Enhanced Expression of TRAP1 Protects Mitochondrial Function in Motor Neurons under Conditions of Oxidative Stress
Abstract
1. Introduction
2. Results
2.1. TRAP1 Rescues Oxidative Stress-Induced Loss of Mitochondrial Membrane Potential in Primary Motor Neuron Cultures
2.2. TRAP1 Overexpression Reduces Oxidative Stress-Induced ROS Release and Protects Motor Neurons from Cell Death
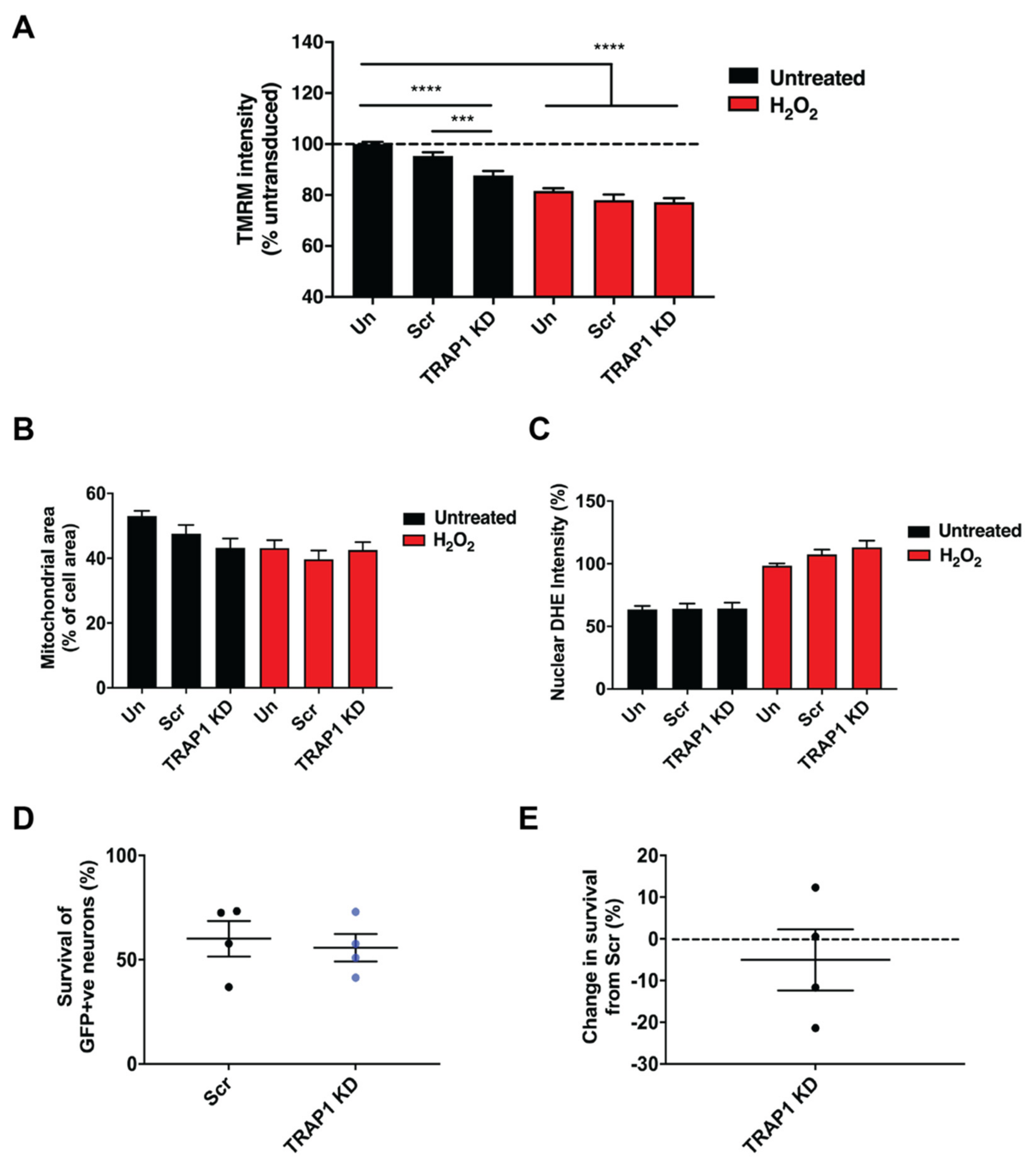
2.3. TRAP1 Knockdown Reduces Mitochondrial Membrane Potential but Does Not Exacerbate Damage to Motor Neurons Induced by Oxidative Stress
3. Discussion
4. Materials and Methods
4.1. Primary Motor Neuron Cultures
4.2. Treatment of Primary Motor Neuron Cultures
4.3. TRAP1 Construct Design
4.4. Generation of Viral Vectors for Gene Delivery
4.5. Mitochondrial Membrane Potential Recordings
4.6. ROS Production Measurements
4.7. Cell Viability Assay
4.8. Western Blotting
4.9. Immunofluorescence
4.10. Image Analysis
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ALS | amyotrophic lateral sclerosis |
| DHE | dihydroethidium |
| DIV | days in vitro |
| Dnase | deoxyribonuclease |
| EGTA | egtazic acid |
| Hsp | heat shock protein |
| PD | Parkinson’s disease |
| RIPA | Radioimmunoprecipitation assay buffer |
| ROS | reactive oxygen species |
| SOD1 | superoxide dismutase 1 |
| TMRM | tetramethylrhodamine |
| TRAP1 | TNF-receptor associated protein |
| TUJ1 | neuron-specific class III beta-tubulin |
References
- Kalmar, B.; Greensmith, L. Cellular Chaperones as Therapeutic Targets in ALS to Restore Protein Homeostasis and Improve Cellular Function. Front. Mol. Neurosci. 2017, 10, 251. [Google Scholar] [CrossRef] [PubMed]
- Song, H.Y.; Dunbar, J.D.; Zhang, Y.X.; Guo, D.; Donner, D.B. Identification of a protein with homology to hsp90 that binds the type 1 tumor necrosis factor receptor. J. Biol. Chem. 1995, 270, 3574–3581. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.F.; Chen, Y.; Dai, K.; Chen, P.L.; Riley, D.J.; Lee, W.H. A new member of the hsp90 family of molecular chaperones interacts with the retinoblastoma protein during mitosis and after heat shock. Mol. Cell. Biol. 1996, 16, 4691–4699. [Google Scholar] [CrossRef]
- Cechetto, J.D.; Gupta, R.S. Immunoelectron Microscopy Provides Evidence That Tumor Necrosis Factor Receptor-Associated Protein 1 (TRAP-1) Is a Mitochondrial Protein Which Also Localizes at Specific Extramitochondrial Sites. Exp. Cell Res. 2000, 260, 30–39. [Google Scholar] [CrossRef]
- Pridgeon, J.W.; Olzmann, J.; Chin, L.-S.; Li, L. PINK1 Protects against Oxidative Stress by Phosphorylating Mitochondrial Chaperone TRAP1. PLoS Biol. 2007, 5, e172. [Google Scholar] [CrossRef] [PubMed]
