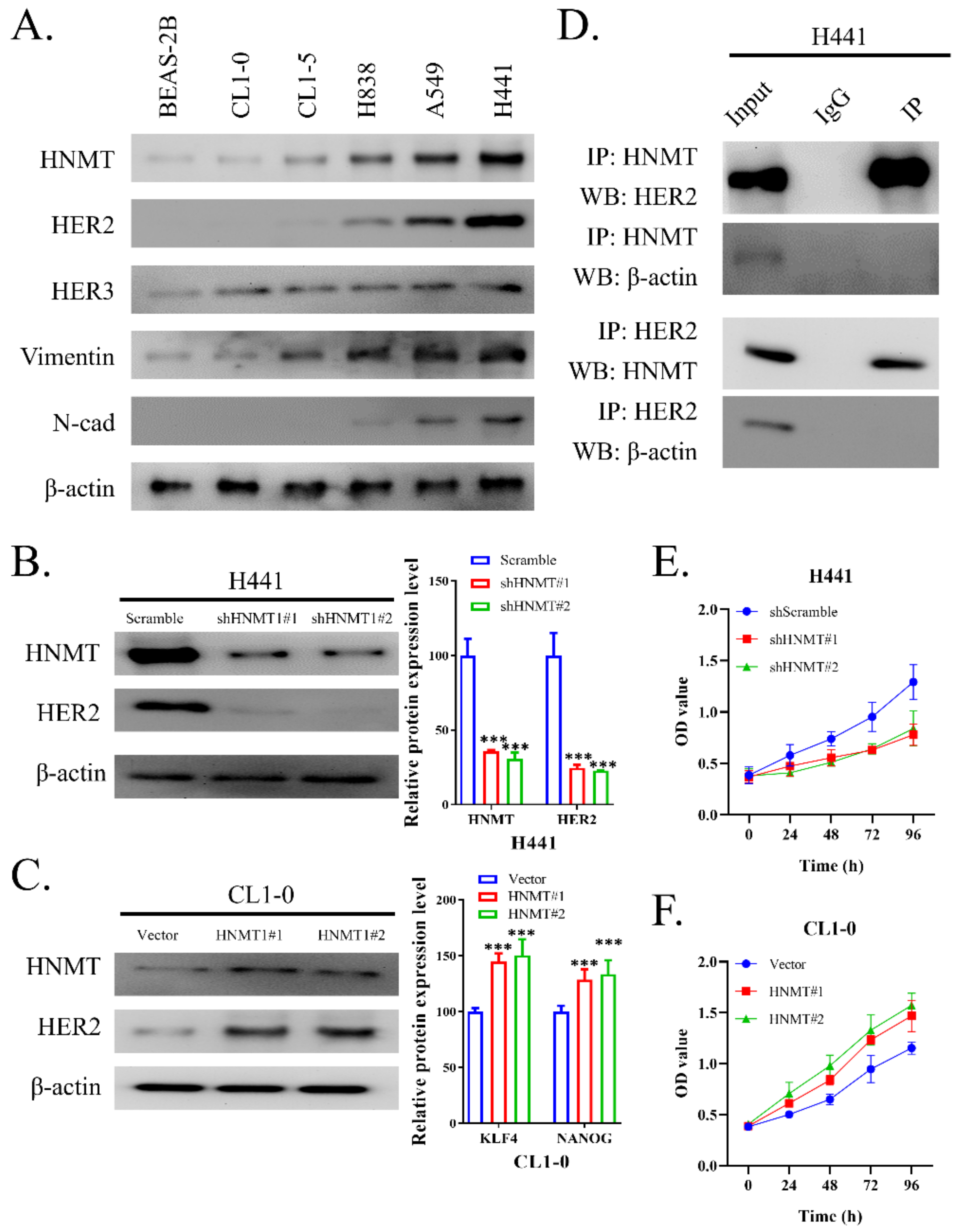
2.2. HNMT Interacts with HER2 and Affects NSCLC Cell Line Development
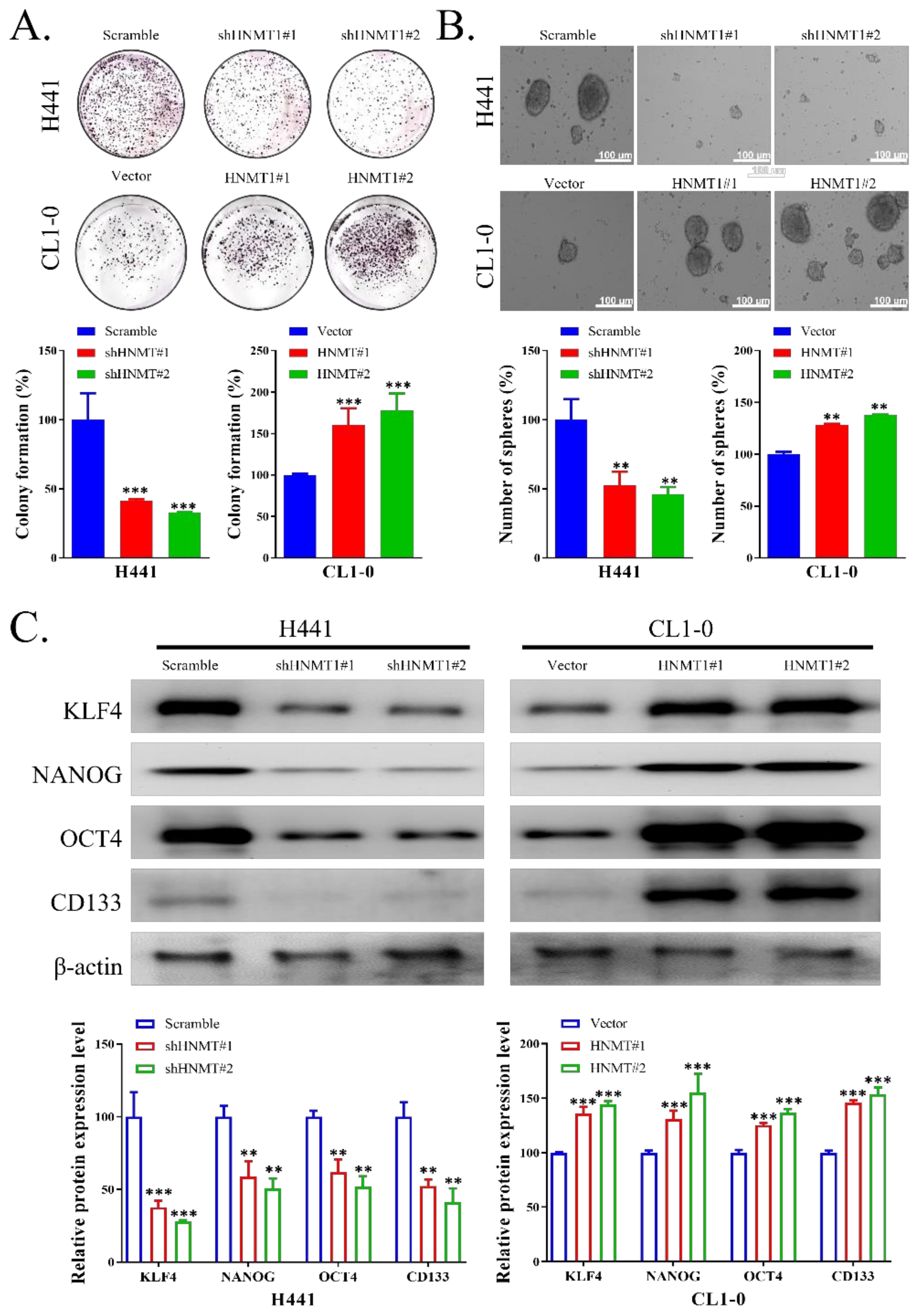
2.3. HNMT Promotes CSC Development in NSCLC Cell Lines
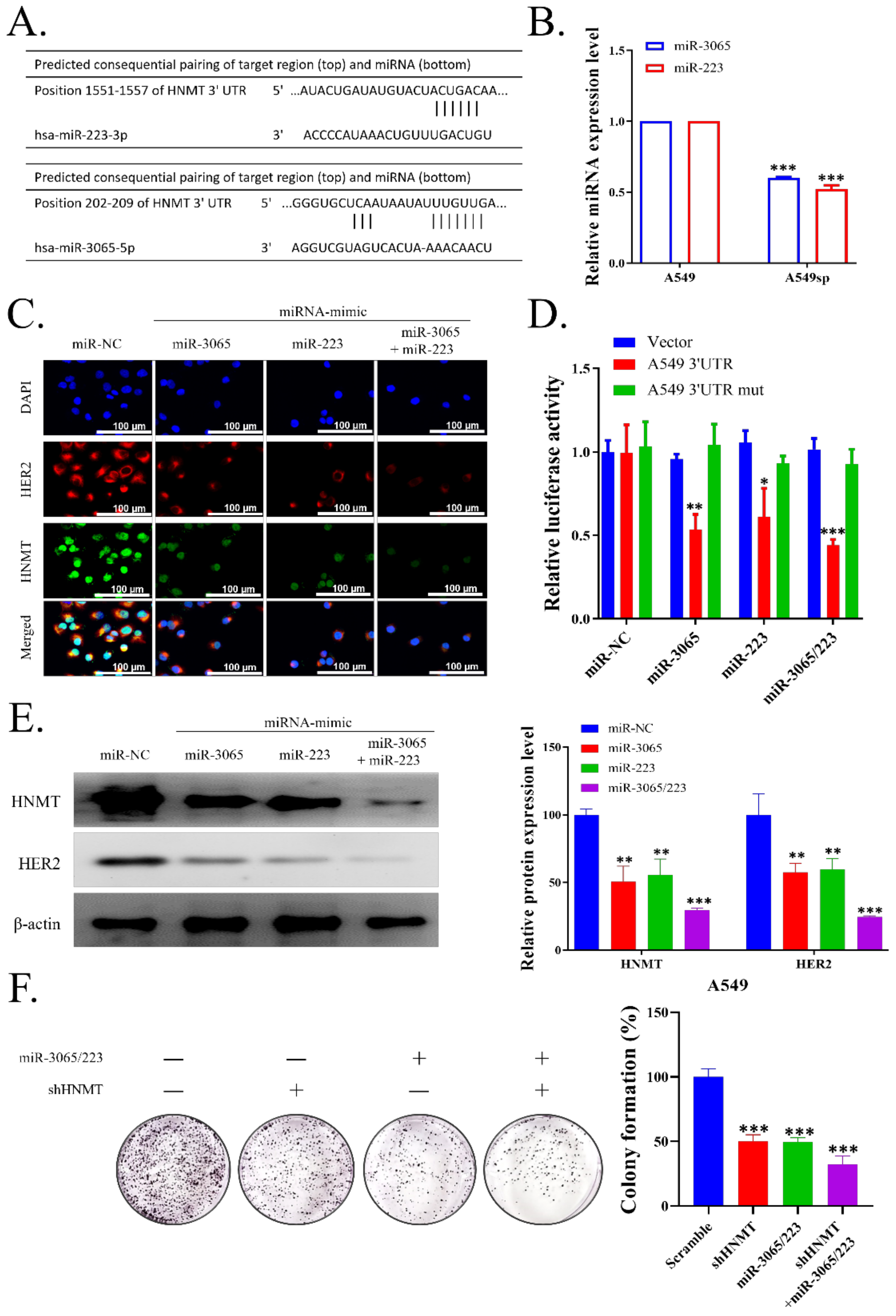
2.4. HNMT Is the Gene Target of miR-223 and miR-3065-5p
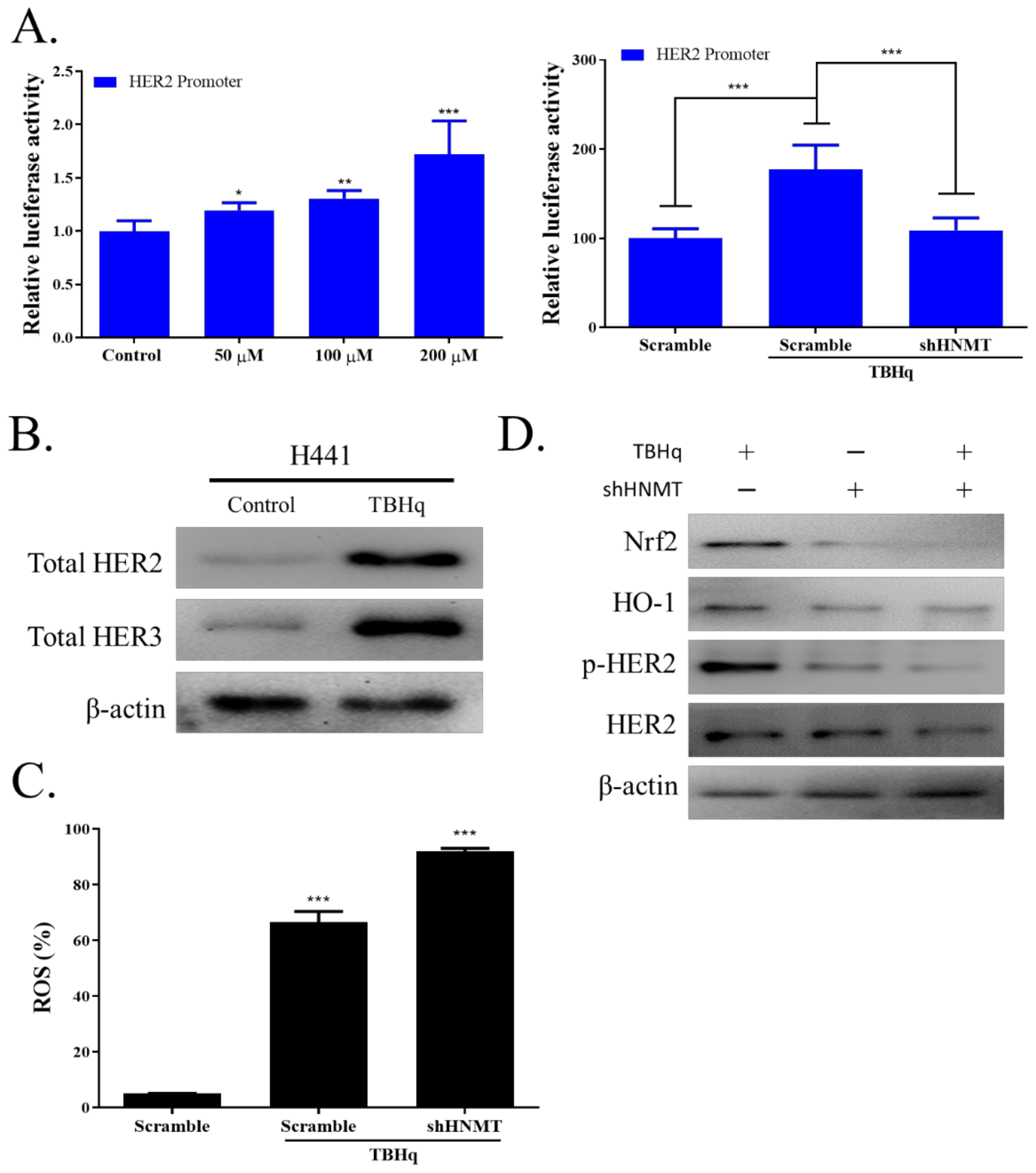
2.5. HNMT Regulates the Antioxidative Stress Pathway in NSCLC Cells
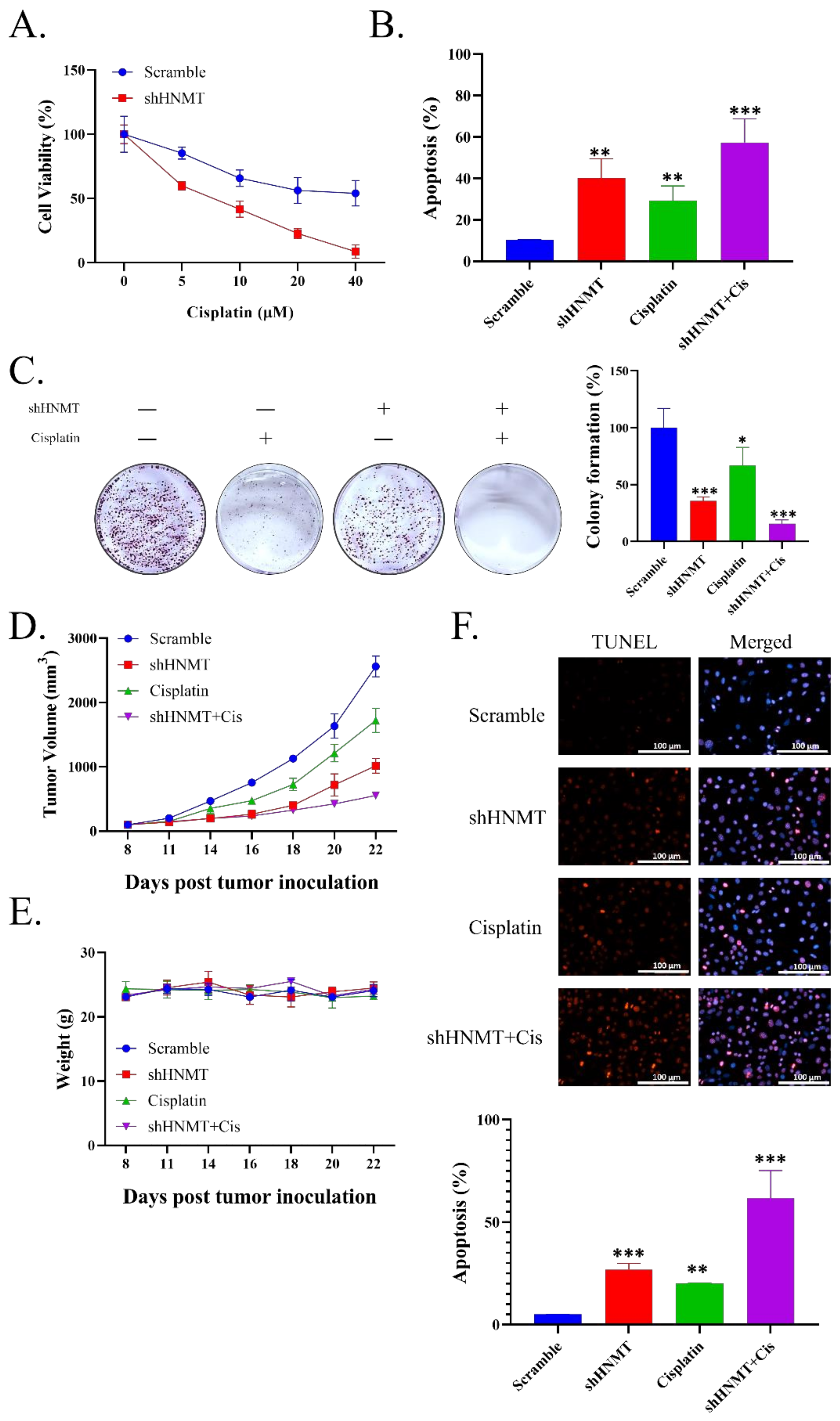
2.6. HNMT Inhibition Sensitizes NSCLC to Cisplatin Chemotherapy Both at In Vivo and In Vitro
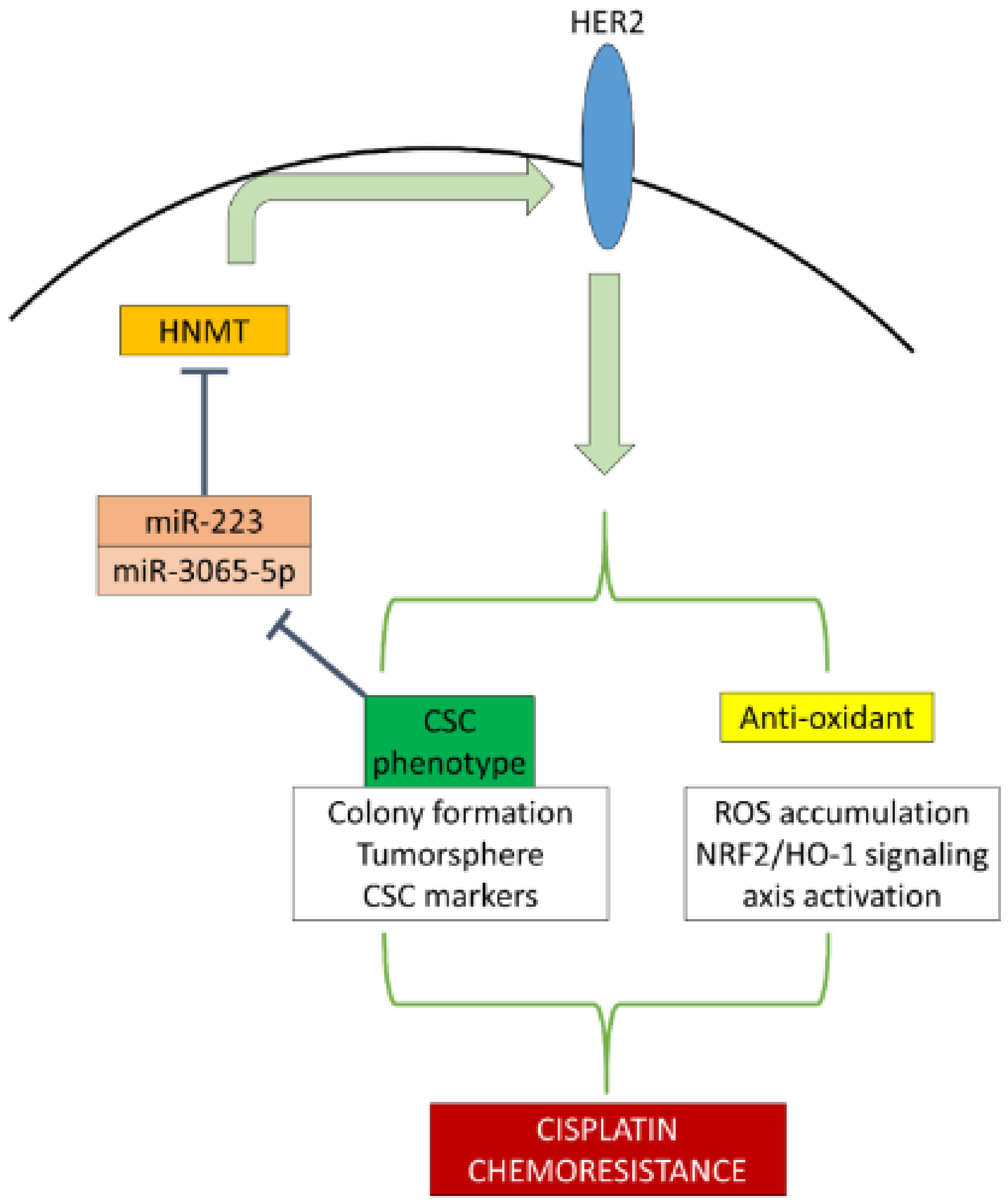
3. Discussion
4. Methods
4.1. Clinical Samples
4.2. Cells and Culture Medium
4.3. Cell Stable Transfection
4.4. Co-Immunoprecipitation (Co-IP)
4.5. Western Blotting
4.6. Total RNA Isolation and Qunatitiave Reverse-Transcription Polymerase Chain Reaction (qRT-PCR)
4.7. Clonogenic Assay
4.8. Sulforhodamine B Assay
4.9. Immunofluorescence Assay
4.10. ROS Production Measurement
4.11. Tumor Xenograft Study
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Subramanian, J.; Morgensztern, D.; Goodgame, B.; Baggstrom, M.Q.; Gao, F.; Piccirillo, J.; Govindan, R. Distinctive characteristics of non-small cell lung cancer (NSCLC) in the young: A surveillance, epidemiology, and end results (SEER) analysis. J. Thorac. Oncol. 2010, 5, 23–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, A. Chemotherapy, chemoresistance and the changing treatment landscape for NSCLC. Lung Cancer 2011, 71, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Zhong, G.; Qin, S.; Townsend, D.; Schulte, B.A.; Tew, K.D.; Wang, G.Y. Oxidative stress induces senescence in breast cancer stem cells. Biochem. Biophys. Res. Commun. 2019, 514, 1204–1209. [Google Scholar] [CrossRef] [PubMed]
