Neuronal Phenotype of col4a1 and col25a1: An Intriguing Hypothesis in Vertebrates Brain Aging
Abstract
:1. Introduction
2. Results
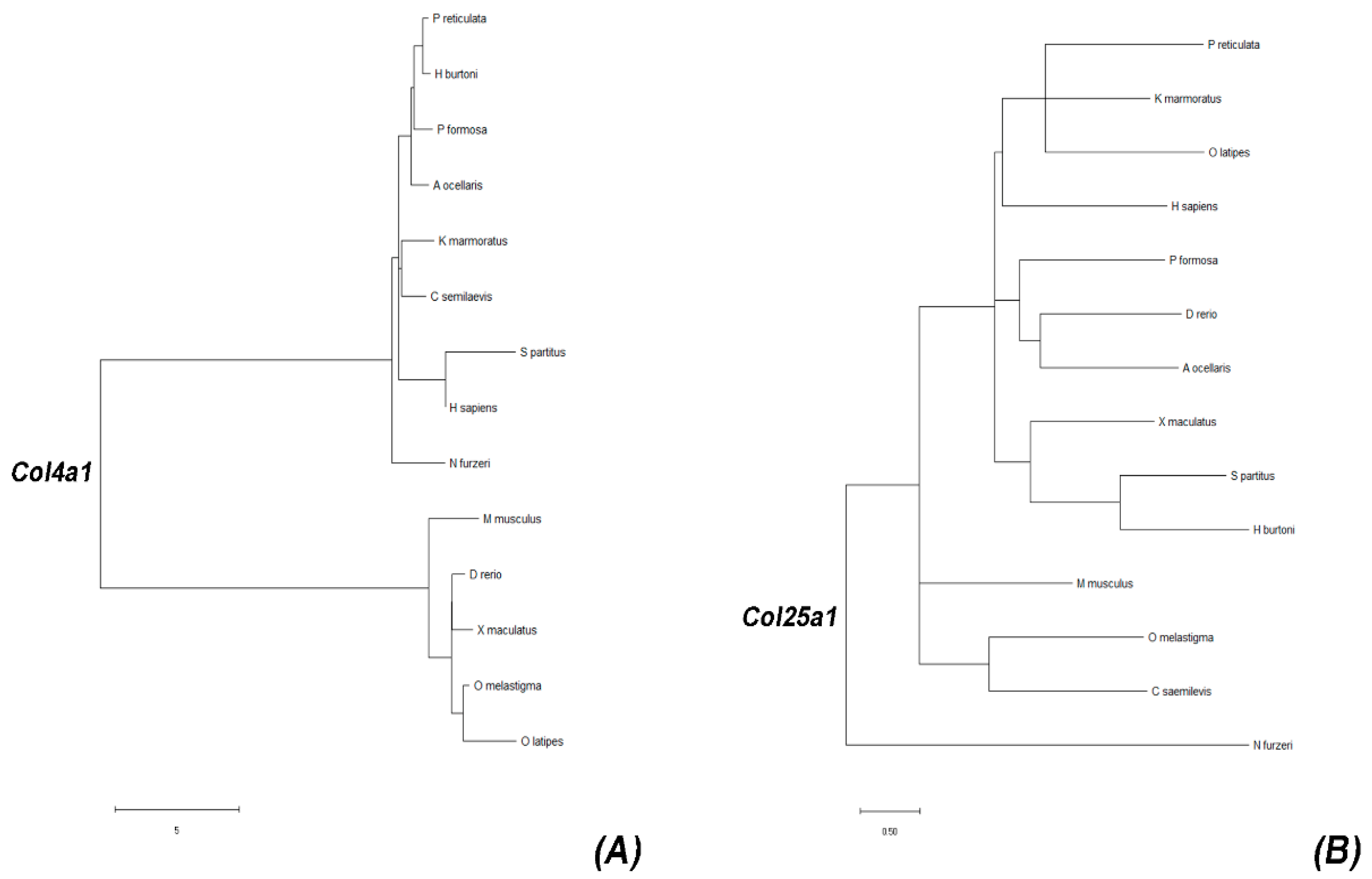
2.1. Identification and Evolutionary History of col4a1 and col25a1
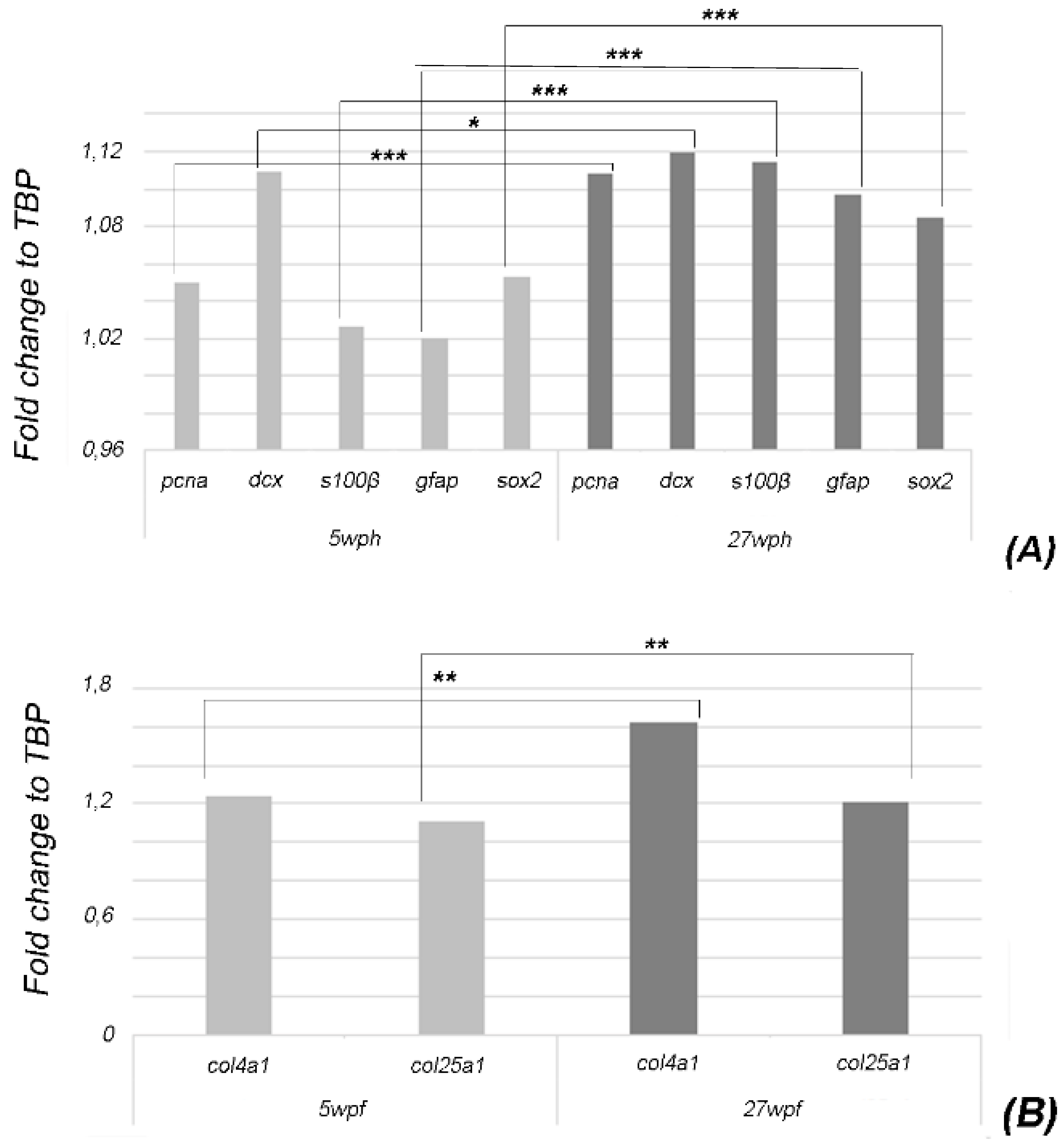
2.2. Age-Related Expression of Brain Cell Lineage Markers and col4a1 and col25a1
2.3. Neuroanatomical Localization of col4a1 and col25a1 mRNAs in the Brain of Young and Old Animals
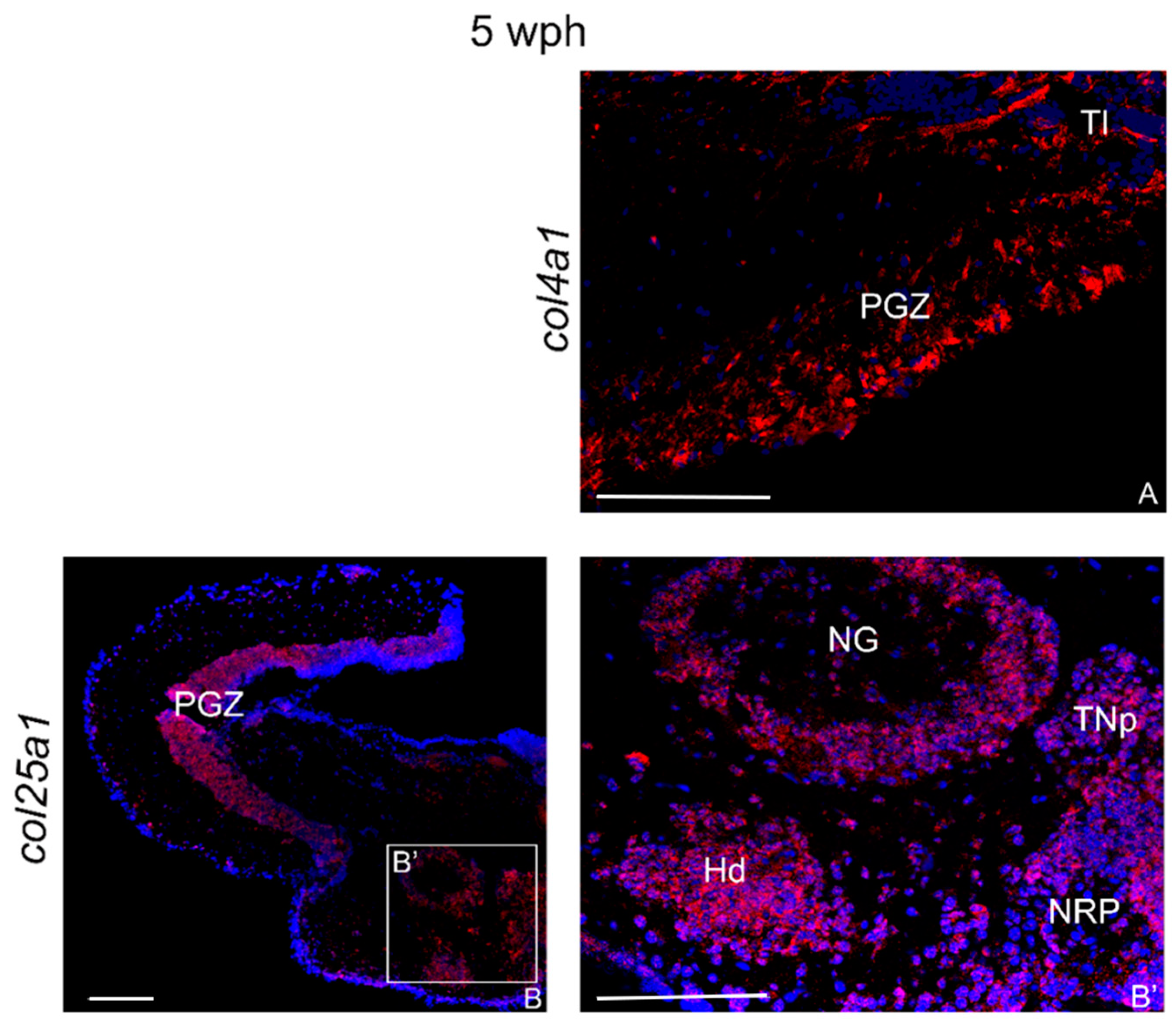
2.3.1. col4a1 and col25a1 mRNAs in the Brain of Young Animals
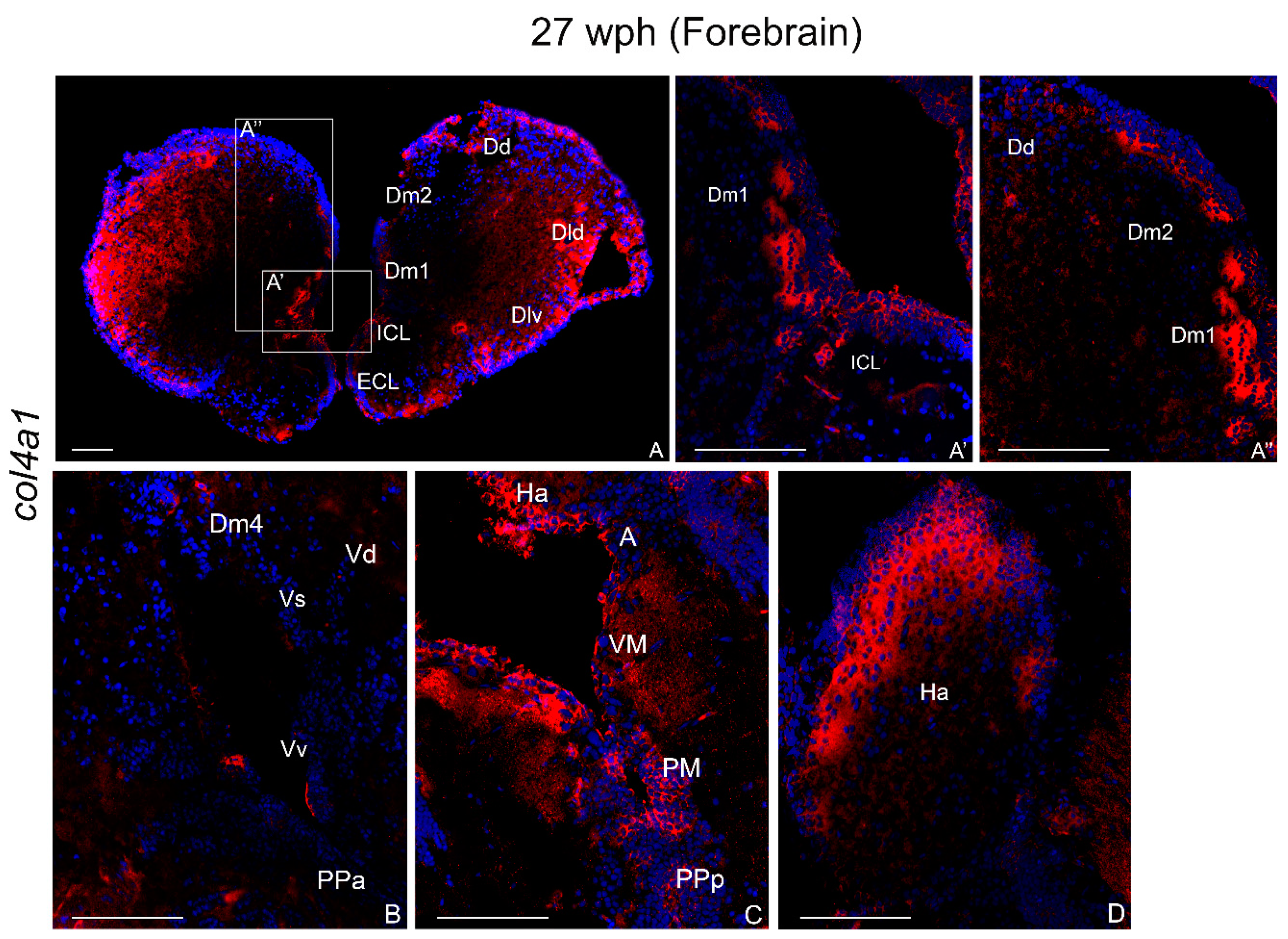
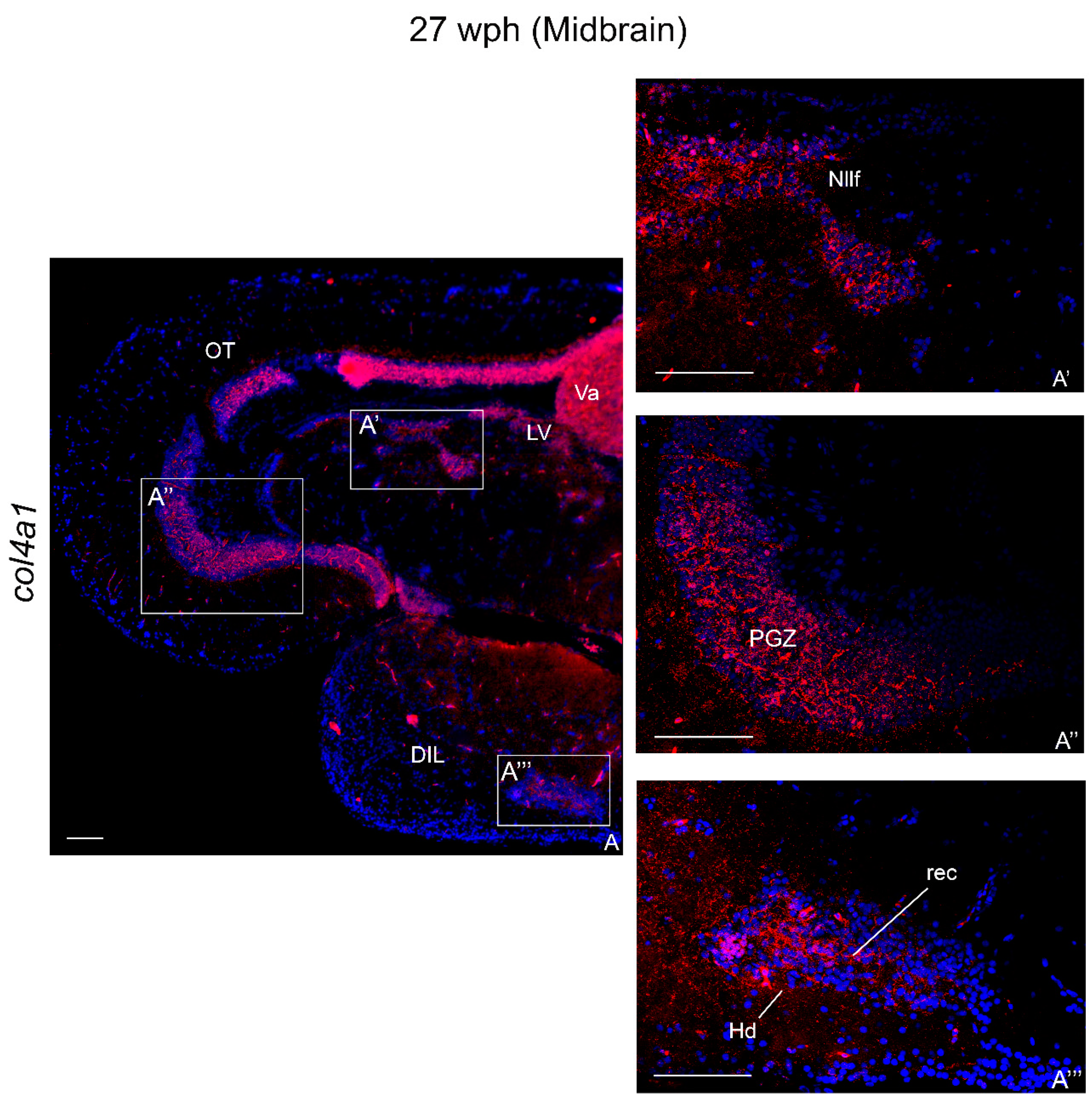
2.3.2. col4a1 mRNA in the Brain of Old Animals
Forebrain
Midbrain
Hindbrain
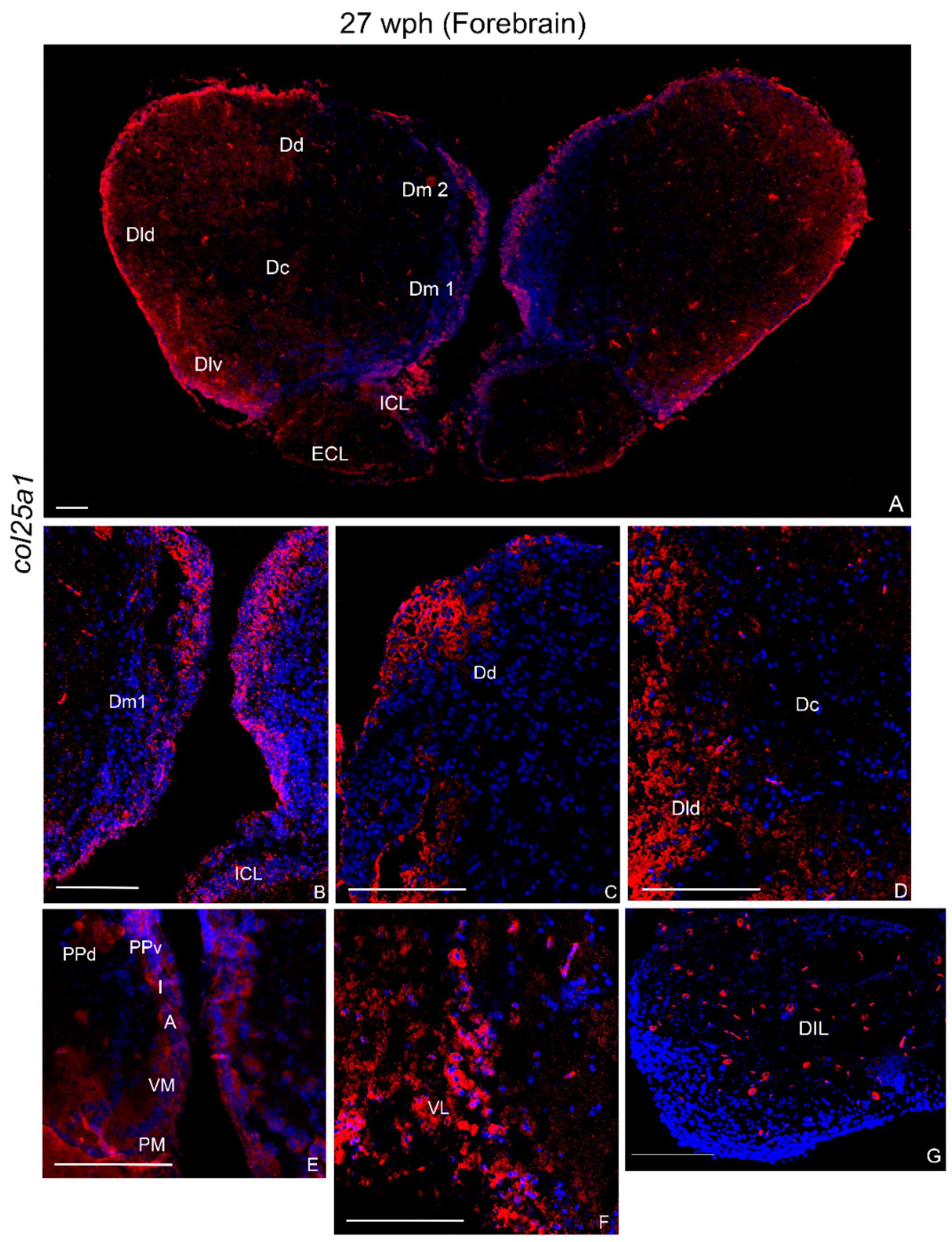
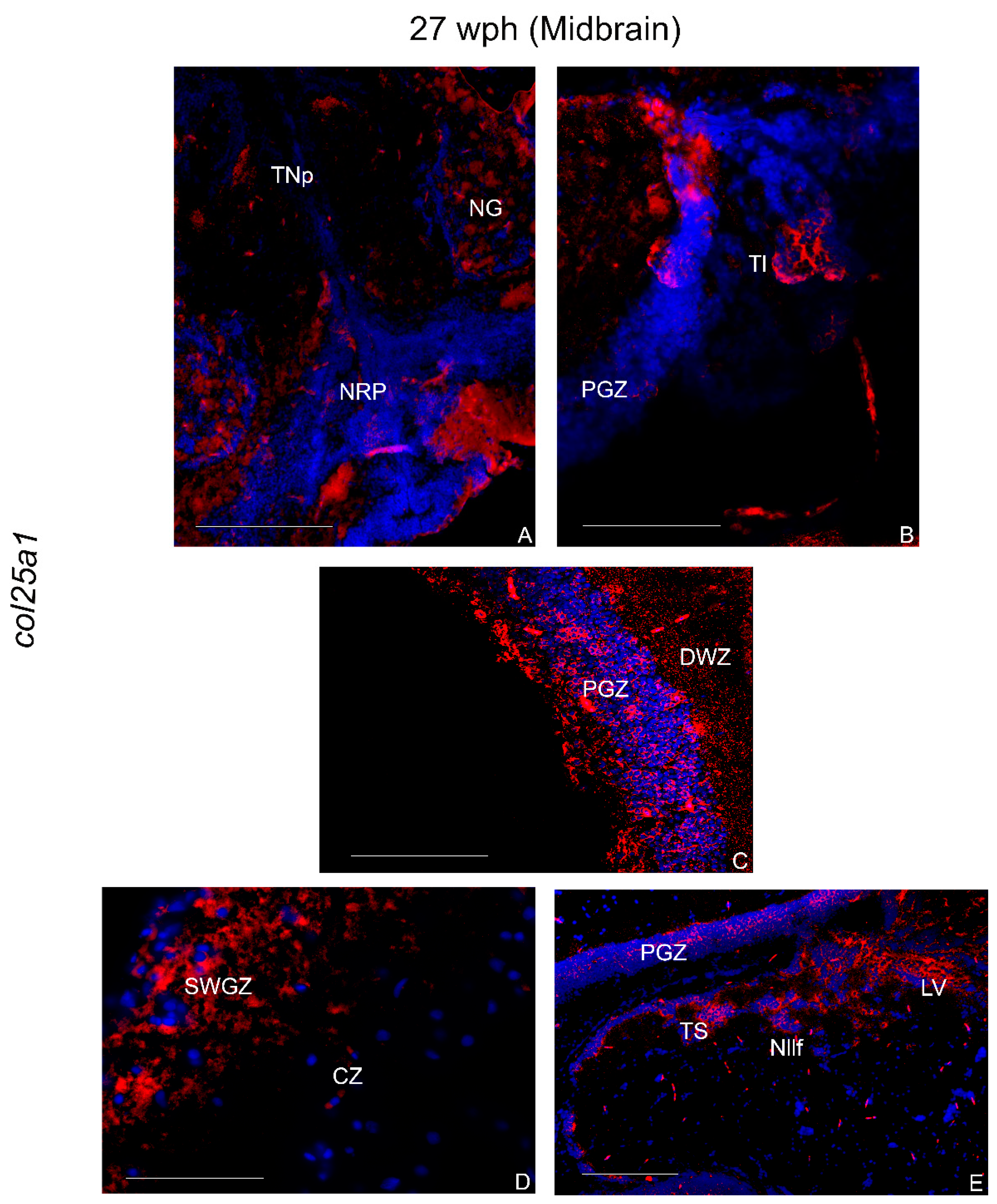
2.3.3. col25a1 mRNA in the Brain of Old Animals
Forebrain
Midbrain
Hindbrain
2.4. Characterization of col4a1 and col25a1 Expressing Cells in the Brain of Old Animals
2.4.1. S100β
2.4.2. DCX, HuC/HuD, MAP2
3. Discussion
4. Materials and Methods
4.1. Protocols
4.2. Phylogenetic Analysis and Protein Alignment
4.3. Animals and Tissue Sampling
4.4. RNA Isolation and cDNA Synthesis
4.5. Identification of col4a1 and col25a1
4.6. Phylogenetic Analysis and Protein Alignment
4.7. Quantitative Real Time-PCR (qPCR)
4.8. Statistical Analysis
4.9. Riboprobe Synthesis
4.10. Fluorescence In Situ Hybridization
4.11. Combined FISH with IF
4.12. Image Acquisition and Processing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MDPI | Multidisciplinary Digital Publishing Institute |
| DOAJ | Directory of open access journals |
| TLA | Three letter acronym |
| LD | linear dichroism |
References
- Hou, Y.; Dan, X.; Babbar, M.; Wei, Y.; Hasselbalch, S.G.; Croteau, D.L.; Bohr, V.A. Ageing as a risk factor for neurodegenerative disease. Nat. Rev. Neurol. 2019, 15, 565–581. [Google Scholar] [CrossRef]