- Kang, B.-H. TRAP1 regulation of mitochondrial life or death decision in cancer cells and mitochondria-targeted TRAP1 inhibitors. BMB Rep. 2012, 45, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Kang, B.H.; Plescia, J.; Dohi, T.; Rosa, J.; Doxsey, S.J.; Altieri, D.C. Regulation of Tumor Cell Mitochondrial Homeostasis by an Organelle-Specific Hsp90 Chaperone Network. Cell 2007, 131, 257–270. [Google Scholar] [CrossRef] [PubMed]
- Agorreta, J.; Hu, J.; Liu, D.; Delia, M.; Turley, H.; Ferguson, D.J.P.; Iborra, F.; Pajares, M.J.; Larrayoz, M.; Zudaire, I.; et al. TRAP1 regulates proliferation, mitochondrial function, and has prognostic significance in NSCLC. Mol. Cancer Res. 2014, 12, 660–669. [Google Scholar] [CrossRef]
- Guzzo, G.; Sciacovelli, M.; Bernardi, P.; Rasola, A. Inhibition of succinate dehydrogenase by the mitochondrial chaperone TRAP1 has anti-oxidant and anti-apoptotic effects on tumor cells. Oncotarget 2014, 5, 11897–11908. [Google Scholar] [CrossRef]
- Basit, F.; Van Oppen, L.M.P.E.; Schöckel, L.; Bossenbroek, H.M.; Van Emst-de Vries, S.E.; Hermeling, J.C.W.; Grefte, S.; Kopitz, C.; Heroult, M.; Willems, P.H.; et al. Mitochondrial complex I inhibition triggers a mitophagy-dependent ROS increase leading to necroptosis and ferroptosis in melanoma cells. Cell Death Dis. 2017, 8, e2716. [Google Scholar] [CrossRef]
- Abramov, A.Y.; Berezhnov, A.V.; Fedotova, E.I.; Zinchenko, V.P.; Dolgacheva, L.P. Interaction of misfolded proteins and mitochondria in neurodegenerative disorders. Biochem. Soc. Trans. 2017, 45, 1025–1033. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, J.C.; Zimprich, A.; Berrio, D.A.C.; Schindler, K.M.; Maurer, B.; Schulte, C.; Bus, C.; Hauser, A.-K.; Kübler, M.; Lewin, R.; et al. Metformin reverses TRAP1 mutation-associated alterations in mitochondrial function in Parkinson’s disease. Brain 2017, 140, 2444–2459. [Google Scholar] [CrossRef]
- Butler, E.K.; Voigt, A.; Lutz, A.K.; Toegel, J.P.; Gerhardt, E.; Karsten, P.; Falkenburger, B.; Reinartz, A.; Winklhofer, K.F.; Schulz, J.B. The Mitochondrial Chaperone Protein TRAP1 Mitigates α-Synuclein Toxicity. PLoS Genet. 2012, 8, e1002488. [Google Scholar] [CrossRef] [PubMed]
- Hardiman, O.; Al-Chalabi, A.; Chio, A.; Corr, E.M.; Logroscino, G.; Robberecht, W.; Shaw, P.J.; Simmons, Z.; van den Berg, L.H. Amyotrophic lateral sclerosis. Nat. Rev. Dis. Primers 2017, 3, 17071. [Google Scholar] [CrossRef] [PubMed]