- Kharkar, P.S. Cancer Stem Cell (CSC) Inhibitors in Oncology-A Promise for a Better Therapeutic Outcome: State of the Art and Future Perspectives. J. Med. Chem. 2020, 63, 15279–15307. [Google Scholar] [CrossRef] [PubMed]
- Pirpour Tazehkand, A.; Akbarzadeh, M.; Velaie, K.; Sadeghi, M.R.; Samadi, N. The role of Her2-Nrf2 axis in induction of oxaliplatin resistance in colon cancer cells. Biomed. Pharm. 2018, 103, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Chio, I.I.C.; Tuveson, D.A. ROS in Cancer: The Burning Question. Trends Mol. Med. 2017, 23, 411–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zipper, L.M.; Mulcahy, R.T. Inhibition of ERK and p38 MAP kinases inhibits binding of Nrf2 and induction of GCS genes. Biochem. Biophys. Res. Commun. 2000, 278, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.J.; Yi, Y.W.; Hong, Y.B.; Kim, H.J.; Jang, Y.J.; Seong, Y.S.; Bae, I. HER2 confers drug resistance of human breast cancer cells through activation of NRF2 by direct interaction. Sci. Rep. 2014, 4, 7201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, R.E.; Stevens, D.R.; Haas, H.L. The physiology of brain histamine. Prog. Neurobiol. 2001, 63, 637–672. [Google Scholar] [CrossRef]
- Haas, H.L.; Panula, P. Histamine receptors. Neuropharmacology 2016, 106, 1–2. [Google Scholar] [CrossRef] [Green Version]
- Leurs, R.; Smit, M.J.; Timmerman, H. Molecular pharmacological aspects of histamine receptors. Pharmacol. Ther. 1995, 66, 413–463. [Google Scholar] [CrossRef]
- von Mach-Szczypinski, J.; Stanosz, S.; Sieja, K.; Stanosz, M. Histamine and its metabolizing enzymes in tissues of primary ductal breast cancer. Eur. J. Gynaecol. Oncol. 2009, 30, 509–511. [Google Scholar] [PubMed]