- Mayne, K.; White, J.A.; McMurran, C.E.; Rivera, F.J.; de la Fuente, A.G. Aging and Neurodegenerative Disease: Is the Adaptive Immune System a Friend or Foe? Front. Aging Neurosci. 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Tony Wyss, C. Ageing, neurodegeneration and brain rejuvenation. Nature 2016, 10, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Podtelezhnikov, A.A.; Tanis, K.Q.; Nebozhyn, M.; Ray, W.J.; Stone, D.J.; Loboda, A.P. Molecular insights into the pathogenesis of Alzheimer’s disease and its relationship to normal aging. PLoS ONE 2011, 13, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wakabayashi, T. Transmembrane Collagens in Neuromuscular Development and Disorders. Front. Mol. Neurosci. 2021, 13, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Monavarfeshani, A.; Knill, C.N.; Sabbagh, U.; Su, J.; Fox, M.A.; Pantazopoulos, H.; Hedin-Pereira, C.; Lucas, G. Region- and Cell-Specific Expression of Transmembrane Collagens in Mouse Brain. Front. Integr. Neurosci. 2017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urabe, N.; Naito, I.; Saito, K.; Yonezawa, T.; Sado, Y.; Yoshioka, H.; Kusachi, S.; Tsuji, T.; Ohtsuka, A.; Taguchi, T.; et al. Basement membrane type IV collagen molecules in the choroid plexus, pia mater and capillaries in the mouse brain. Arch. Histol. Cytol. 2002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuo, D.S.; Labelle-dumais, C.; Gould, D.B. COL4A1 and COL4A2 mutations and disease: Insights into pathogenic mechanisms and potential therapeutic targets. Hum. Mol. Genet. 2012, 21. [Google Scholar] [CrossRef] [Green Version]
- Uspenskaia, O.; Liebetrau, M.; Herms, J.; Danek, A.; Hamann, G.F. Aging is associated with increased collagen type IV accumulation in the basal lamina of human cerebral microvessels. BMC Neurosci. 2004, 6, 37. [Google Scholar] [CrossRef] [Green Version]
- Kalaria, R.N.; Pax, A.B. Increased collagen content of cerebral microvessels in Alzheimer’s disease. Brain Res. 1995, 705, 349–352. [Google Scholar] [CrossRef]
