Intraspecific Diversity of Microbial Anti-Inflammatory Molecule (MAM) from Faecalibacterium prausnitzii
Abstract
:1. Introduction
2. Results
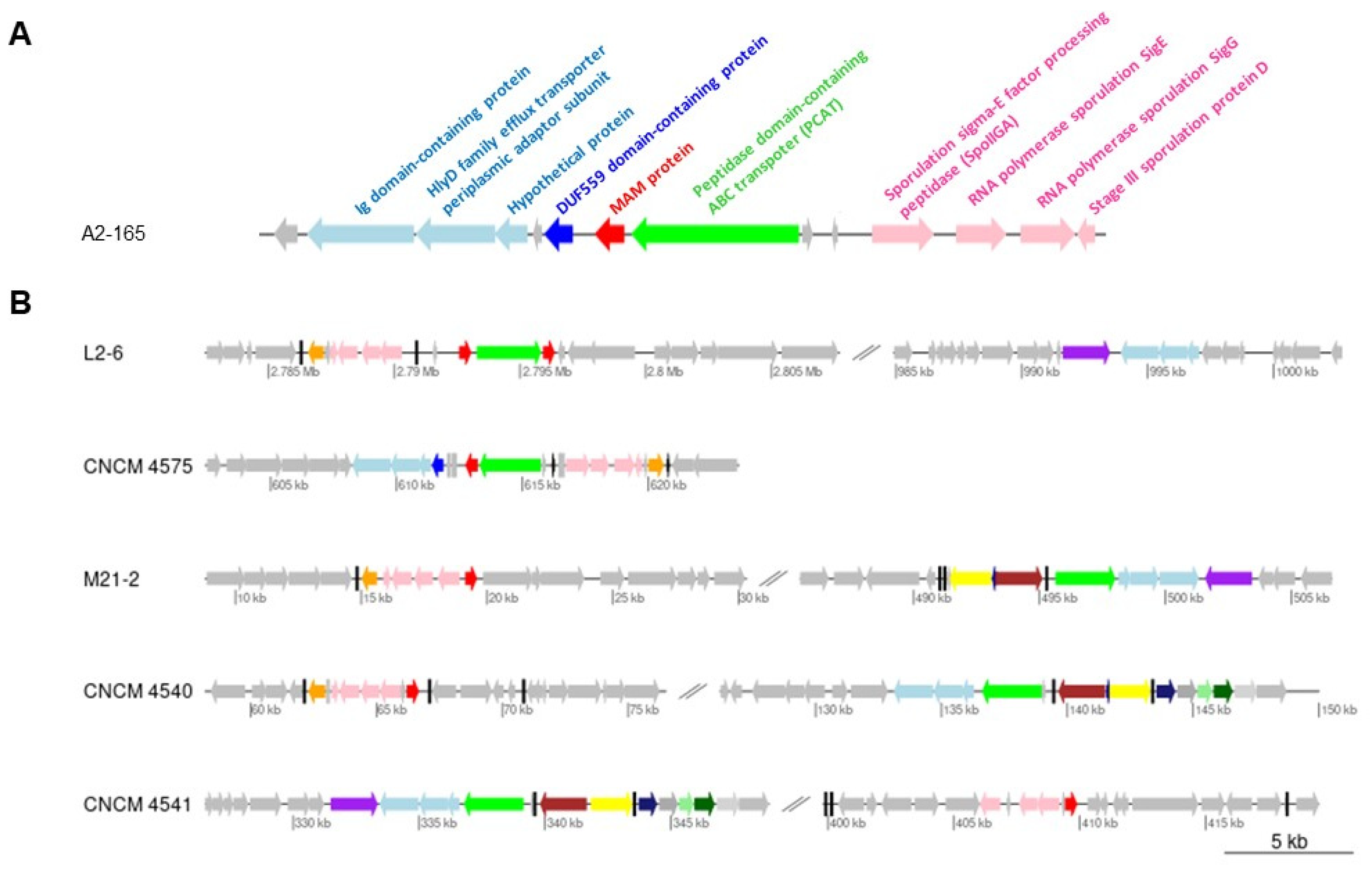
2.1. Gene Context Analysis
2.2. Analysis of the Promoter Regions
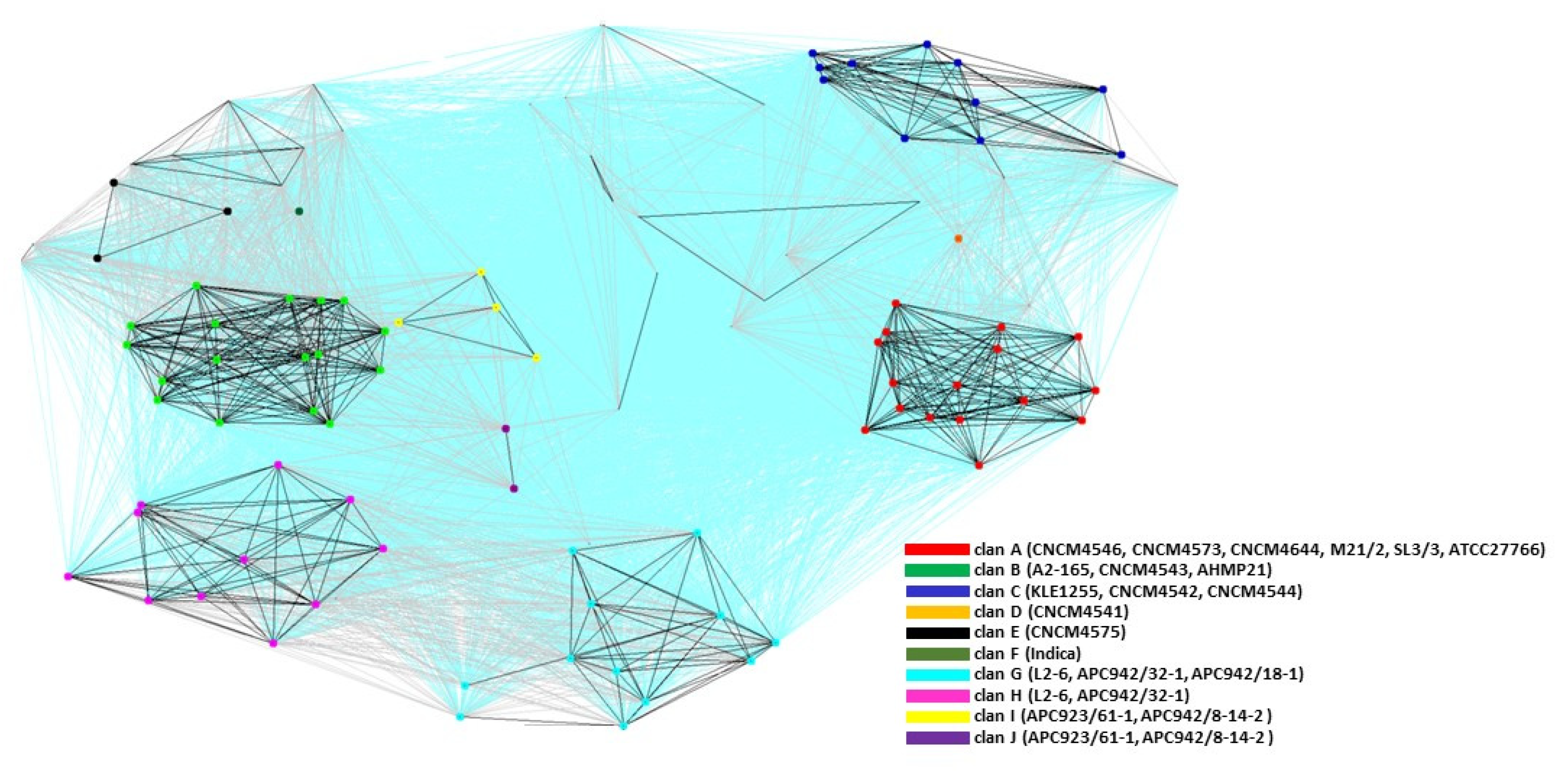
2.3. Cluster Analysis of the MAM Proteins
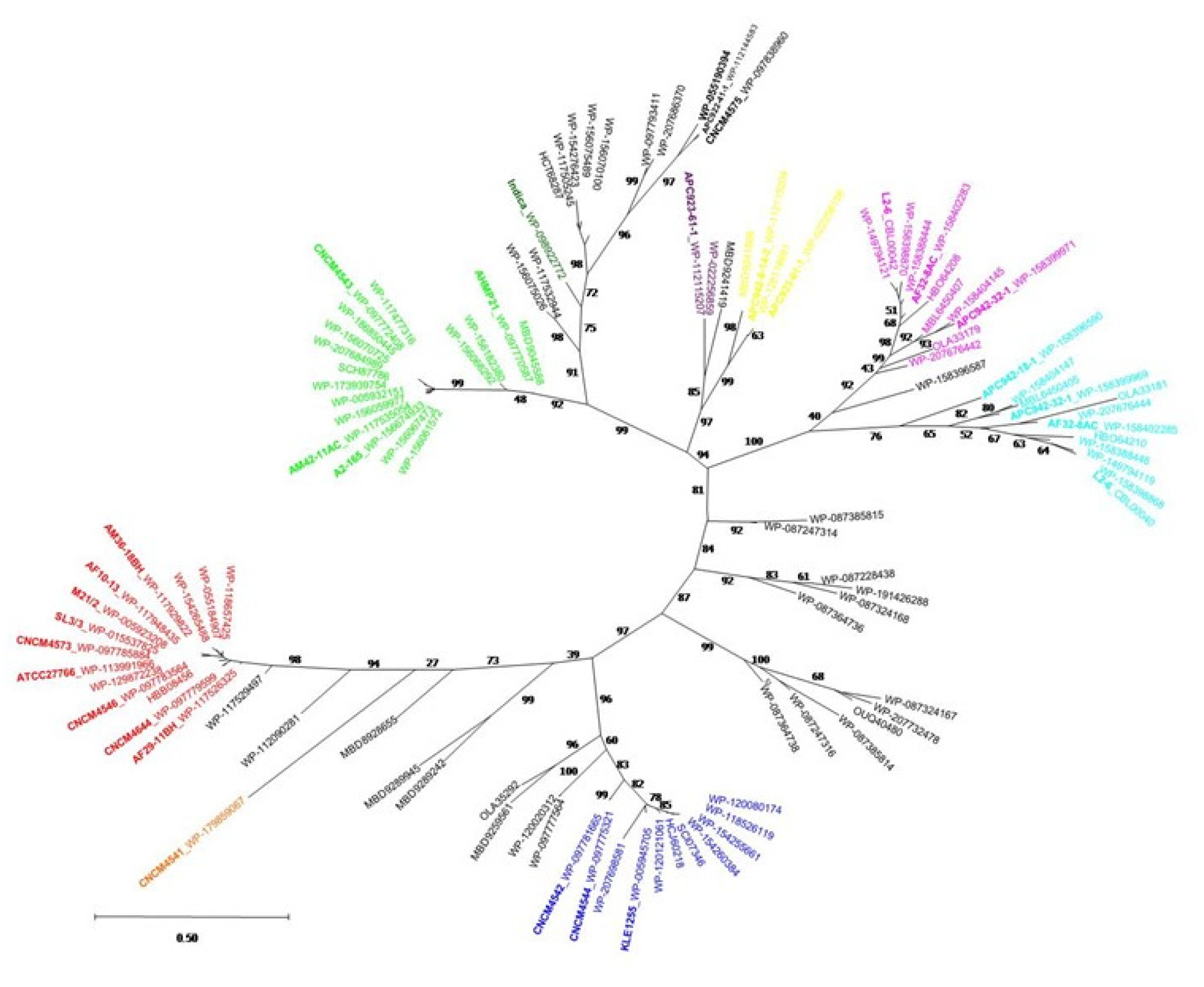
2.4. Phylogenetic Analysis of the MAM Protein Family
2.5. In Vitro Anti-Inflammatory Activity of MAMs from Different Clans
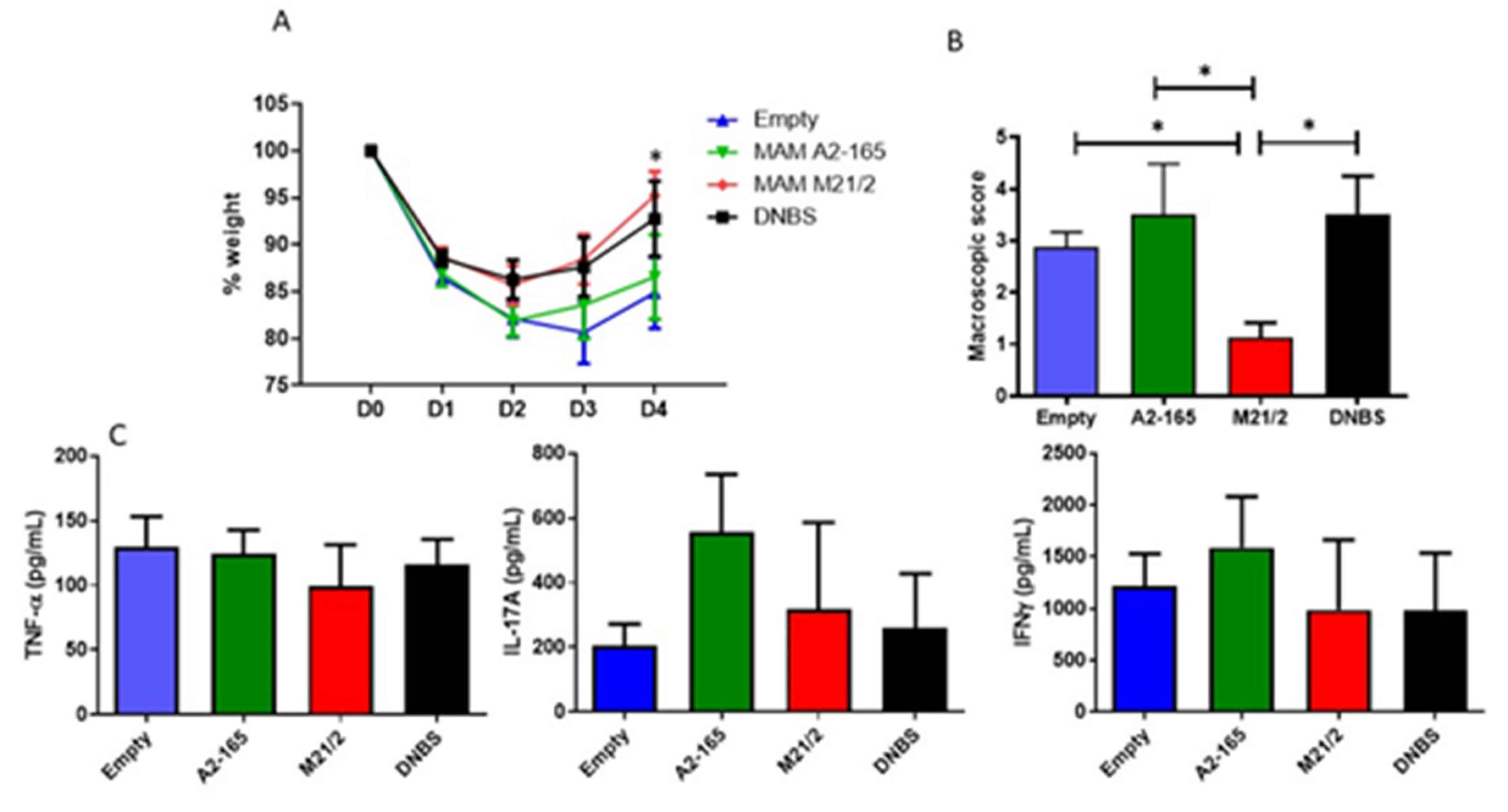
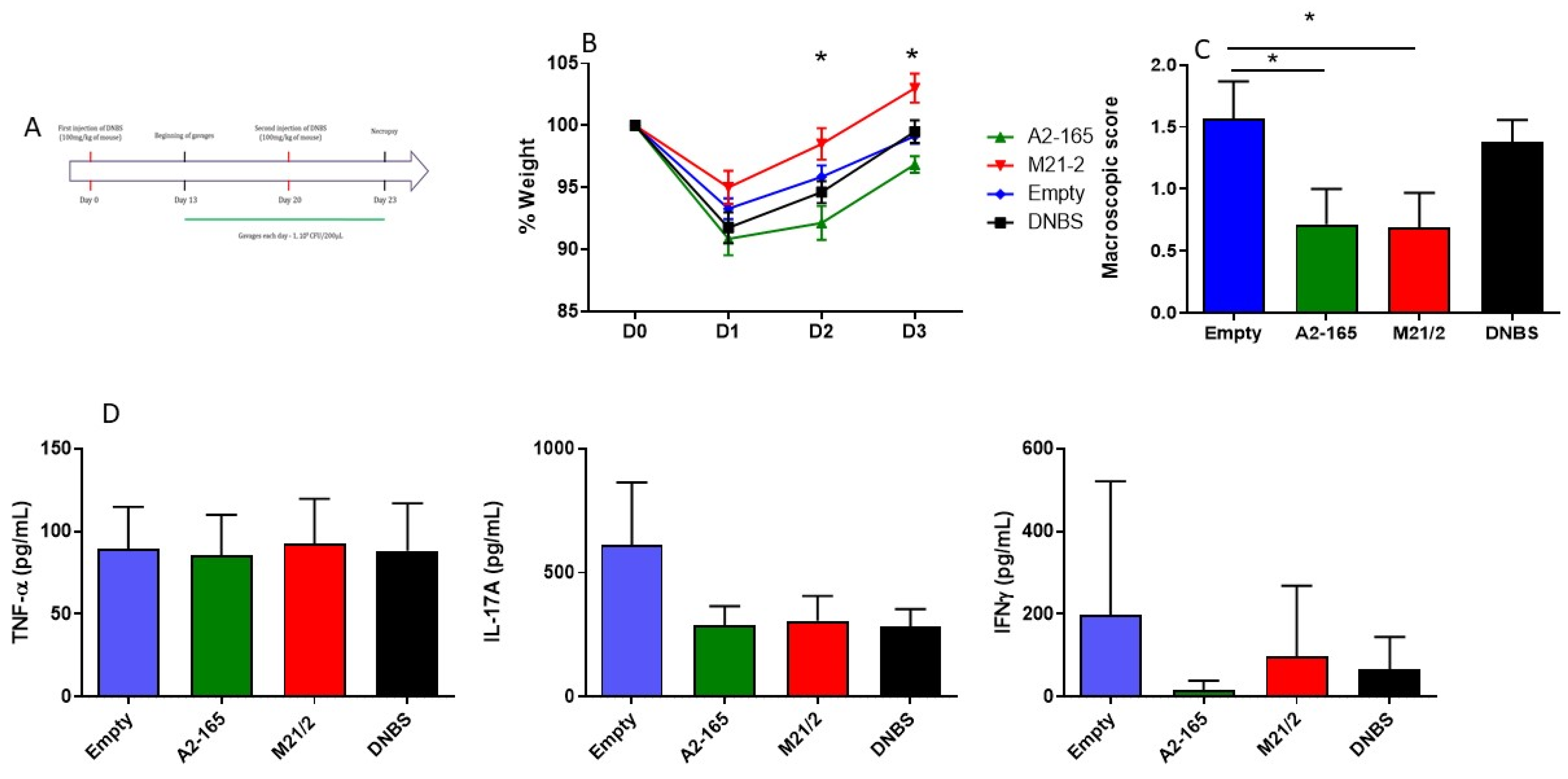
2.6. In Vivo Comparison of Anti-Inflammatory Activity of MAM from F. prausnitzii A2-165 and M21-2 in a Model of Acute Colitis
2.7. In Vivo Comparison of Anti-Inflammatory Activity of MAM from F. prausnitzii A2-165 and M21-2 in a Model of Chronic Colitis
3. Discussion
4. Materials and Methods
4.1. Data Collection, Sequence and Phylogenetic Analysis
4.2. Plasmid and Strains
4.3. HEK-293 Culture Conditions, Transfected Cell Line, NF-κB Reporter Assay and MAM Immunoblotting
4.4. Animals and Experimental Design
4.5. Lymphocyte Isolation and Measurement of Cytokine Production