- Tanaka, S.; Sakaguchi, M.; Yoneyama, H.; Usami, Y.; Harusawa, S. Histamine H3 receptor antagonist OUP-186 attenuates the proliferation of cultured human breast cancer cell lines. Biochem. Biophys. Res. Commun. 2016, 480, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, H.; Kitada, K.; Nakai, K.; Sarai, A. PrognoScan: A new database for meta-analysis of the prognostic value of genes. BMC Med. Genom. 2009, 2, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ricciardi, G.R.; Russo, A.; Franchina, T.; Ferraro, G.; Zanghi, M.; Picone, A.; Scimone, A.; Adamo, V. NSCLC and HER2: Between lights and shadows. J. Thorac. Oncol. 2014, 9, 1750–1762. [Google Scholar] [CrossRef] [Green Version]
- Koster, J. R2: Genomics Analysis and Visualization Platform. Available online: http://r2.amc.nl (accessed on 21 November 2021).
- Guda, R.; Kumar, G.; Korra, R.; Balaji, S.; Dayakar, G.; Palabindela, R.; Myadaraveni, P.; Yellu, N.R.; Kasula, M. EGFR, HER2 target based molecular docking analysis, in vitro screening of 2, 4, 5-trisubstituted imidazole derivatives as potential anti-oxidant and cytotoxic agents. J. Photochem. Photobiol. B 2017, 176, 69–80. [Google Scholar] [CrossRef]
- Pino-Angeles, A.; Reyes-Palomares, A.; Melgarejo, E.; Sanchez-Jimenez, F. Histamine: An undercover agent in multiple rare diseases? J. Cell. Mol. Med. 2012, 16, 1947–1960. [Google Scholar] [CrossRef] [Green Version]
- Francis, H.; DeMorrow, S.; Venter, J.; Onori, P.; White, M.; Gaudio, E.; Francis, T.; Greene, J.F., Jr.; Tran, S.; Meininger, C.J.; et al. Inhibition of histidine decarboxylase ablates the autocrine tumorigenic effects of histamine in human cholangiocarcinoma. Gut 2012, 61, 753–764. [Google Scholar] [CrossRef] [Green Version]
- Shan, L.; Bao, A.M.; Swaab, D.F. Changes in Histidine Decarboxylase, Histamine N-Methyltransferase and Histamine Receptors in Neuropsychiatric Disorders. Handb. Exp. Pharmacol. 2017, 241, 259–276. [Google Scholar] [CrossRef]