- Farkas, E.; De Jong, G.I.; de Vos, R.A.; Jansen Steur, E.N.; Luiten, P.G. Pathological features of cerebral cortical capillaries are doubled in Alzheimer’s disease and Parkinson’s disease. Acta Neuropathol. 2000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeuchi, M.; Yamaguchi, S.; Yonemura, S.; Kakiguchi, K.; Sato, Y.; Higashiyama, T.; Shimizu, T.; Hibi, M. Type IV Collagen Controls the Axogenesis of Cerebellar Granule Cells by Regulating Basement Membrane Integrity in Zebrafish. PLoS Genet. 2015, 11, e1005587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, T.; Baier, H. Lamina-specific axonal projections in the zebrafish tectum require the type IV collagen Dragnet. Nat. Neurosci. 2007, 10, 1529–1537. [Google Scholar] [CrossRef] [PubMed]
- Xiao, T.; Staub, W.; Robles, E.; Gosse, N.J.; Cole, G.J.; Baier, H. Assembly of lamina-specific neuronal connections by slit bound to type IV collagen. Cell 2011, 146, 164–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hashimoto, T.; Wakabayashi, T.; Watanabe, A.; Kowa, H.; Hosoda, R.; Nakamura, A.; Kanazawa, I.; Arai, T.; Takio, K.; Mann, D.M.A.; et al. CLAC: A novel Alzheimer amyloid plaque component derived from a transmembrane precursor, CLAC-P/collagen type XXV. EMBO J. 2002, 21, 1524–1534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, T.; Wakabayashi, T.; Oizumi, H.; Nishio, S.; Sato, T.; Harada, A.; Fujii, D.; Matsuo, Y.; Hashimoto, T.; Iwatsubo, T. CLAC-P/Collagen Type XXV Is Required for the Intramuscular Innervation of Motoneurons during Neuromuscular Development. J. Neurosci. 2014, 34, 1370–1379. [Google Scholar] [CrossRef] [Green Version]
- Osada, Y.; Hashimoto, T.; Nishimura, A.; Matsuo, Y.; Wakabayashi, T.; Iwatsubo, T. CLAC binds to amyloid β peptides through the positively charged amino acid cluster within the collagenous domain 1 and inhibits formation of amyloid fibrils. J. Biol. Chem. 2005, 280, 8596–8605. [Google Scholar] [CrossRef] [Green Version]