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Quevrain, E.; Maubert, M.A.; Michon, C.; Chain, F.; Marquant, R.; Tailhades, J.; Miquel, S.; Carlier, L.; Bermudez-Humaran, L.G.; Pigneur, B.; et al. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn’s disease. Gut 2016, 65, 415–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermúdez-Humarán, L.G.; Gratadoux, J.J.; Blugeon, S.; Bridonneau, C.; Furet, J.P.; Corthier, G.; et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA 2008, 105, 16731–16736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, J.; Tang, H.; Li, M.; Pang, X.; Wang, L.; Zhang, M.; Zhao, Y.; Zhang, X.; Shen, J. The abundance of fecal Faecalibacterium prausnitzii in relation to obesity and gender in Chinese adults. Arch. Microbiol. 2014, 196, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Saitoh, S.; Noda, S.; Aiba, Y.; Takagi, A.; Sakamoto, M.; Benno, Y.; Koga, Y. Bacteroides ovatus as the predominant commensal intestinal microbe causing a systemic antibody response in inflammatory bowel disease. Clin. Diagn. Lab. Immunol. 2002, 9, 54–59. [Google Scholar] [CrossRef] [Green Version]
- Zhou, L.; Zhang, M.; Wang, Y.; Dorfman, R.G.; Liu, H.; Yu, T.; Chen, X.; Tang, D.; Xu, L.; Yin, Y.; et al. Faecalibacterium prausnitzii Produces Butyrate to Maintain Th17/Treg Balance and to Ameliorate Colorectal Colitis by Inhibiting Histone Deacetylase 1. Inflamm. Bowel Dis. 2018, 24, 1926–1940. [Google Scholar] [CrossRef] [Green Version]
