Epithelial Cell-Derived Extracellular Vesicles Trigger the Differentiation of Two Epithelial Cell Lines
Abstract
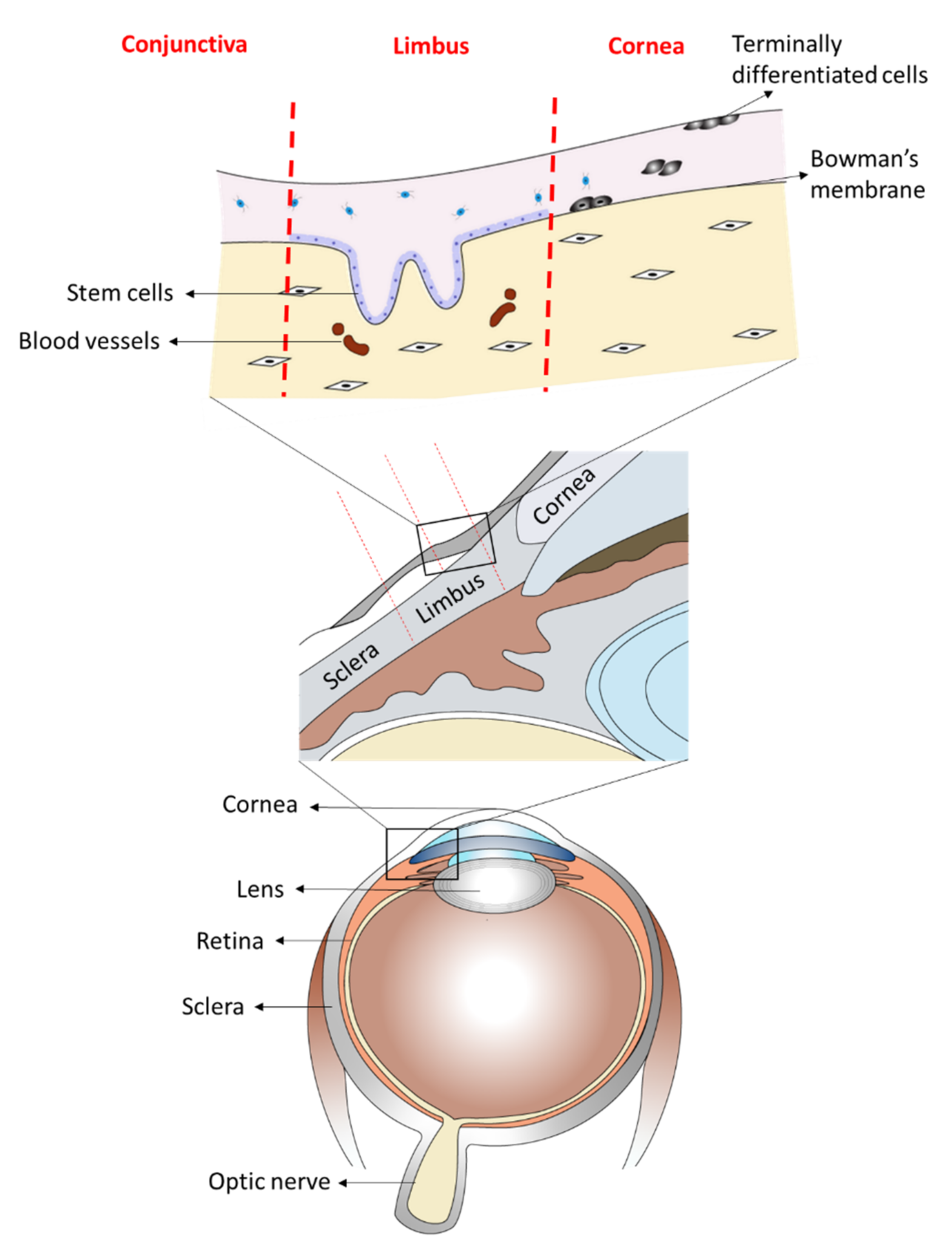
:1. Introduction
2. Results
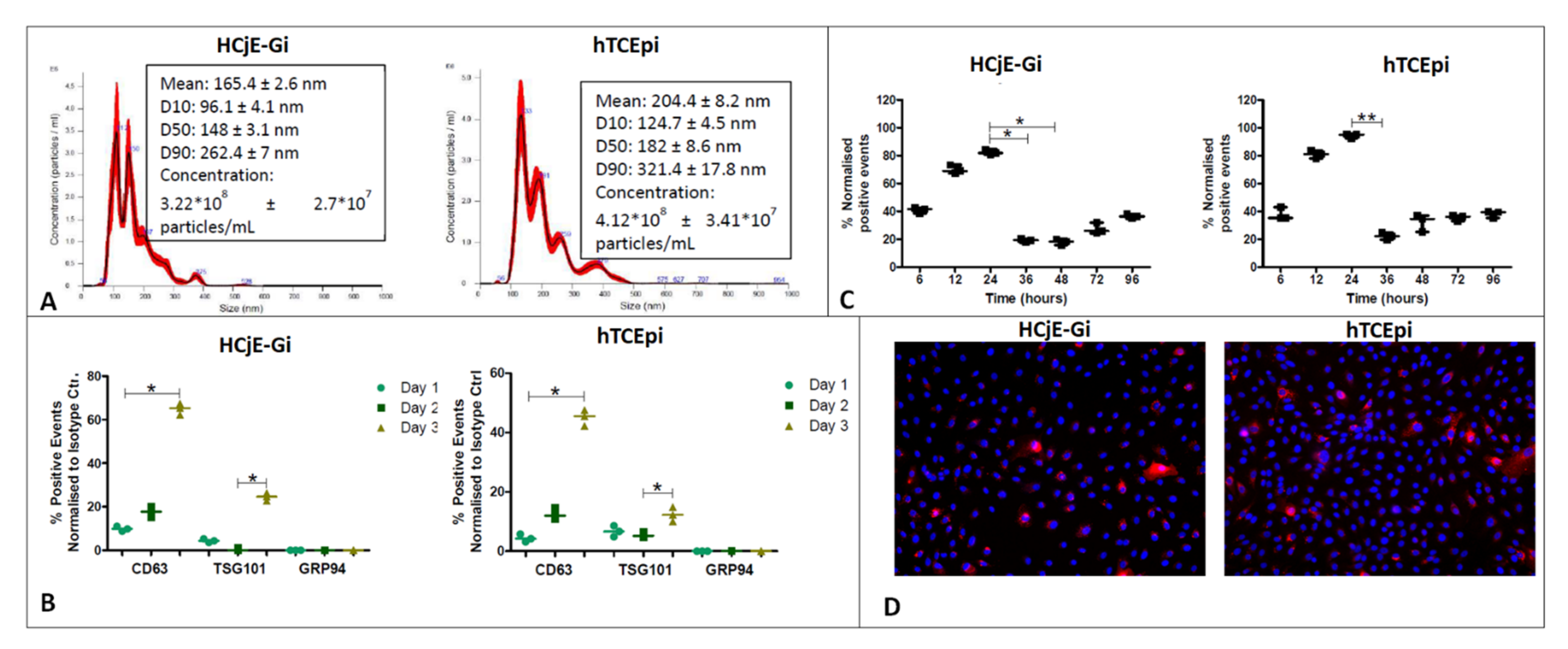
2.1. Isolation, Characterization and Uptake of Corneal and Conjunctival Epithelial Cells-Derived Extracellular Vesicles
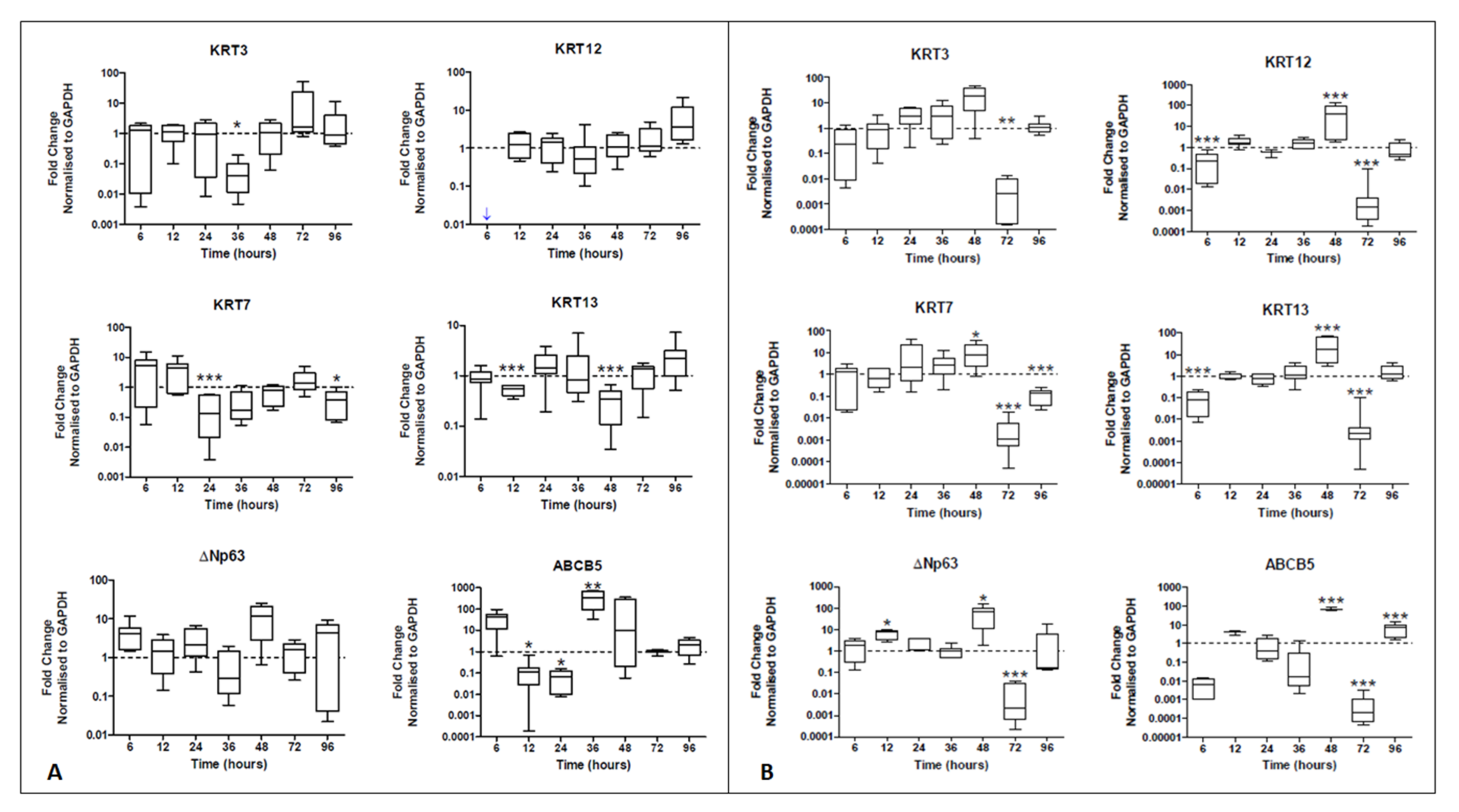
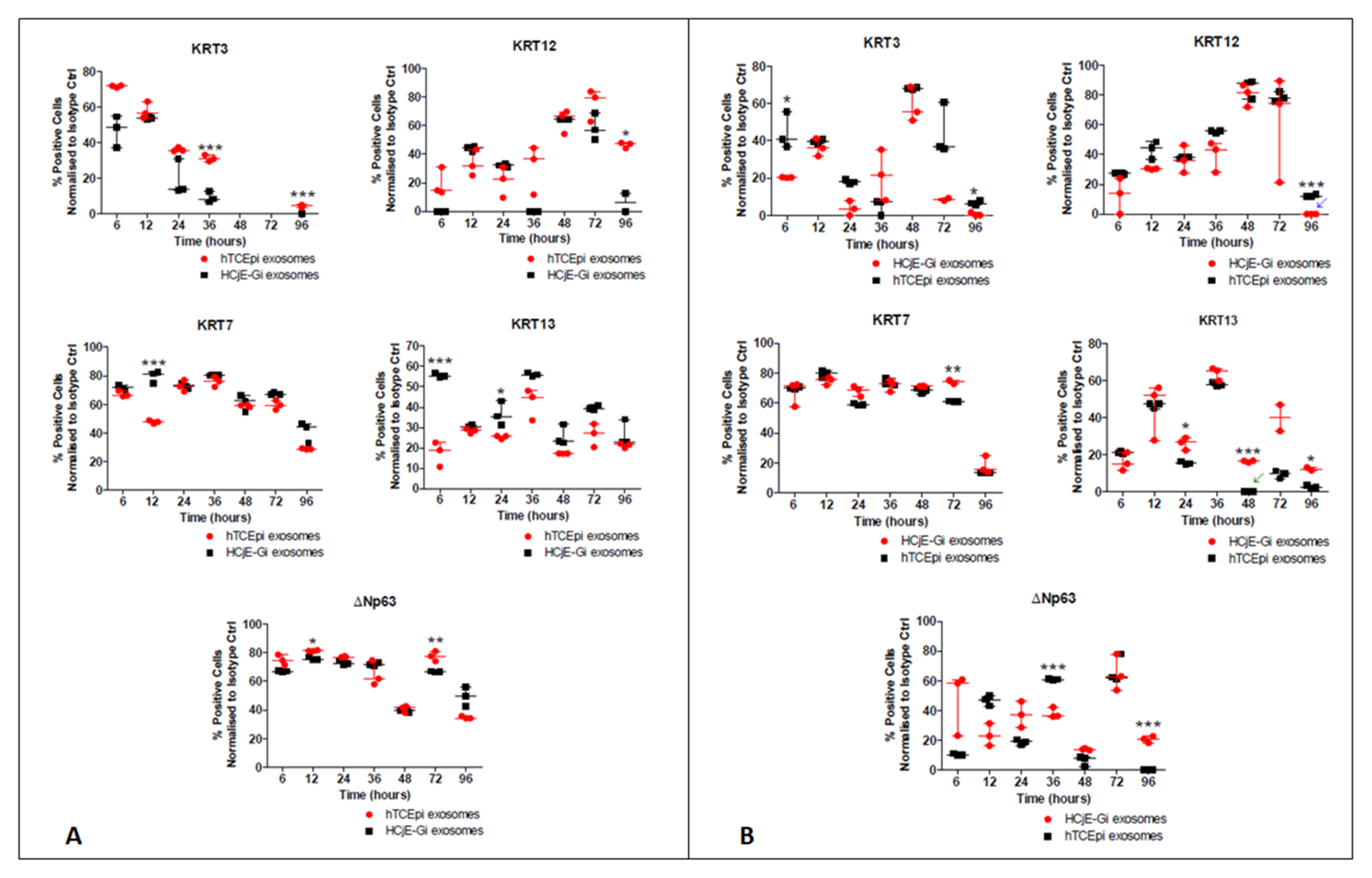
2.2. HCjE-Gi Cells Cultured with hTCEpi-Derived EVs Lose Conjunctival Epithelial Marker Expression and Gain Corneal Epithelial Marker Expression and hTCEpi Cells Cultured in HCjE-Gi-Derived EVs Lose Corneal Epithelial Marker Expression and Gain Conjunctival Epithelial Marker Expression
2.2.1. Real-Time PCR
2.2.2. Flow Cytometry
2.3. Distinct Small RNA Profile of HCjE-Gi and hTCEpi-Derived Extracellular Vesicles
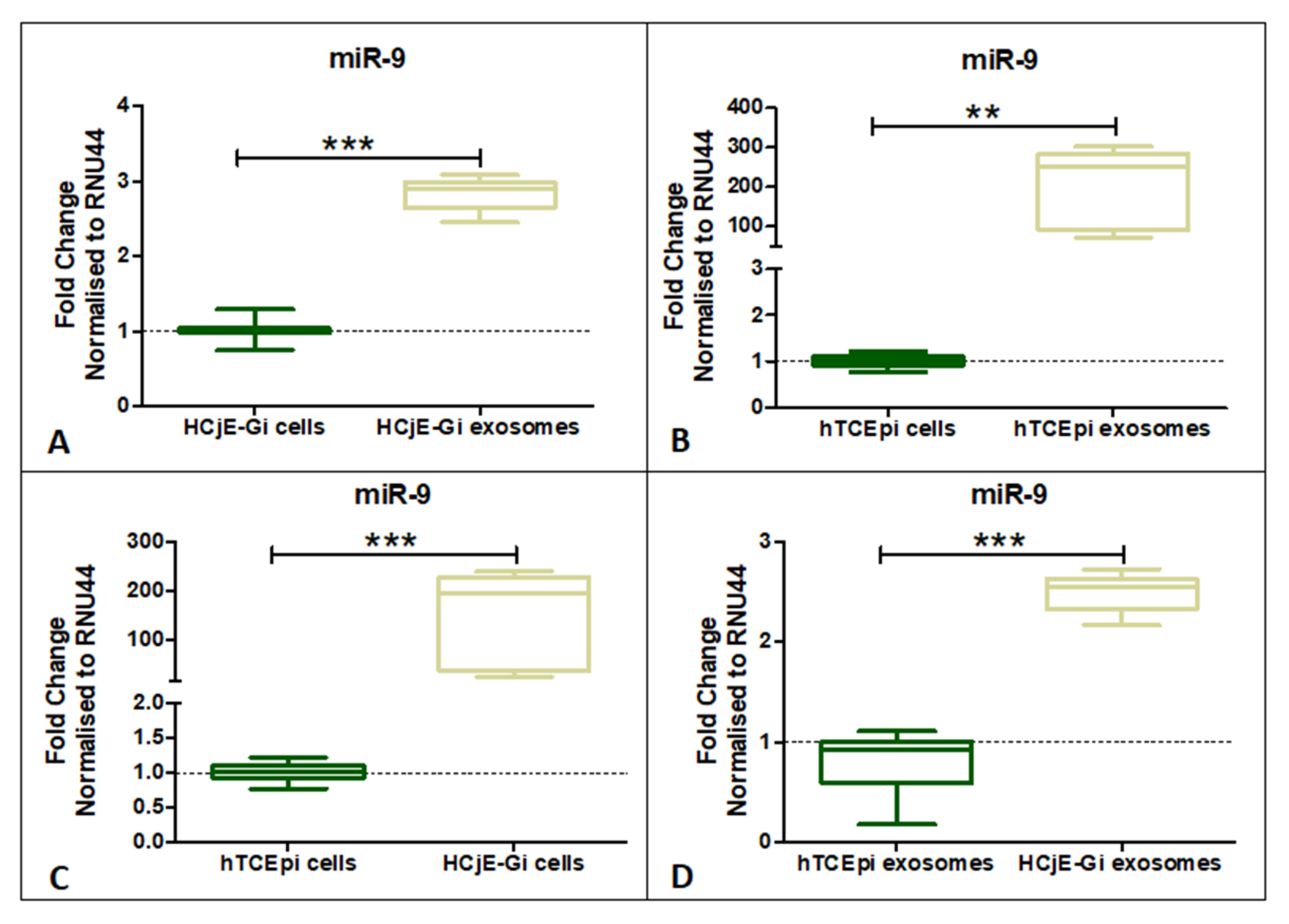
2.4. miR-9-5p Expression Profile on HCjE-Gi, hTCEpi Cells and Their Derived Extracellular Vesicles
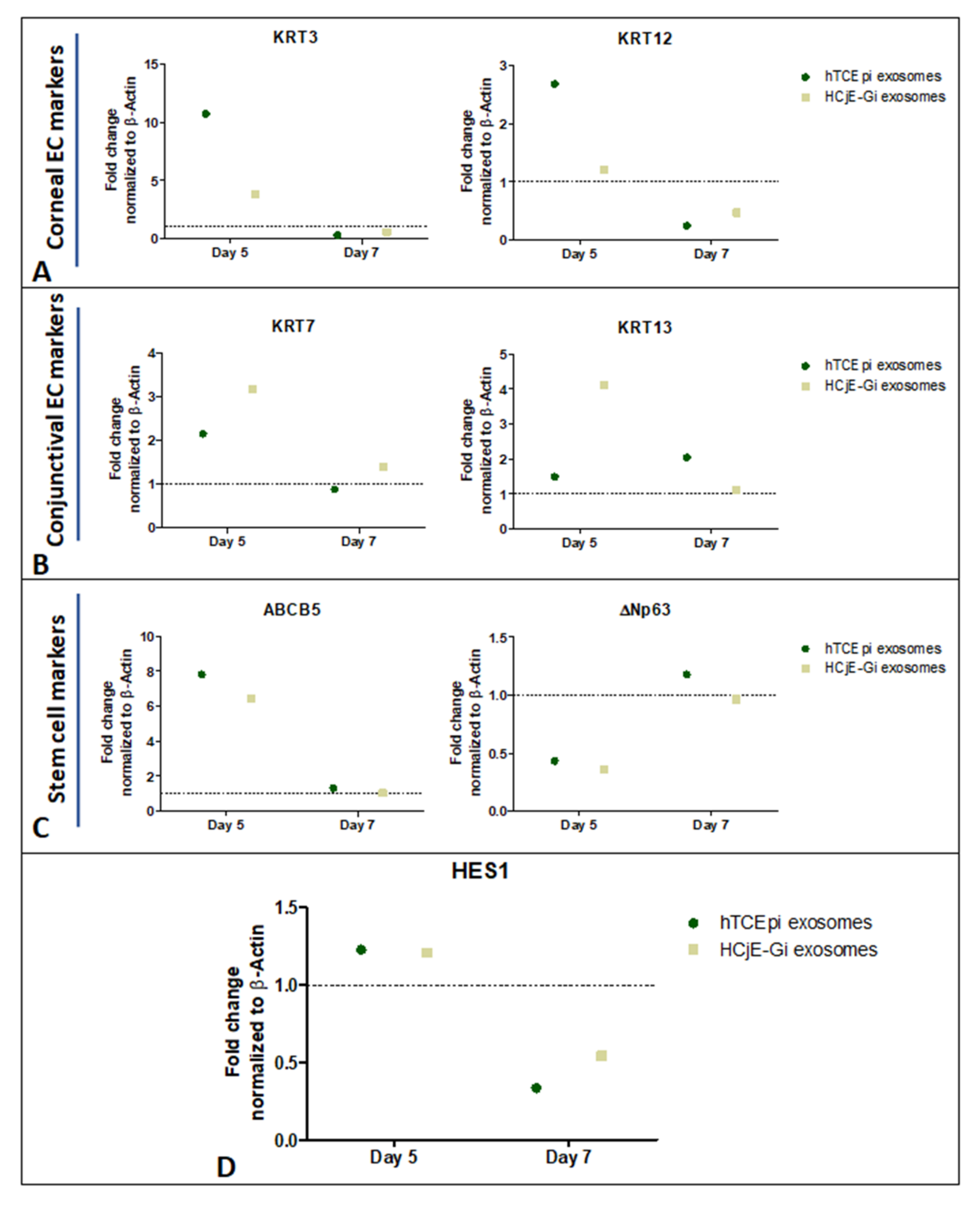
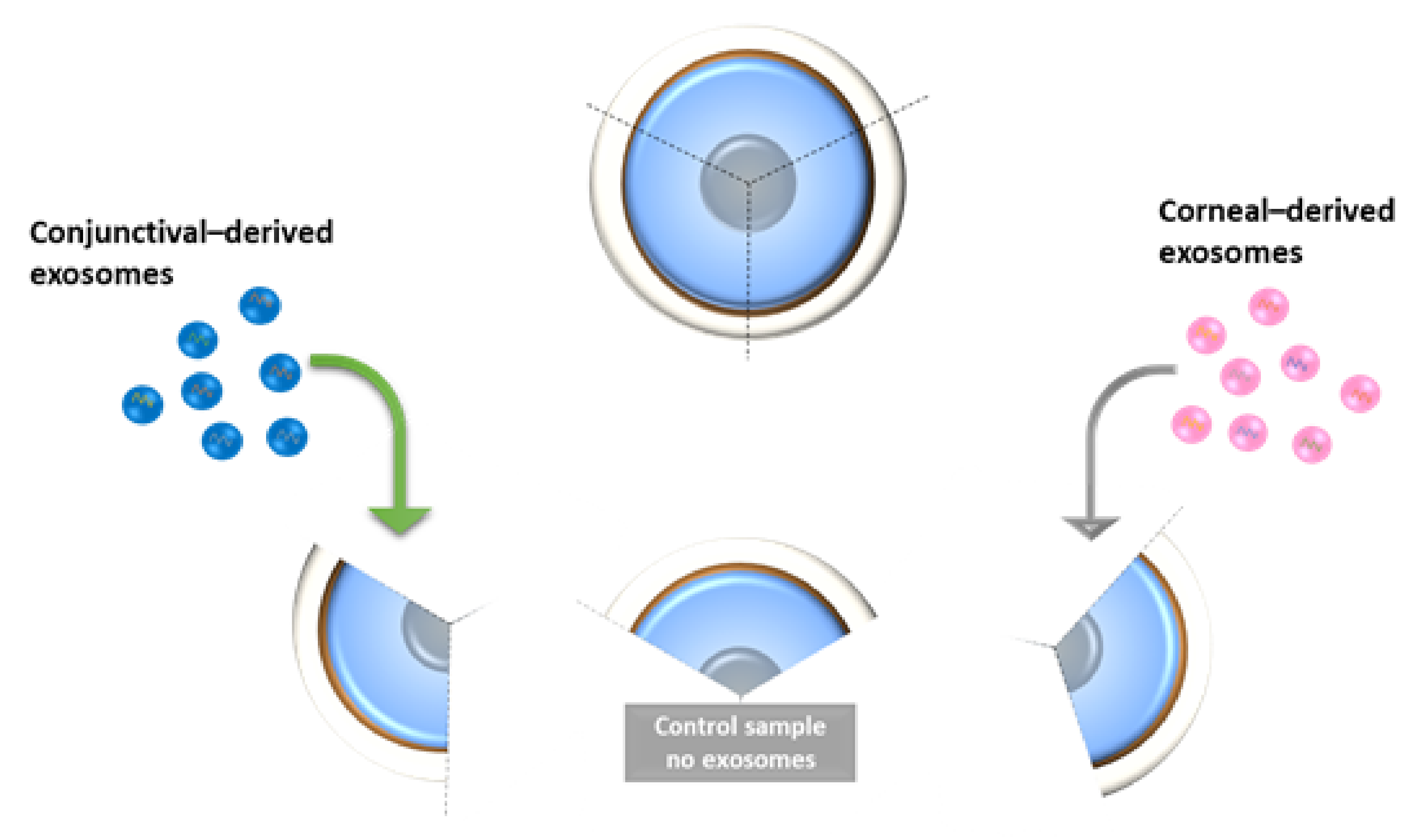
2.5. Expression of Corneal and Conjunctival Biomarkers and HES-1 on Corneal Tissues Following Treatment with HCjE-Gi and hTCEpi-Derived Extracellular Vesicles in an Ex Vivo Study
3. Discussion
4. Materials and Methods
4.1. Ethical Statement
4.2. Cell Culture
4.3. Extraction, Quantification and Sizing, Characterisation and Cellular Uptake of the EVs
4.3.1. Extraction Using Ultracentrifugation
4.3.2. Quantification and Sizing by NanoSight
4.3.3. Characterization by Flow Cytometry
4.3.4. XenoLight DiR Labelling
4.3.5. Cellular Uptake of the EVs—DiL Labelling
4.4. Cell Culture with Extracellular Vesicles and Characterisation
4.4.1. Cell Culture with EVs
4.4.2. Characterization Using Real-Time qPCR
4.4.3. Analysis Using Flow Cytometry
4.5. Exosome Cargo Characterization
4.5.1. End-Point PCR
4.5.2. Exosome Cargo Characterization by Next Generation Sequencing
4.5.3. RNA Isolation and Quantitative Real-Time PCR for miRNA-9-5p
4.5.4. Ex Vivo Study on Human Cadaveric Donor Corneal Tissues—A Pilot Study
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Davanger, M.; Evensen, A. Role of the Pericorneal Papillary Structure in Renewal of Corneal Epithelium. Nature 1971, 229, 560–561. [Google Scholar] [CrossRef] [PubMed]
- Ebato, B.; Friend, J.; Thoft, R.A. Comparison of limbal and peripheral human corneal epithelium in tissue culture. Investig. Ophthalmol. Vis. Sci. 1988, 29, 1533–1537. [Google Scholar]