- Forsell, C.; Bj, B.F.; Lilius, L.; Axelman, K.; Froelich, S.; Fratiglioni, L.; Winblad, B.; Graff, C. Genetic association to the amyloid plaque associated protein gene COL25A1 in Alzheimer’s disease. Neurobiol. Aging 2010, 31, 409–415. [Google Scholar] [CrossRef]
- Tong, Y.; Xu, Y.; Scearce-levie, K.; Ptá, L.J.; Fu, Y. COL25A1 triggers and promotes Alzheimer ’ s disease-like pathology in vivo. Neurogenetics 2010, 41–52. [Google Scholar] [CrossRef] [Green Version]
- Platzer, M.; Englert, C. Nothobranchius furzeri: A Model for Aging Research and More. Trends Genet. 2016, 32, 543–552. [Google Scholar] [CrossRef]
- D’Angelo, L.; Lossi, L.; Merighi, A.; de Girolamo, P. Anatomical features for the adequate choice of experimental animal models in biomedicine: I. Fishes. Ann. Anat. 2016, 205, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.K.; Brunet, A. The African turquoise killifish: A research organism to study vertebrate aging and diapause. Aging Cell 2018, 17, e12757. [Google Scholar] [CrossRef] [PubMed]
- Leggieri, A.; Attanasio, C.; Palladino, A.; Cellerino, A.; Lucini, C.; Paolucci, M.; Terzibasi Tozzini, E.; de Girolamo, P.; D’Angelo, L. Identification and Expression of Neurotrophin-6 in the Brain of Nothobranchius furzeri: One More Piece in Neurotrophin Research. J. Clin. Med. 2019, 8, 595. [Google Scholar] [CrossRef] [Green Version]
- De Girolamo, P.; Leggieri, A.; Palladino, A.; Lucini, C.; Attanasio, C.; D’angelo, L. Cholinergic system and NGF receptors: Insights from the brain of the short-lived fish nothobranchius furzeri. Brain Sci. 2020, 10, 394. [Google Scholar] [CrossRef] [PubMed]
