Cerebrospinal Fluid Metabolome in Parkinson’s Disease and Multiple System Atrophy
Abstract
1. Introduction
2. Characteristic Differences between PD and MSA
3. Metabolomics Approaches for PD and MSA in CSF
3.1. Analysis of Metabolites
3.2. Metabolomic Differences in the CSF in PD and MSA
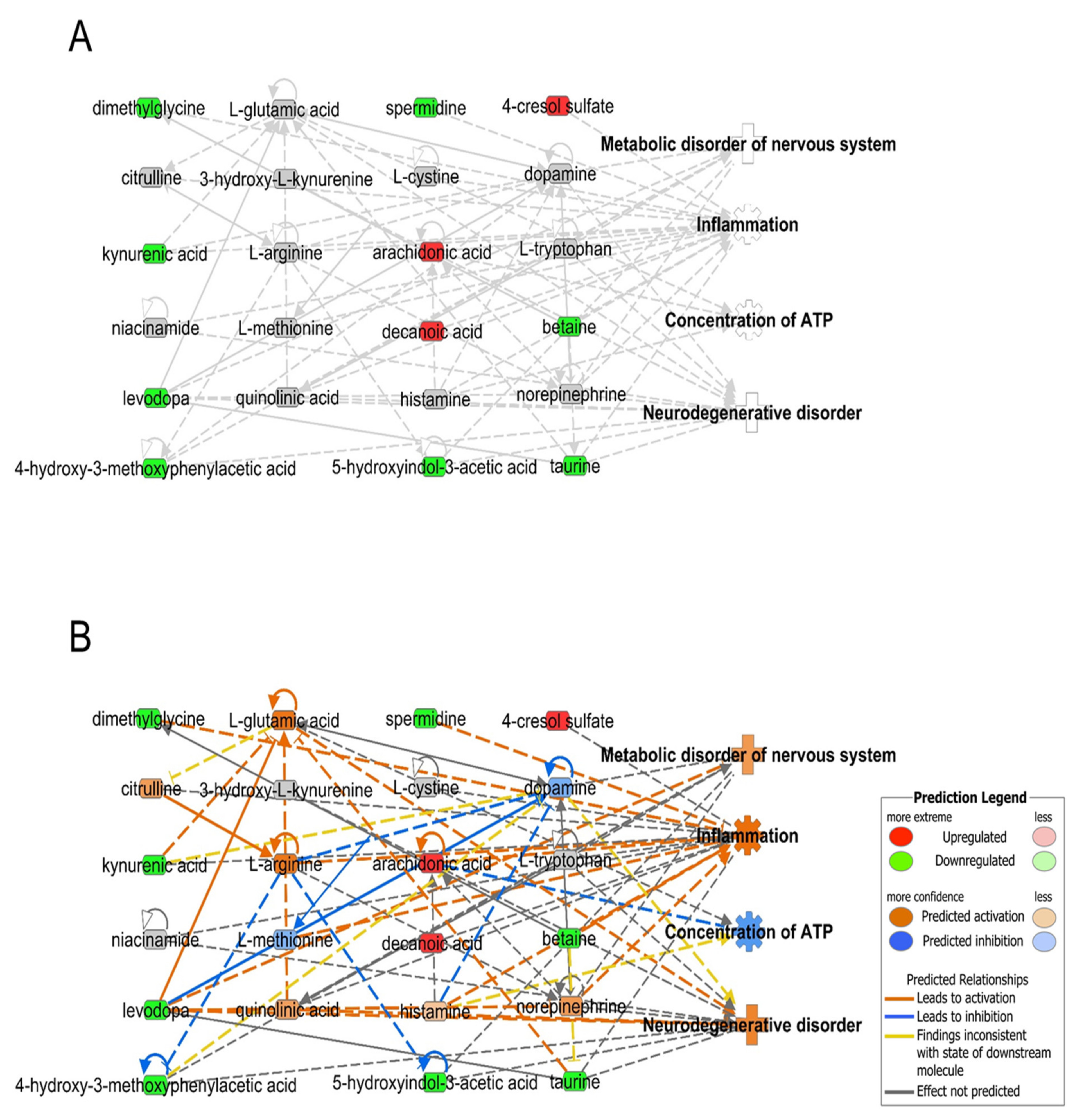
3.3. Application of Metabotypes
4. Proteomics and MicroRNA Analysis in PD and MSA
4.1. Proteomics
4.2. MicroRNAs
4.3. Integrated Omics
5. Summary
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Spillantini, M.G.; Schmidt, M.L.; Lee, V.M.; Trojanowski, J.Q.; Jakes, R.; Goedert, M. Alpha-synuclein in Lewy bodies. Nature 1997, 388, 839–840. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, T.R.; Holmes, B.B.; Furman, J.L.; Dhavale, D.D.; Su, B.W.; Song, E.S.; Cairns, N.J.; Kotzbauer, P.T.; Diamond, M.I. Parkinson’s disease and multiple system atrophy have distinct α-synuclein seed characteristics. J. Biol. Chem. 2019, 294, 1045–1058. [Google Scholar] [CrossRef] [PubMed]
- Jeromin, A.; Bowser, R. Biomarkers in Neurodegenerative Diseases. Adv. NeuroBiol. 2017, 15, 491–528. [Google Scholar]
- Koulman, A.; Lane, G.A.; Harrison, S.J.; Volmer, D.A. From differentiating metabolites to biomarkers. Anal. Bioanal. Chem. 2009, 394, 663–670. [Google Scholar] [CrossRef] [PubMed]
- Al-Aama, J.Y.; Al Mahdi, H.B.; Salama, M.A.; Bakur, K.H.; Alhozali, A.; Mosli, H.H.; Bahijri, S.M.; Bahieldin, A.; Willmitzer, L.; Edris, S. Detection of Secondary Metabolites as Biomarkers for the Early Diagnosis and Prevention of Type 2 Diabetes. Diabetes Metab. Syndr. Obes. 2019, 12, 2675–2684. [Google Scholar] [CrossRef]
- Gallelli, L.; Mannino, G.C.; Luciani, F.; de Sire, A.; Mancuso, E.; Gangemi, P.; Cosco, L.; Monea, G.; Averta, C.; Minchella, P.; et al. Vitamin D Serum Levels in Subjects Tested for SARS-CoV-2: What Are the Differences among Acute, Healed, and Negative COVID-19 Patients? A Multicenter Real-Practice Study. Nutrients 2021, 13, 3932. [Google Scholar] [CrossRef] [PubMed]
- Martignoni, E.; Blandini, F.; Petraglia, F.; Pacchetti, C.; Bono, G.; Nappi, G. Cerebrospinal fluid norepinephrine, 3-methoxy-4-hydroxyphenylglycol and neuropeptide Y levels in Parkinson’s disease, multiple system atrophy and dementia of the Alzheimer type. J. Neural. Transm. Park Dis. Dement. Sect. 1992, 4, 191–205. [Google Scholar] [CrossRef]
- Paik, M.-J.; Ahn, Y.-H.; Lee, P.H.; Kang, H.; Park, C.B.; Choi, S.; Lee, G. Polyamine patterns in the cerebrospinal fluid of patients with Parkinson’s disease and multiple system atrophy. Clin. Chim. Acta 2010, 411, 1532–1535. [Google Scholar] [CrossRef]