- Cotsarelis, G.; Cheng, S.Z.; Dong, G.; Sun, T.T.; Lavker, R.M. Existence of slow-cycling limbal epithelial basal cells that can be preferentially stimulated to proliferate: Implications on epithelial stem cells. Cell 1989, 57, 201–209. [Google Scholar] [CrossRef]
- Kenyon, K.R.; Tseng, S.C. Limbal autograft transplantation for ocular surface disorders. Ophthalmology 1989, 96, 709–722, discussion 722–703. [Google Scholar]
- Friedenwald, J. Growth pressure and metaplasia of conjunctival and corneal epithelium. Doc. Ophthalmol. 1951, 5, 184–192. [Google Scholar] [CrossRef]
- Mann, I.; Pullinger, B.D. A Study of Mustard Gas Lesions of the Eyes of Rabbits and Men: (Section of Ophthalmology). J. R. Soc. Med. 1942, 35, 229–244. [Google Scholar] [CrossRef] [Green Version]
- Thoft, R.A.; Friend, J. Biochemical transformation of regenerating ocular surface epithelium. Investig. Ophthalmol. Vis. Sci. 1977, 16, 14–20. [Google Scholar]
- Kinoshita, S.; Friend, J.; Thoft, R.A. Biphasic cell proliferation in transdifferentiation of conjunctival to corneal epithelium in rabbits. Invest. Ophthalmol. Vis. Sci. 1983, 24, 1008–1014. [Google Scholar]
- Harris, T.M.; Berry, E.R.; Pakurar, A.S.; Sheppard, L.B. Biochemical transformation of bulbar conjunctiva into corneal epithelium: An electrophoretic analysis. Exp. Eye Res. 1985, 41, 597–605. [Google Scholar] [CrossRef]
- Kurpakus, M.A.; Stock, E.L.; Jones, J.C. The role of the basement membrane in differential expression of keratin proteins in epithelial cells. Dev. Biol. 1992, 150, 243–255. [Google Scholar] [CrossRef]
- Shapiro, M.S.; Friend, J.; Thoft, R.A. Corneal re-epithelialization from the conjunctiva. Invest. Ophthalmol. Vis. Sci. 1981, 21, 135–142. [Google Scholar]
- Moyer, P.D.; Kaufman, A.H.; Zhang, Z.; Kao, C.W.; Spaulding, A.G.; Kao, W.W. Conjunctival epithelial cells can resurface denuded cornea, but do not transdifferentiate to express cornea-specific keratin 12 following removal of limbal epithelium in mouse. Differentiation 1996, 60, 31–38. [Google Scholar] [CrossRef]
- Shen, C.N.; Horb, M.E.; Slack, J.M.; Tosh, D. Transdifferentiation of pancreas to liver. Mech. Dev. 2003, 120, 107–116. [Google Scholar] [CrossRef]
- Lassar, A.B.; Paterson, B.M.; Weintraub, H. Transfection of a DNA locus that mediates the conversion of 10T1/2 fibroblasts to myoblasts. Cell 1986, 47, 649–656. [Google Scholar] [CrossRef]
- Song, K.; Nam, Y.-J.; Luo, X.; Qi, X.; Tan, W.; Huang, G.N.; Acharya, A.; Smith, C.L.; Tallquist, M.D.; Neilson, E.G.; et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature 2012, 485, 599. [Google Scholar] [CrossRef] [Green Version]