- Cellerino, A.; Valenzano, D.R.; Reichard, M. From the bush to the bench: The annual Nothobranchius fishes as a new model system in biology. Biol. Rev. Camb. Philos. Soc. 2016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Cicco, E.; Tozzini, E.T.; Rossi, G.; Cellerino, A. The short-lived annual fish Nothobranchius furzeri shows a typical teleost aging process reinforced by high incidence of age-dependent neoplasias. Exp. Gerontol. 2011, 46, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Tozzini, E.T.; Baumgart, M.; Battistoni, G.; Cellerino, A. Adult neurogenesis in the short-lived teleost Nothobranchius furzeri: Localization of neurogenic niches, molecular characterization and effects of aging. Aging Cell 2012, 11, 241–251. [Google Scholar] [CrossRef] [Green Version]
- Guvatova, Z.G.; Fedorova, M.S.; Vershinina, Y.S.; Pudova, E.A.; Lipatova, A.V.; Volodin, V.V.; Gladysh, N.S.; Tokarev, A.T.; Kornev, A.B.; Pavlov, V.S.; et al. De Novo Transcriptome Profiling of Brain Tissue from the Annual Killifish Nothobranchius guentheri. Life 2021, 11, 137. [Google Scholar] [CrossRef]
- Leggieri, A.; Palladino, A.; Attanasio, C.; Avallone, L.; de Girolamo, P.; D’Angelo, L.; Lucini, C. Id(entifying) the inhibitor of DNA binding 3 in the brain of Nothobranchius furzeri upon aging. J. Anat. 2021, 238, 1106–1115. [Google Scholar] [CrossRef]
- Marschallinger, J.; Schäffner, I.; Klein, B.; Gelfert, R.; Rivera, F.J.; Illes, S.; Grassner, L.; Janssen, M.; Rotheneichner, P.; Schmuckermair, C.; et al. Structural and functional rejuvenation of the aged brain by an approved anti-asthmatic drug. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [Green Version]
- Carrasco-garcia, E.; Moreno-cugnon, L.; Garcia, I.; Borras, C.; Revuelta, M.; Izeta, A.; Lopez-lluch, G.; Pancorbo, M.M. De SOX2 expression diminishes with ageing in several tissues in mice and humans. Mech. Ageing Dev. 2019, 177, 30–36. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, L.; De Girolamo, P.; Cellerino, A.; Tozzini, E.T.; Varricchio, E.; Castaldo, L.; Lucini, C. Immunolocalization of S100-like protein in the brain of an emerging model organism: Nothobranchius furzeri. Microsc. Res. Tech. 2012. [Google Scholar] [CrossRef] [PubMed]