- Kieran, D.; Kalmar, B.; Dick, J.R.T.; Riddoch-Contreras, J.; Burnstock, G.; Greensmith, L. Treatment with arimoclomol, a coinducer of heat shock proteins, delays disease progression in ALS mice. Nat. Med. 2004, 10, 402–405. [Google Scholar] [CrossRef]
- Sharp, P.S.; Akbar, M.T.; Bouri, S.; Senda, A.; Joshi, K.; Chen, H.-J.; Latchman, D.S.; Wells, D.J.; de Belleroche, J. Protective effects of heat shock protein 27 in a model of ALS occur in the early stages of disease progression. Neurobiol. Dis. 2008, 30, 42–55. [Google Scholar] [CrossRef]
- Perry, S.W.; Norman, J.P.; Barbieri, J.; Brown, E.B.; Gelbard, H.A. Mitochondrial membrane potential probes and the proton gradient: A practical usage guide. BioTechniques 2011, 50, 98–115. [Google Scholar] [CrossRef]
- Baqri, R.M.; Pietron, A.V.; Gokhale, R.H.; Turner, B.A.; Kaguni, L.S.; Shingleton, A.W.; Kunes, S.; Miller, K.E. Mitochondrial chaperone TRAP1 activates the mitochondrial UPR and extends healthspan in Drosophila. Mech. Ageing Dev. 2014, 141–142, 35–45. [Google Scholar] [CrossRef][Green Version]
- Costa, A.C.; Loh, S.H.Y.; Martins, L.M. Drosophila Trap1 protects against mitochondrial dysfunction in a PINK1/parkin model of Parkinson’s disease. Cell Death Dis. 2013, 4, e467. [Google Scholar] [CrossRef]
- Serapian, S.A.; Sanchez-Martín, C.; Moroni, E.; Rasola, A.; Colombo, G. Targeting the mitochondrial chaperone TRAP1: Strategies and therapeutic perspectives. Trends Pharmacol. Sci. 2021, 42, 566–576. [Google Scholar] [CrossRef]
- Sciacovelli, M.; Guzzo, G.; Morello, V.; Frezza, C.; Zheng, L.; Nannini, N.; Calabrese, F.; Laudiero, G.; Esposito, F.; Landriscina, M.; et al. The Mitochondrial Chaperone TRAP1 Promotes Neoplastic Growth by Inhibiting Succinate Dehydrogenase. Cell Metab. 2013, 17, 988–999. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, S.; Tsutsumi, S.; Muhlebach, G.; Sourbier, C.; Lee, M.-J.; Lee, S.; Vartholomaiou, E.; Tatokoro, M.; Beebe, K.; Miyajima, N.; et al. Molecular chaperone TRAP1 regulates a metabolic switch between mitochondrial respiration and aerobic glycolysis. Proc. Natl. Acad. Sci. USA 2013, 110, E1604–E1612. [Google Scholar] [CrossRef] [PubMed]
- Joshi, A.; Dai, L.; Liu, Y.; Lee, J.; Ghahhari, N.M.; Segala, G.; Beebe, K.; Jenkins, L.M.; Lyons, G.C.; Bernasconi, L.; et al. The mitochondrial HSP90 paralog TRAP1 forms an OXPHOS-regulated tetramer and is involved in mitochondrial metabolic homeostasis. BMC Biol. 2020, 18, 10. [Google Scholar] [CrossRef]
- Masgras, I.; Sanchez-Martin, C.; Colombo, G.; Rasola, A. The Chaperone TRAP1 As a Modulator of the Mitochondrial Adaptations in Cancer Cells. Front. Oncol. 2017, 7, 58. [Google Scholar] [CrossRef] [PubMed]