- Wei, Z.G.; Sun, T.T.; Lavker, R.M. Rabbit conjunctival and corneal epithelial cells belong to two separate lineages. Investig. Ophthalmol. Vis. Sci. 1996, 37, 523–533. [Google Scholar]
- Ramirez-Miranda, A.; Nakatsu, M.N.; Zarei-Ghanavati, S.; Nguyen, C.V.; Deng, S.X. Keratin 13 is a more specific marker of conjunctival epithelium than keratin 19. Mol. Vis. 2011, 17, 1652–1661. [Google Scholar]
- Schermer, A.; Galvin, S.; Sun, T.T. Differentiation-related expression of a major 64K corneal keratin in vivo and in culture suggests limbal location of corneal epithelial stem cells. J. Cell Biol. 1986, 103, 49–62. [Google Scholar] [CrossRef]
- Merjava, S.; Neuwirth, A.; Tanzerova, M.; Jirsova, K. The spectrum of cytokeratins expressed in the adult human cornea, limbus and perilimbal conjunctiva. Histol. Histopathol. 2011, 26, 323–331. [Google Scholar]
- Ramaekers, F.; Huysmans, A.; Schaart, G.; Moesker, O.; Vooijs, P. Tissue distribution of keratin 7 as monitored by a monoclonal antibody. Exp. Cell Res. 1987, 170, 235–249. [Google Scholar] [CrossRef]
- Poli, M.; Burillon, C.; Auxenfans, C.; Rovere, M.R.; Damour, O. Immunocytochemical Diagnosis of Limbal Stem Cell Deficiency: Comparative Analysis of Current Corneal and Conjunctival Biomarkers. Cornea 2015, 34, 817–823. [Google Scholar] [CrossRef]
- Gipson, I.K. The epithelial basement membrane zone of the limbus. Eye 1989, 3, 132–140. [Google Scholar] [CrossRef] [Green Version]
- Di Girolamo, N. Stem cells of the human cornea. Br. Med. Bull. 2011, 100, 191–207. [Google Scholar] [CrossRef]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [Green Version]
- Evans-Osses, I.; Reichembach, L.H.; Ramirez, M.I. Exosomes or microvesicles? Two kinds of extracellular vesicles with different routes to modify protozoan-host cell interaction. Parasitol. Res. 2015, 114, 3567–3575. [Google Scholar] [CrossRef]
- Thery, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef]
- Clayton, A.; Court, J.; Navabi, H.; Adams, M.; Mason, M.D.; Hobot, J.A.; Newman, G.R.; Jasani, B. Analysis of antigen presenting cell derived exosomes, based on immuno-magnetic isolation and flow cytometry. J. Immunol. Methods 2001, 247, 163–174. [Google Scholar] [CrossRef]
- Blanchard, N.; Lankar, D.; Faure, F.; Regnault, A.; Dumont, C.; Raposo, G.; Hivroz, C. TCR activation of human T cells induces the production of exosomes bearing the TCR/CD3/zeta complex. J. Immunol. 2002, 168, 3235–3241. [Google Scholar] [CrossRef] [Green Version]