- Krystal, A.D.; Richelson, E.; Roth, T. Review of the histamine system and the clinical effects of H1 antagonists: Basis for a new model for understanding the effects of insomnia medications. Sleep Med. Rev. 2013, 17, 263–272. [Google Scholar] [CrossRef]
- Schwelberger, H.G.; Feurle, J.; Houen, G. Mapping of the binding sites of human histamine N-methyltransferase (HNMT) monoclonal antibodies. Inflamm. Res. 2017, 66, 1021–1029. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.Q.; Zhu, S.T.; Wang, M.; Guo, S.L.; Sun, X.J.; Cheng, R.; Xing, J.; Wang, W.H.; Shao, L.L.; Zhang, S.T. Pathway analysis of differentially expressed genes in human esophageal squamous cell carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2015, 19, 1652–1661. [Google Scholar] [PubMed]
- Gao, H.Y.; Luo, X.G.; Chen, X.; Wang, J.H. Identification of key genes affecting disease free survival time of pediatric acute lymphoblastic leukemia based on bioinformatic analysis. Blood Cells Mol. Dis. 2015, 54, 38–43. [Google Scholar] [CrossRef] [PubMed]
- Roh, T.; Kwak, M.Y.; Kwak, E.H.; Kim, D.H.; Han, E.Y.; Bae, J.Y.; Bang du, Y.; Lim, D.S.; Ahn, I.Y.; Jang, D.E.; et al. Chemopreventive mechanisms of methionine on inhibition of benzo(a)pyrene-DNA adducts formation in human hepatocellular carcinoma HepG2 cells. Toxicol. Lett. 2012, 208, 232–238. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.F.; Cheng, J.; Wang, J.W.; Song, Y.; Gu, S.X. Histamine and lung cancer. II. Human lung cancer cell line and histamine. Zhonghua Zhong Liu Za Zhi 1988, 10, 19–22. [Google Scholar] [PubMed]
- Kondratenko, T.Y.; Zacharova, I.V.; Katukov, V.; Kuzina, N.V.; Severin, E.S.; Kornilova, Z.; Perelman, M.I. The study of histamine H1- and H2-receptors in human lung cancer. Biochem. Mol. Biol. Int. 1993, 31, 399–404. [Google Scholar]