- Goldstein, D.S.; Holmes, C.; Sharabi, Y. Cerebrospinal fluid biomarkers of central catecholamine deficiency in Parkinson’s disease and other synucleinopathies. Brain 2012, 135, 1900–1913. [Google Scholar] [CrossRef]
- Goldstein, D.S.; Sullivan, P.; Holmes, C.; Lamotte, G.; Lenka, A.; Sharabi, Y. Differential abnormalities of cerebrospinal fluid dopaminergic versus noradrenergic indices in synucleinopathies. J. NeuroChem. 2021, 158, 554–568. [Google Scholar] [CrossRef]
- Hillesheim, E.; Ryan, M.F.; Gibney, E.; Roche, H.M.; Brennan, L. Optimisation of a metabotype approach to deliver targeted dietary advice. Nutr. Metab. 2020, 17, 82. [Google Scholar] [CrossRef] [PubMed]
- Walsh, M.J.; Limos, L.; Tourtellotte, W.W. Two-Dimensional Electrophoresis of Cerebrospinal Fluid and Ventricular Fluid Proteins, Identification of Enriched and Unique Proteins, and Comparison with Serum. J. Neurochem. 1984, 43, 1277–1285. [Google Scholar] [CrossRef]
- Marques, T.M.; Kuiperij, H.B.; Bruinsma, I.B.; van Rumund, A.; Aerts, M.B.; Esselink, R.A.J.; Bloem, B.R.; Verbeek, M.M. MicroRNAs in Cerebrospinal Fluid as Potential Biomarkers for Parkinson’s Disease and Multiple System Atrophy. Mol. Neurobiol. 2017, 54, 7736–7745. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.H.; Park, K.W.; Heo, K.O.; Chung, S.J.; Choo, M.S. Urodynamic study for distinguishing multiple system atrophy from Parkinson disease. Neurology 2019, 93, e946–e953. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Deng, N.; Lu, K.; Liao, Q.; Long, X.; Gou, D.; Bi, F.; Zhou, J. Elevated plasma miR-133b and miR-221-3p as biomarkers for early Parkinson’s disease. Sci. Rep. 2021, 11, 15268. [Google Scholar] [CrossRef]
- Marques, T.M.; van Rumund, A.; Kersten, I.; Bruinsma, I.B.; Wessels, H.; Gloerich, J.; Kaffa, C.; Esselink, R.A.J.; Bloem, B.R.; Kuiperij, H.B.; et al. Identification of cerebrospinal fluid biomarkers for parkinsonism using a proteomics approach. NPJ Parkinsons Dis. 2021, 7, 107. [Google Scholar] [CrossRef]
- Kim, J.S.; Yoon, T.-J.; Yu, K.N.; Kim, B.G.; Park, S.J.; Kim, H.W.; Lee, K.H.; Park, S.B.; Lee, J.-K.; Cho, M.H. Toxicity and Tissue Distribution of Magnetic Nanoparticles in Mice. Toxicol. Sci. 2006, 89, 338–347. [Google Scholar] [CrossRef]
- Shim, W.; Paik, M.J.; Nguyen, D.-T.; Lee, J.-K.; Lee, Y.; Kim, J.-H.; Shin, E.-H.; Kang, J.S.; Jung, H.-S.; Choi, S.; et al. Analysis of Changes in Gene Expression and Metabolic Profiles Induced by Silica-Coated Magnetic Nanoparticles. ACS Nano 2012, 6, 7665–7680. [Google Scholar] [CrossRef]