- Raposo, G.; Nijman, H.W.; Stoorvogel, W.; Liejendekker, R.; Harding, C.V.; Melief, C.J.; Geuze, H.J.B. Lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996, 183, 1161–1172. [Google Scholar] [CrossRef]
- Gruenberg, J.; Maxfield, F.R. Membrane transport in the endocytic pathway. Curr. Opin. Cell Biol. 1995, 7, 552–563. [Google Scholar] [CrossRef]
- Moldovan, L.; Batte, K.; Wang, Y.; Wisler, J.; Piper, M. Analyzing the Circulating MicroRNAs in Exosomes/Extracellular Vesicles from Serum or Plasma by qRT-PCR. In Circulating MicroRNAs; Kosaka, N., Ed.; Humana Press: Totowa, NJ, USA, 2013; Volume 1024, pp. 129–145. [Google Scholar]
- Han, K.Y.; Tran, J.A.; Chang, J.H.; Azar, D.T.; Zieske, J.D. Potential role of corneal epithelial cell-derived exosomes in corneal wound healing and neovascularization. Sci. Rep. 2017, 7, 40548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chowdhury, R.; Webber, J.P.; Gurney, M.; Mason, M.D.; Tabi, Z.; Clayton, A. Cancer exosomes trigger mesenchymal stem cell differentiation into pro-angiogenic and pro-invasive myofibroblasts. Oncotarget 2015, 6, 715–731. [Google Scholar] [CrossRef] [PubMed]
- Lotvall, J.; Hill, A.F.; Hochberg, F.; Buzas, E.I.; Di Vizio, D.; Gardiner, C.; Gho, Y.S.; Kurochkin, I.V.; Mathivanan, S.; Quesenberry, P.; et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the International Society for Extracellular Vesicles. J. Extracell. Vesicles 2014, 3, 26913. [Google Scholar] [CrossRef]
- Di Iorio, E.; Barbaro, V.; Ruzza, A.; Ponzin, D.; Pellegrini, G.; De Luca, M. Isoforms of DeltaNp63 and the migration of ocular limbal cells in human corneal regeneration. Proc. Natl. Acad. Sci. USA 2005, 102, 9523–9528. [Google Scholar] [CrossRef] [Green Version]
- Pellegrini, G.; Dellambra, E.; Golisano, O.; Martinelli, E.; Fantozzi, I.; Bondanza, S.; Ponzin, D.; McKeon, F.; De Luca, M. p63 identifies keratinocyte stem cells. Proc. Natl. Acad. Sci. USA 2001, 98, 3156–3161. [Google Scholar] [CrossRef] [Green Version]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [Green Version]
- Garcia, N.A.; Moncayo-Arlandi, J.; Sepulveda, P.; Diez-Juan, A. Cardiomyocyte exosomes regulate glycolytic flux in endothelium by direct transfer of GLUT transporters and glycolytic enzymes. Cardiovasc. Res. 2016, 109, 397–408. [Google Scholar] [CrossRef] [Green Version]
- Bobrie, A.; Colombo, M.; Raposo, G.; Thery, C. Exosome secretion: Molecular mechanisms and roles in immune responses. Traffic 2011, 12, 1659–1668. [Google Scholar] [CrossRef]