- Yoshioka, Y.; Suzuki, T.; Matsuo, Y.; Tsurita, G.; Watanabe, T.; Dohmae, N.; Nakamura, Y.; Hamamoto, R. Protein lysine methyltransferase SMYD3 is involved in tumorigenesis through regulation of HER2 homodimerization. Cancer Med. 2017, 6, 1665–1672. [Google Scholar] [CrossRef]
- Slamon, D.J.; Clark, G.M.; Wong, S.G.; Levin, W.J.; Ullrich, A.; McGuire, W.L. Human breast cancer: Correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 1987, 235, 177–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, H.; Kawasaki, N.; Taguchi, M.; Kabasawa, K. Association of HER-2 overexpression with prognosis in nonsmall cell lung carcinoma: A metaanalysis. Cancer 2005, 103, 1865–1873. [Google Scholar] [CrossRef]
- Liu, L.; Shao, X.; Gao, W.; Bai, J.; Wang, R.; Huang, P.; Yin, Y.; Liu, P.; Shu, Y. The role of human epidermal growth factor receptor 2 as a prognostic factor in lung cancer: A meta-analysis of published data. J. Thorac. Oncol. 2010, 5, 1922–1932. [Google Scholar] [CrossRef] [Green Version]
- Shitara, K.; Yatabe, Y.; Matsuo, K.; Sugano, M.; Kondo, C.; Takahari, D.; Ura, T.; Tajika, M.; Ito, S.; Muro, K. Prognosis of patients with advanced gastric cancer by HER2 status and trastuzumab treatment. Gastric Cancer 2013, 16, 261–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazieres, J.; Barlesi, F.; Filleron, T.; Besse, B.; Monnet, I.; Beau-Faller, M.; Peters, S.; Dansin, E.; Fruh, M.; Pless, M.; et al. Lung cancer patients with HER2 mutations treated with chemotherapy and HER2-targeted drugs: Results from the European EUHER2 cohort. Ann. Oncol. 2016, 27, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Korkaya, H.; Paulson, A.; Iovino, F.; Wicha, M.S. HER2 regulates the mammary stem/progenitor cell population driving tumorigenesis and invasion. Oncogene 2008, 27, 6120–6130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magnifico, A.; Albano, L.; Campaner, S.; Delia, D.; Castiglioni, F.; Gasparini, P.; Sozzi, G.; Fontanella, E.; Menard, S.; Tagliabue, E. Tumor-initiating