- Rossi, O.; Khan, M.T.; Schwarzer, M.; Hudcovic, T.; Srutkova, D.; Duncan, S.H.; Stolte, E.H.; Kozakova, H.; Flint, H.J.; Samsom, J.N.; et al. Faecalibacterium prausnitzii Strain HTF-F and Its Extracellular Polymeric Matrix Attenuate Clinical Parameters in DSS-Induced Colitis. PLoS ONE 2015, 10, e0123013. [Google Scholar] [CrossRef] [Green Version]
- Benus, R.F.J.; Van Der Werf, T.S.; Welling, G.W.; Judd, P.A.; Taylor, M.A.; Harmsen, H.J.M.; Whelan, K. Association between Faecalibacterium prausnitzii and dietary fibre in colonic fermentation in healthy human subjects. Br. J. Nutr. 2010, 104, 693–700. [Google Scholar] [CrossRef] [Green Version]
- Breyner, N.M.; Michon, C.; de Sousa, C.S.; Vilas Boas, P.B.; Chain, F.; Azevedo, V.A.; Langella, P.; Chatel, J.M. Microbial anti-inflammatory molecule (MAM) from Faecalibacterium prausnitzii shows a protective effect on DNBS and DSS-induced colitis model in mice through inhibition of NF-κB pathway. Front. Microbiol. 2017, 8, 114. [Google Scholar] [CrossRef] [Green Version]
- O’Toole, P.W.; Marchesi, J.R.; Hill, C. Next-generation probiotics: The spectrum from probiotics to live biotherapeutics. Nat. Microbiol. 2017, 2, 17057. [Google Scholar] [CrossRef]
- Miquel, S.; Martin, R.; Rossi, O.; Bermudez-Humaran, L.G.; Chatel, J.M.; Sokol, H.; Thomas, M.; Wells, J.M.; Langella, P. Faecalibacterium prausnitzii and human intestinal health. Curr. Opin. Microbiol. 2013, 16, 255–261. [Google Scholar] [CrossRef]