- Han, K.Y.; Dugas-Ford, J.; Seiki, M.; Chang, J.H.; Azar, D.T. Evidence for the Involvement of MMP14 in MMP2 Processing and Recruitment in Exosomes of Corneal Fibroblasts. Investig. Ophthalmol. Vis. Sci. 2015, 56, 5323–5329. [Google Scholar] [CrossRef] [Green Version]
- Leszczynska, A.; Kulkarni, M.; Ljubimov, A.V.; Saghizadeh, M. Exosomes from normal and diabetic human corneolimbal keratocytes differentially regulate migration, proliferation and marker expression of limbal epithelial cells. Sci. Rep. 2018, 8, 15173. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, T.; Inatomi, T.; Sotozono, C.; Koizumi, N.; Kinoshita, S. Ocular surface reconstruction using stem cell and tissue engineering. Prog. Retin. Eye Res. 2016, 51, 187–207. [Google Scholar] [CrossRef]
- Escudier, B.; Dorval, T.; Chaput, N.; Andre, F.; Caby, M.P.; Novault, S.; Flament, C.; Leboulaire, C.; Borg, C.; Amigorena, S.; et al. Vaccination of metastatic melanoma patients with autologous dendritic cell (DC) derived-exosomes: Results of thefirst phase I clinical trial. J. Transl. Med. 2005, 3, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ratajczak, J.; Miekus, K.; Kucia, M.; Zhang, J.; Reca, R.; Dvorak, P.; Ratajczak, M.Z. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: Evidence for horizontal transfer of mRNA and protein delivery. Leukemia 2006, 20, 847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ratajczak, J.; Wysoczynski, M.; Hayek, F.; Janowska-Wieczorek, A.; Ratajczak, M.Z. Membrane-derived microvesicles: Important and underappreciated mediators of cell-to-cell communication. Leukemia 2006, 20, 1487–1495. [Google Scholar] [CrossRef]
- Katsman, D.; Stackpole, E.J.; Domin, D.R.; Farber, D.B. Embryonic stem cell-derived microvesicles induce gene expression changes in Muller cells of the retina. PLoS ONE 2012, 7, e50417. [Google Scholar] [CrossRef]
- Mead, B.; Tomarev, S. Bone Marrow-Derived Mesenchymal Stem Cells-Derived Exosomes Promote Survival of Retinal Ganglion Cells Through miRNA-Dependent Mechanisms. Stem. Cells Transl. Med. 2017, 6, 1273–1285. [Google Scholar] [CrossRef]
- Blazquez, R.; Sanchez-Margallo, F.M.; de la Rosa, O.; Dalemans, W.; Alvarez, V.; Tarazona, R.; Casado, J.G. Immunomodulatory Potential of Human Adipose Mesenchymal Stem Cells Derived Exosomes on in vitro Stimulated T Cells. Front. Immunol. 2014, 5, 556. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Yin, Y.; Lai, R.C.; Tan, S.S.; Choo, A.B.; Lim, S.K. Mesenchymal stem cells secrete immunologically active exosomes. Stem. Cells Dev. 2014, 23, 1233–1244. [Google Scholar] [CrossRef]