cells of HER2-positive carcinoma cell lines express the highest oncoprotein levels and are sensitive to trastuzumab. Clin. Cancer Res. 2009, 15, 2010–2021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Honkanen, T.; Wilenius, E.; Koivunen, P.; Koivunen, J.P. HER2 regulates cancer stem-like cell phenotype in ALK translocated NSCLC. Int. J. Oncol. 2017, 51, 599–606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Gao, Y.; Hai, J.; Yang, J.; Duan, S. HER2 decreases drug sensitivity of ovarian cancer cells via inducing stem cell-like property in an NFkappaB-dependent way. Biosci. Rep. 2019, 39, BSR20180829. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, S.; Okada, M.; Shibuya, K.; Seino, M.; Sato, A.; Takeda, H.; Seino, S.; Yoshioka, T.; Kitanaka, C. JNK suppression of chemotherapeutic agents-induced ROS confers chemoresistance on pancreatic cancer stem cells. Oncotarget 2015, 6, 458–470. [Google Scholar] [CrossRef] [Green Version]
- Jia, H.Z.; Liu, S.L.; Zou, Y.F.; Chen, X.F.; Yu, L.; Wan, J.; Zhang, H.Y.; Chen, Q.; Xiong, Y.; Yu, B.; et al. MicroRNA-223 is involved in the pathogenesis of atopic dermatitis by affecting histamine-N-methyltransferase. Cell Mol. Biol. 2018, 64, 103–107. [Google Scholar] [CrossRef]
- Favero, A.; Segatto, I.; Perin, T.; Belletti, B. The many facets of miR-223 in cancer: Oncosuppressor, oncogenic driver, therapeutic target, and biomarker of response. Wiley Interdiscip. Rev. RNA 2021, 12, e1659. [Google Scholar] [CrossRef]
- Ding, S.; Huang, X.; Zhu, J.; Xu, B.; Xu, L.; Gu, D.; Zhang, W. ADH7, miR-3065 and LINC01133 are associated with cervical cancer progression in different age groups. Oncol. Lett. 2020, 19, 2326–2338. [Google Scholar] [CrossRef]