- Martin, R.; Bermudez-Humaran, L.G.; Langella, P. Searching for the Bacterial Effector: The Example of the Multi-Skilled Commensal Bacterium Faecalibacterium prausnitzii. Front. Microbiol. 2018, 9, 346. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.; Miquel, S.; Benevides, L.; Bridonneau, C.; Robert, V.; Hudault, S.; Chain, F.; Berteau, O.; Azevedo, V.; Chatel, J.M.; et al. Functional Characterization of Novel Faecalibacterium prausnitzii Strains Isolated from Healthy Volunteers: A Step Forward in the Use of F. prausnitzii as a Next-Generation Probiotic. Front. Microbiol. 2017, 8, 1226. [Google Scholar] [CrossRef] [PubMed]
- Benevides, L.; Burman, S.; Martin, R.; Robert, V.; Thomas, M.; Miquel, S.; Chain, F.; Sokol, H.; Bermudez-Humaran, L.G.; Morrison, M.; et al. New Insights into the Diversity of the Genus Faecalibacterium. Front. Microbiol. 2017, 8, 1790. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Siles, M.; Khan, T.M.; Duncan, S.H.; Harmsen, H.J.M.; Garcia-Gil, L.J.; Flint, H.J. Cultured representatives of two major phylogroups of human colonic Faecalibacterium prausnitzii can utilize pectin, uronic acids, and host-derived substrates for growth. Appl. Environ. Microbiol. 2012, 78, 420–428. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Siles, M.; Martinez-Medina, M.; Surís-Valls, R.; Aldeguer, X.; Sabat-Mir, M.; Duncan, S.H.; Flint, H.J.; Garcia-Gil, L.J. Changes in the Abundance of Faecalibacterium prausnitzii Phylogroups I and II in the Intestinal Mucosa of Inflammatory Bowel Disease and Patients with Colorectal Cancer. Inflamm. Bowel Dis. 2016, 22, 28–41. [Google Scholar] [CrossRef] [Green Version]
- Fitzgerald, C.B.; Shkoporov, A.N.; Sutton, T.D.S.; Chaplin, A.V.; Velayudhan, V.; Ross, R.P.; Hill, C. Comparative analysis of Faecalibacterium prausnitzii genomes shows a high level of genome plasticity and warrants separation into new species-level taxa. BMC Genom. 2018, 19, 931. [Google Scholar] [CrossRef] [Green Version]
- Zou, Y.; Lin, X.; Xue, W.; Tuo, L.; Chen, M.S.; Chen, X.H.; Sun, C.H.; Li, F.; Liu, S.W.; Dai, Y.; et al. Characterization and description of Faecalibacterium butyricigenerans sp. nov. and F. longum sp. nov., isolated from human faeces. Sci. Rep. 2021, 11, 11340. [Google Scholar] [CrossRef]
- De Filippis, F.; Pasolli, E.; Ercolini, D. Newly Explored Faecalibacterium Diversity Is Connected to Age, Lifestyle, Geography, and Disease. Curr. Biol. 2020, 30, 4932–4943. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [Green Version]