- Grigor’eva, A.E.; Tamkovich, S.N.; Eremina, A.V.; Tupikin, A.E.; Kabilov, M.R.; Chernykh, V.V.; Vlassov, V.V.; Laktionov, P.P.; Ryabchikova, E.I. Exosomes in tears of healthy individuals: Isolation, identification, and characterization. Biochem. Suppl. Ser. B Biomed. Chem. 2016, 10, 165–172. [Google Scholar] [CrossRef]
- Schlotzer-Schrehardt, U.; Dietrich, T.; Saito, K.; Sorokin, L.; Sasaki, T.; Paulsson, M.; Kruse, F.E. Characterization of extracellular matrix components in the limbal epithelial stem cell compartment. Exp. Eye Res. 2007, 85, 845–860. [Google Scholar] [CrossRef]
- Tseng, S.C.; Hirst, L.W.; Farazdaghi, M.; Green, W.R. Goblet cell density and vascularization during conjunctival transdifferentiation. Invest. Ophthalmol. Vis. Sci. 1984, 25, 1168–1176. [Google Scholar] [PubMed]
- Thoft, R.A.; Friend, J.; Murphy, H.S. Ocular surface epithelium and corneal vascularization in rabbits. I. The role of wounding. Invest. Ophthalmol. Vis. Sci. 1979, 18, 85–92. [Google Scholar] [PubMed]
- Tseng, S.C.; Farazdaghi, M.; Rider, A.A. Conjunctival transdifferentiation induced by systemic vitamin A deficiency in vascularized rabbit corneas. Investig. Ophthalmol. Vis. Sci. 1987, 28, 1497–1504. [Google Scholar]
- Aitken, D.; Friend, J.; Thoft, R.A.; Lee, W.R. An ultrastructural study of rabbit ocular surface transdifferentiation. Investig. Ophthalmol. Vis. Sci. 1988, 29, 224–231. [Google Scholar]
- Friend, J.; Thoft, R.A. Functional competence of regenerating ocular surface epithelium. Investig. Ophthalmol. Vis. Sci. 1978, 17, 134–139. [Google Scholar]
- Fang, S.; Xu, C.; Zhang, Y.; Xue, C.; Yang, C.; Bi, H.; Qian, X.; Wu, M.; Ji, K.; Zhao, Y.; et al. Umbilical Cord-Derived Mesenchymal Stem Cell-Derived Exosomal MicroRNAs Suppress Myofibroblast Differentiation by Inhibiting the Transforming Growth Factor-beta/SMAD2 Pathway During Wound Healing. Stem. Cells Transl. Med. 2016, 5, 1425–1439. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, X.L.; Zhang, D.; Liu, F.; Cheng, Y.J.; Ma, Y.; Zhou, Y.J.; Zhao, Y.X. Platelet-derived exosomes affect the proliferation and migration of human umbilical vein endothelial cells via miR-126. Curr. Vasc. Pharmacol. 2019, 17, 379–387. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, H.; Chen, Y.; Qiao, G.; Jiang, W.; Ni, P.; Liu, X.; Ma, L. miR-598 inhibits metastasis in colorectal cancer by suppressing JAG1/Notch2 pathway stimulating EMT. Exp. Cell Res. 2017, 352, 104–112. [Google Scholar] [CrossRef]
- Zhang, S.; Yu, M.; Liu, C.; Wang, L.; Hu, Y.; Bai, Y.; Hua, J. MIR-34c regulates mouse embryonic stem cells differentiation into male germ-like cells through RARg. Cell Biochem. Funct. 2012, 30, 623–632. [Google Scholar] [CrossRef]