- Gesualdi, N.M.; Chirico, G.; Pirozzi, G.; Costantino, E.; Landriscina, M.; Esposito, F. Tumor necrosis factor-associated protein 1 (TRAP-1) protects cells from oxidative stress and apoptosis. Stress 2007, 10, 342–350. [Google Scholar] [CrossRef] [PubMed]
- Cunha-Oliveira, T.; Montezinho, L.; Mendes, C.; Firuzi, O.; Saso, L.; Oliveira, P.J.; Silva, F.S.G. Oxidative Stress in Amyotrophic Lateral Sclerosis: Pathophysiology and Opportunities for Pharmacological Intervention. Oxidative Med. Cell. Longev. 2020, 2020, 5021694. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Tai, P.W.L.; Gao, G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discov. 2019, 18, 358–378. [Google Scholar] [CrossRef] [PubMed]
- Camu, W.; Henderson, C.E. Purification of embryonic rat motoneurons by panning on a monoclonal antibody to the low-affinity NGF receptor. J. Neurosci. Methods 1992, 44(1), 59–70. [Google Scholar] [CrossRef]
- Bilsland, L.G.; Nirmalananthan, N.; Yip, J.; Greensmith, L.; Duchen, M.R. Expression of mutant SOD1G93Ain astrocytes induces functional deficits in motoneuron mitochondria. J. Neurochem. 2008, 107, 1271–1283. [Google Scholar] [CrossRef]
- Lou, E.; Fujisawa, S.; Barlas, A.; Romin, Y.; Manova-Todorova, K.; Moore, M.A.; Subramanian, S. Tunneling Nanotubes. Commun. Integr. Biol. 2012, 5, 399–403. [Google Scholar] [CrossRef]


| Name | shRNA Sequence |
|---|---|
| trap1-1 | GGTTGAAGTCTATTCTCGATC |
| trap1-2 | GGATGTTCTACAACAGAGATT |
| Scrambled control | GCTTCGCGCCGTAGTCTTA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Clarke, B.E.; Kalmar, B.; Greensmith, L. Enhanced Expression of TRAP1 Protects Mitochondrial Function in Motor Neurons under Conditions of Oxidative Stress. Int. J. Mol. Sci. 2022, 23, 1789. https://doi.org/10.3390/ijms23031789
Clarke BE, Kalmar B, Greensmith L. Enhanced Expression of TRAP1 Protects Mitochondrial Function in Motor Neurons under Conditions of Oxidative Stress. International Journal of Molecular Sciences. 2022; 23(3):1789. https://doi.org/10.3390/ijms23031789
Chicago/Turabian StyleClarke, Benjamin E., Bernadett Kalmar, and Linda Greensmith. 2022. "Enhanced Expression of TRAP1 Protects Mitochondrial Function in Motor Neurons under Conditions of Oxidative Stress" International Journal of Molecular Sciences 23, no. 3: 1789. https://doi.org/10.3390/ijms23031789
APA StyleClarke, B. E., Kalmar, B., & Greensmith, L. (2022). Enhanced Expression of TRAP1 Protects Mitochondrial Function in Motor Neurons under Conditions of Oxidative Stress. International Journal of Molecular Sciences, 23(3), 1789. https://doi.org/10.3390/ijms23031789