- Lebas, M.; Garault, P.; Carrillo, D.; Codoner, F.M.; Derrien, M. Metabolic Response of Faecalibacterium prausnitzii to Cell-Free Supernatants from Lactic Acid Bacteria. Microorganisms 2020, 8, 1528. [Google Scholar] [CrossRef]
- Frickey, T.; Lupas, A. CLANS: A Java application for visualizing protein families based on pairwise similarity. Bioinformatics 2004, 20, 3702–3704. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, P.H.; Touchon, M.; Cury, J.; Rocha, E.P.C. The chromosomal organization of horizontal gene transfer in bacteria. Nat. Commun. 2017, 8, 841. [Google Scholar] [CrossRef] [PubMed]
- Havarstein, L.S.; Diep, D.B.; Nes, I.F. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol. Microbiol. 1995, 16, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Chikindas, M.L.; Weeks, R.; Drider, D.; Chistyakov, V.A.; Dicks, L.M. Functions and emerging applications of bacteriocins. Curr. Opin. Biotechnol. 2018, 49, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.Y.; Medlin, J.S.; Nguyen, D.R.; Disbennett, W.M.; Dawid, S. Molecular Determinants of Substrate Selectivity of a Pneumococcal Rgg-Regulated Peptidase-Containing ABC Transporter. mBio 2020, 11, e02502-19. [Google Scholar] [CrossRef] [Green Version]
- Fusunyan, R.D.; Quinn, J.J.; Fujimoto, M.; MacDermott, R.P.; Sanderson, I.R. Butyrate switches the pattern of chemokine secretion by intestinal epithelial cells through histone acetylation. Mol. Med. 1999, 5, 631–640. [Google Scholar] [CrossRef] [Green Version]