- Bae, Y.; Yang, T.; Zeng, H.C.; Campeau, P.M.; Chen, Y.; Bertin, T.; Dawson, B.C.; Munivez, E.; Tao, J.; Lee, B.H. miRNA-34c regulates Notch signaling during bone development. Hum. Mol. Genet. 2012, 21, 2991–3000. [Google Scholar] [CrossRef] [Green Version]
- Winkler, M.A.; Dib, C.; Ljubimov, A.V.; Saghizadeh, M. Targeting miR-146a to treat delayed wound healing in human diabetic organ-cultured corneas. PLoS ONE 2014, 9, e114692. [Google Scholar] [CrossRef] [Green Version]
- Teng, Y.; Wong, H.K.; Jhanji, V.; Chen, J.H.; Young, A.L.; Zhang, M.; Choy, K.W.; Mehta, J.S.; Pang, C.P.; Yam, G.H. Signature microRNAs in human cornea limbal epithelium. Funct. Integr. Genomics. 2015, 15, 277–294. [Google Scholar] [CrossRef]
- Tan, S.L.; Ohtsuka, T.; Gonzalez, A.; Kageyama, R. MicroRNA9 regulates neural stem cell differentiation by controlling Hes1 expression dynamics in the developing brain. Genes Cells 2012, 17, 952–961. [Google Scholar] [CrossRef]
- Nakamura, T.; Ohtsuka, T.; Sekiyama, E.; Cooper, L.J.; Kokubu, H.; Fullwood, N.J.; Barrandon, Y.; Kageyama, R.; Kinoshita, S. Hes1 regulates corneal development and the function of corneal epithelial stem/progenitor cells. Stem. Cells 2008, 26, 1265–1274. [Google Scholar] [CrossRef]
- Nolte-‘t Hoen, E.N.M.; Buermans, H.P.J.; Waasdorp, M.; Stoorvogel, W.; Wauben, M.H.M.; ‘t Hoen, P.A.C. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012, 40, 9272–9285. [Google Scholar] [CrossRef] [Green Version]
- Goldie, B.J.; Dun, M.D.; Lin, M.; Smith, N.D.; Verrills, N.M.; Dayas, C.V.; Cairns, M.J. Activity-associated miRNA are packaged in Map1b-enriched exosomes released from depolarized neurons. Nucleic Acids Res. 2014, 42, 9195–9208. [Google Scholar] [CrossRef] [Green Version]
- Villarroya-Beltri, C.; Gutierrez-Vazquez, C.; Sanchez-Cabo, F.; Perez-Hernandez, D.; Vazquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sanchez-Madrid, F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013, 4, 2980. [Google Scholar] [CrossRef] [Green Version]
- Gipson, I.K. Mucin gene expression in immortalized human corneal-limbal and conjunctival epithelial cell lines. Invest. Ophthalmol. Vis. Sci. 2003, 44, 2496–2506. [Google Scholar] [CrossRef]