- Ayanlaja, A.A.; Xiong, Y.; Gao, Y.; Ji, G.; Tang, C.; Abdullah, Z.; Gao, D. Distinct features of doublecortin as a marker of neuronal migration and its implications in cancer cell mobility. Front. Mol. Neurosci. 2017, 10, 199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magalhães, J.P.; De Curado, J.; Church, G.M. Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics 2009, 25, 875–881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahm, A.; Bens, M.; Platzer, M.; Cellerino, A. Parallel evolution of genes controlling mitonuclear balance in short-lived annual fishes. Aging Cell 2017, 16, 488–496. [Google Scholar] [CrossRef] [Green Version]
- Ewald, C.Y.; Landis, J.N.; Porter Abate, J.; Murphy, C.T.; Blackwell, T.K. Dauer-independent insulin/IGF-1-signalling implicates collagen remodelling in longevity. Nature 2015. [Google Scholar] [CrossRef] [Green Version]
- Fushan, A.A.; Turanov, A.A.; Lee, S.G.; Kim, E.B.; Lobanov, A.V.; Yim, S.H.; Buffenstein, R.; Lee, S.R.; Chang, K.T.; Rhee, H.; et al. Gene expression defines natural changes in mammalian lifespan. Aging Cell 2015, 14, 352–365. [Google Scholar] [CrossRef]
- Thomas, M.H.; Gui, Y.; Garcia, P.; Karout, M.; Gomez Ramos, B.; Jaeger, C.; Michelucci, A.; Gaigneaux, A.; Kollmus, H.; Centeno, A.; et al. Quantitative trait locus mapping identifies a locus linked to striatal dopamine and points to collagen IV alpha-6 chain as a novel regulator of striatal axonal branching in mice. Genes Brain Behav. 2021, 20, e12769. [Google Scholar] [CrossRef]
- Soltani, M.H.; Pichardo, R.; Song, Z.; Sangha, N.; Camacho, F.; Satyamoorthy, K.; Sangueza, O.P.; Setaluri, V. Microtubule-associated protein 2, a marker of neuronal differentiation, induces mitotic defects, inhibits growth of melanoma cells, and predicts metastatic potential of cutaneous melanoma. Am. J. Pathol. 2005, 166, 1841–1850. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Chen, K.; Sloan, S.A.; Bennett, M.L.; Scholze, A.R.; O’Keefee, S.; Phatnani, H.P.; Guarnieri, P.; Caneda, C.; Ruderisch, N.; et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 2014. [Google Scholar] [CrossRef]