- Kamitani, H.; Ikawa, H.; Hsi, L.C.; Watanabe, T.; DuBois, R.N.; Eling, T.E. Regulation of 12-lipoxygenase in rat intestinal epithelial cells during differentiation and apoptosis induced by sodium butyrate. Arch. Biochem. Biophys. 1999, 368, 45–55. [Google Scholar] [CrossRef]
- Laval, L.; Martin, R.; Natividad, J.N.; Chain, F.; Miquel, S.; Desclee de Maredsous, C.; Capronnier, S.; Sokol, H.; Verdu, E.F.; van Hylckama Vlieg, J.E.; et al. Lactobacillus rhamnosus CNCM I-3690 and the commensal bacterium Faecalibacterium prausnitzii A2-165 exhibit similar protective effects to induced barrier hyper-permeability in mice. Gut Microbes 2015, 6, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Sarrabayrouse, G.; Bossard, C.; Chauvin, J.M.; Jarry, A.; Meurette, G.; Quevrain, E.; Bridonneau, C.; Preisser, L.; Asehnoune, K.; Labarriere, N.; et al. CD4CD8alphaalpha lymphocytes, a novel human regulatory T cell subset induced by colonic bacteria and deficient in patients with inflammatory bowel disease. PLoS Biol. 2014, 12, e1001833. [Google Scholar] [CrossRef] [Green Version]
- Hall, T. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Auger, S.; Kropp, C.; Borras-Nogues, E.; Chanput, W.; Andre-Leroux, G.; Gitton-Quent, O.; Benevides, L.; Breyner, N.; Azevedo, V.; Langella, P.; et al. Intraspecific Diversity of Microbial Anti-Inflammatory Molecule (MAM) from Faecalibacterium prausnitzii. Int. J. Mol. Sci. 2022, 23, 1705. https://doi.org/10.3390/ijms23031705