- Robertson, D.M.; Li, L.; Fisher, S.; Pearce, V.P.; Shay, J.W.; Wright, W.E.; Cavanagh, H.D.; Jester, J.V. Characterization of growth and differentiation in a telomerase-immortalized human corneal epithelial cell line. Invest. Ophthalmol. Vis. Sci. 2005, 46, 470–478. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Kuhn, R.M.; Haussler, D.; Kent, W.J. The UCSC genome browser and associated tools. Brief. Bioinform. 2013, 14, 144–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marcel, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.J. 2011, 17, 10–12. [Google Scholar]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| miRNA | Abundance HCjE-Gi | Abundance hTCEpi | p-Value | Log2 Fold Change | Role |
|---|---|---|---|---|---|
| miR-598 | Residual | 111.74 | 0.048 | infinity | Downregulated in cancer tissues |
| miR-34c-3p | 46.56 | 1548.96 | 0.011 | 5.1 | Upregulated in cell differentiation |
| mir-34c | 116.39 | 2713.78 | 0.029 | 4.5 | Upregulated in cell differentiation |
| mir-146a | 10080.83 | 510.48 | 0.048 | −4.3 | Upregulated in stem cell maintenance homeostasis |
| miR-146a-5p | 10080.83 | 510.48 | 0.048 | −4.3 | Upregulated in stem cell maintenance homeostasis |
| mir-155 | 703.82 | 4.38 | 0.0001 | −7.3 | Upregulation represses cell differentiation |
| miR-155-5p | 703.82 | 4.38 | 0.0001 | −7.3 | Upregulation represses cell differentiation |
| miR-9-5p | 86.27 | Residual | 0.03 | infinity | Probably involved in cell differentiation |
| mir-9-1 | 98.59 | Residual | 0.018 | infinity | Probably involved in cell differentiation |
| mir-9-2 | 98.59 | Residual | 0.018 | infinity | Probably involved in cell differentiation |
| mir-9-3 | 98.59 | Residual | 0.018 | infinity | Probably involved in cell differentiation |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramos, T.; Parekh, M.; Kaye, S.B.; Ahmad, S. Epithelial Cell-Derived Extracellular Vesicles Trigger the Differentiation of Two Epithelial Cell Lines. Int. J. Mol. Sci. 2022, 23, 1718. https://doi.org/10.3390/ijms23031718
Ramos T, Parekh M, Kaye SB, Ahmad S. Epithelial Cell-Derived Extracellular Vesicles Trigger the Differentiation of Two Epithelial Cell Lines. International Journal of Molecular Sciences. 2022; 23(3):1718. https://doi.org/10.3390/ijms23031718
Chicago/Turabian StyleRamos, Tiago, Mohit Parekh, Stephen B. Kaye, and Sajjad Ahmad. 2022. "Epithelial Cell-Derived Extracellular Vesicles Trigger the Differentiation of Two Epithelial Cell Lines" International Journal of Molecular Sciences 23, no. 3: 1718. https://doi.org/10.3390/ijms23031718
APA StyleRamos, T., Parekh, M., Kaye, S. B., & Ahmad, S. (2022). Epithelial Cell-Derived Extracellular Vesicles Trigger the Differentiation of Two Epithelial Cell Lines. International Journal of Molecular Sciences, 23(3), 1718. https://doi.org/10.3390/ijms23031718