- Muller, S.; Nowak, K. Exploring the miRNA-mRNA regulatory network in clear cell renal cell carcinomas by next-generation sequencing expression profiles. BioMed Res. Int. 2014, 2014, 948408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Clinicopathological Variables | N = 60 | HNMT | x2 | p-Value | |
|---|---|---|---|---|---|
| High Expression | Low Expression | ||||
| Age, years | |||||
| ≤60 | 29 | 11 | 18 | 1.133 | 0.287 |
| >60 | 31 | 16 | 15 | ||
| Gender | |||||
| Male | 37 | 18 | 19 | 0.519 | 0.471 |
| Female | 23 | 9 | 14 | ||
| Differentiation | |||||
| Well/Moderately | 38 | 13 | 25 | 4.875 | 0.027 |
| Poor | 22 | 14 | 8 | ||
| Tumor Size (cm) | |||||
| ≤5 | 39 | 12 | 27 | 9.118 | 0.003 |
| >5 | 21 | 15 | 6 | ||
| Lymph node metastasis | |||||
| N0 | 36 | 11 | 25 | 7.587 | 0.006 |
| N1-N2 | 24 | 16 | 8 | ||
| Clinical Stage | |||||
| Early-stage (I-II) | 30 | 7 | 23 | 11.38 | <0.001 |
| Late-stage (III-IV) | 30 | 20 | 10 | ||
| Subtypes | |||||
| Adeno | 37 | 20 | 17 | 3.197 | 0.074 |
| Squamous | 23 | 7 | 16 | ||
| HER2 Q score | |||||
| Low | 35 | 11 | 24 | 6.251 | 0.012 |
| High | 25 | 16 | 9 | ||
| Parameter | Univariate | Multivariate | ||||||
|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | p | HR | 95% CI | p | |||
| Gender (Female vs. male) | 0.679 | 0.274 | 1.683 | 0.403 | 0.594 | 0.179 | 1.971 | 0.395 |
| age_60 (<60 vs. >60) | 0.857 | 0.360 | 2.037 | 0.726 | 0.624 | 0.211 | 1.840 | 0.392 |
| tumor_50 (<50 vs. >50) | 0.721 | 0.297 | 1.753 | 0.471 | 0.296 | 0.054 | 1.632 | 0.162 |
| LN_12 (1,2 vs. 0) | 0.675 | 0.278 | 1.637 | 0.384 | 0.427 | 0.125 | 1.458 | 0.174 |
| Subtype_c(squa vs. adeno) | 0.999 | 0.413 | 2.412 | 0.998 | 0.738 | 0.224 | 2.431 | 0.618 |
| stage (late vs. early) | 1.140 | 0.479 | 2.711 | 0.768 | 1.011 | 0.316 | 3.235 | 0.986 |