Auger S, Kropp C, Borras-Nogues E, Chanput W, Andre-Leroux G, Gitton-Quent O, Benevides L, Breyner N, Azevedo V, Langella P, et al. Intraspecific Diversity of Microbial Anti-Inflammatory Molecule (MAM) from Faecalibacterium prausnitzii. International Journal of Molecular Sciences. 2022; 23(3):1705. https://doi.org/10.3390/ijms23031705
Chicago/Turabian StyleAuger, Sandrine, Camille Kropp, Esther Borras-Nogues, Wasaporn Chanput, Gwenaelle Andre-Leroux, Oscar Gitton-Quent, Leandro Benevides, Natalia Breyner, Vasco Azevedo, Philippe Langella, and et al. 2022. "Intraspecific Diversity of Microbial Anti-Inflammatory Molecule (MAM) from Faecalibacterium prausnitzii" International Journal of Molecular Sciences 23, no. 3: 1705. https://doi.org/10.3390/ijms23031705
APA StyleAuger, S., Kropp, C., Borras-Nogues, E., Chanput, W., Andre-Leroux, G., Gitton-Quent, O., Benevides, L., Breyner, N., Azevedo, V., Langella, P., & Chatel, J.-M. (2022). Intraspecific Diversity of Microbial Anti-Inflammatory Molecule (MAM) from Faecalibacterium prausnitzii. International Journal of Molecular Sciences, 23(3), 1705. https://doi.org/10.3390/ijms23031705