- Hagmeyer, S.; Romão, M.A.; Cristóvão, J.S.; Vilella, A.; Zoli, M.; Gomes, C.M.; Grabrucker, A.M. Distribution and relative abundance of S100 proteins in the brain of the APP23 Alzheimer’s disease model mice. Front. Neurosci. 2019, 13, 1–10. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the Number of Nucleotide Substitutions in the Control Region of Mitochondrial DNA in Humans and Chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [PubMed]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montesano, A.; Baumgart, M.; Avallone, L.; Castaldo, L.; Lucini, C.; Tozzini, E.T.; Cellerino, A.; D’Angelo, L.; de Girolamo, P. Age-related central regulation of orexin and NPY in the short-lived African killifish Nothobranchius furzeri. J. Comp. Neurol. 2019, 527, 1508–1526. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Nijveen, H.; Rao, X.; Bisseling, T.; Geurts, R.; Leunissen, J.A.M. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007, 35, 71–74. [Google Scholar] [CrossRef] [Green Version]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: Berlin/Heidelberg, Germany, 2009; Volume 35, ISBN 978-0-387-98140-6. [Google Scholar]
- D’angelo, L. Brain atlas of an emerging teleostean model: Nothobranchius furzeri. Anat Rec. (Hoboken) 2013, 296, 681–691. [Google Scholar] [CrossRef]


| Gene | Primer | Sequence (5′-3′) |
|---|---|---|
| dcx | Forward | AGGTGCTCACTGACATCACA |
| dcx | Reverse | CGCCGAAGAAATCCTGAAGG |
| S100β | Forward | GCGTGTCTACTTGTGTGCAT |
| S100β | Reverse | TTCTCGCCATCTCTCTCGTC |
| sox2 | Forward | GAACGGCACCAACCAGAAAA |
| sox2 | Reverse | TCGGAGTTGTGCATTTTGGG |
| col4a1 | Forward | CTGGAATCCCTGGAGAGCC |
| col4a1 | Reverse | CCTGGAGGCCTTGTGTACC |
| col25a1 | Forward | AACCTGAAACGCATGCAGTT |