| Differentiation_c(well, moderately vs. poor) | 0.952 | 0.400 | 2.267 | 0.911 | 0.472 | 0.099 | 2.255 | 0.347 |
| HNMT (low vs. high) | 0.294 | 0.108 | 0.806 | 0.017 | 0.152 | 0.047 | 0.495 | 0.002 |
| HER2(low vs. high) | 1.061 | 0.442 | 2.546 | 0.894 | 1.346 | 0.408 | 4.445 | 0.626 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuo, K.-T.; Lin, C.-H.; Wang, C.-H.; Pikatan, N.W.; Yadav, V.K.; Fong, I.-H.; Yeh, C.-T.; Lee, W.-H.; Huang, W.-C. HNMT Upregulation Induces Cancer Stem Cell Formation and Confers Protection against Oxidative Stress through Interaction with HER2 in Non-Small-Cell Lung Cancer. Int. J. Mol. Sci. 2022, 23, 1663. https://doi.org/10.3390/ijms23031663
Kuo K-T, Lin C-H, Wang C-H, Pikatan NW, Yadav VK, Fong I-H, Yeh C-T, Lee W-H, Huang W-C. HNMT Upregulation Induces Cancer Stem Cell Formation and Confers Protection against Oxidative Stress through Interaction with HER2 in Non-Small-Cell Lung Cancer. International Journal of Molecular Sciences. 2022; 23(3):1663. https://doi.org/10.3390/ijms23031663
Chicago/Turabian StyleKuo, Kuang-Tai, Cheng-Hsin Lin, Chun-Hua Wang, Narpati Wesa Pikatan, Vijesh Kumar Yadav, Iat-Hang Fong, Chi-Tai Yeh, Wei-Hwa Lee, and Wen-Chien Huang. 2022. "HNMT Upregulation Induces Cancer Stem Cell Formation and Confers Protection against Oxidative Stress through Interaction with HER2 in Non-Small-Cell Lung Cancer" International Journal of Molecular Sciences 23, no. 3: 1663. https://doi.org/10.3390/ijms23031663
APA StyleKuo, K.-T., Lin, C.-H., Wang, C.-H., Pikatan, N. W., Yadav, V. K., Fong, I.-H., Yeh, C.-T., Lee, W.-H., & Huang, W.-C. (2022). HNMT Upregulation Induces Cancer Stem Cell Formation and Confers Protection against Oxidative Stress through Interaction with HER2 in Non-Small-Cell Lung Cancer. International Journal of Molecular Sciences, 23(3), 1663. https://doi.org/10.3390/ijms23031663