| col25a1 | Reverse | TCACGGCAGGACACTCAG |
| TBP | Forward | CGGTTGGAGGGTTTAGTCCT |
| TBP | Reverse | GCAAGACGATTCTGGGTTTG |
| Dilution Number | Final Concentration |
|---|---|
| 1 | 2 ng/µL |
| 2 | 500 pg/µL |
| 3 | 100 pg/µL |
| 4 | 50 pg/µL |
| 5 | 25 pg/µL |
| 6 | 10 pg/µL |
| 7 | 5 pg/µL |
| 8 | 2.5 pg/µL |
| 9 | 1 pg/µL |
| 10 | 0.1 pg/µL |
| Primary Antiserum | Source | Host | Dilution |
|---|---|---|---|
| anti-S100β | Agilent Dako Z0311 | Rabbit Polyclonal | 1:200 |
| anti-DCX | Abcam (18723) | Rabbit polyclonal | 1:100 |
| anti-HuC/HuD | Invitrogen by Thermo Fisher Scientific (A21272) | Mouse IgG2b, biotin-XX conjugate | 1:50 |
| anti-MAP2 | Santa Cruz Biotechnology (A8): sc-74422 | Mouse Monoclonal | 1:100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leggieri, A.; Attanasio, C.; Palladino, A.; de Girolamo, P.; Lucini, C.; D’Angelo, L. Neuronal Phenotype of col4a1 and col25a1: An Intriguing Hypothesis in Vertebrates Brain Aging. Int. J. Mol. Sci. 2022, 23, 1778. https://doi.org/10.3390/ijms23031778
Leggieri A, Attanasio C, Palladino A, de Girolamo P, Lucini C, D’Angelo L. Neuronal Phenotype of col4a1 and col25a1: An Intriguing Hypothesis in Vertebrates Brain Aging. International Journal of Molecular Sciences. 2022; 23(3):1778. https://doi.org/10.3390/ijms23031778
Chicago/Turabian StyleLeggieri, Adele, Chiara Attanasio, Antonio Palladino, Paolo de Girolamo, Carla Lucini, and Livia D’Angelo. 2022. "Neuronal Phenotype of col4a1 and col25a1: An Intriguing Hypothesis in Vertebrates Brain Aging" International Journal of Molecular Sciences 23, no. 3: 1778. https://doi.org/10.3390/ijms23031778
APA StyleLeggieri, A., Attanasio, C., Palladino, A., de Girolamo, P., Lucini, C., & D’Angelo, L. (2022). Neuronal Phenotype of col4a1 and col25a1: An Intriguing Hypothesis in Vertebrates Brain Aging. International Journal of Molecular Sciences, 23(3), 1778. https://doi.org/10.3390/ijms23031778