- Guo, J.; Sun, Z.; Xiao, S.; Liu, D.; Jin, G.; Wang, E.; Zhou, J.; Zhou, J. Proteomic analysis of the cerebrospinal fluid of Parkinson’s disease patients. Cell Res. 2009, 19, 1401–1403. [Google Scholar] [CrossRef][Green Version]
- Shi, M.; Bradner, J.; Hancock, A.M.; Chung, K.A.; Quinn, J.F.; Peskind, E.R.; Galasko, D.; Jankovic, J.; Zabetian, C.P.; Kim, H.M. Cerebrospinal fluid biomarkers for Parkinson disease diagnosis and progression. Ann. Neurol. 2011, 69, 570–580. [Google Scholar] [CrossRef]
- Shi, M.; Movius, J.; Dator, R.; Aro, P.; Zhao, Y.; Pan, C.; Lin, X.; Bammler, T.K.; Stewart, T.; Zabetian, C.P. Cerebrospinal Fluid Peptides as Potential Parkinson Disease Biomarkers: A Staged Pipeline for Discovery and Validation. Mol. Cell. Proteom. 2015, 14, 544–555. [Google Scholar] [CrossRef]
- Burgos, K.; Malenica, I.; Metpally, R.; Courtright, A.; Rakela, B.; Beach, T.; Shill, H.; Adler, C.; Sabbagh, M.; Villa, S.; et al. Profiles of extracellular microRNA in cerebrospinal fluid and serum from patients with Alzheimer’s and Parkinson’s diseases correlate with disease status and features of pathology. PLoS ONE 2014, 9, e94839. [Google Scholar] [CrossRef] [PubMed]
- Conway, K.A.; Harper, J.D.; Lansbury, P.T. Accelerated in vitro fibril formation by a mutant alpha-synuclein linked to early-onset Parkinson disease. Nat. Med. 1998, 4, 1318–1320. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.D.; Allen, N.E.; Canning, C.G.; Fung, V.S.C. Parkinson disease. Handb Clin. Neurol. 2018, 159, 173–193. [Google Scholar] [PubMed]
- von Campenhausen, S.; Bornschein, B.; Wick, R.; Botzel, K.; Sampaio, C.; Poewe, W.; Oertel, W.; Siebert, U.; Berger, K.; Dodel, R. Prevalence and incidence of Parkinson’s disease in Europe. Eur. Neuropsychopharm. 2005, 15, 473–490. [Google Scholar] [CrossRef] [PubMed]
- Tibar, H.; El Bayad, K.; Bouhouche, A.; Ait Ben Haddou, E.H.; Benomar, A.; Yahyaoui, M.; Benazzouz, A.; Regragui, W. Non-Motor Symptoms of Parkinson’s Disease and Their Impact on Quality of Life in a Cohort of Moroccan Patients. Front. Neurol. 2018, 9, 170. [Google Scholar] [CrossRef]
- Berganzo, K.; Tijero, B.; Gonzalez-Eizaguirre, A.; Somme, J.; Lezcano, E.; Gabilondo, I.; Fernandez, M.; Zarranz, J.J.; Gomez-Esteban, J.C. Motor and non-motor symptoms of Parkinson’s disease and their impact on quality of life and on different clinical subgroups. Neurologia 2016, 31, 585–591. [Google Scholar] [CrossRef]
- Lee, H.-J.; Ricarte, D.; Ortiz, D.; Lee, S.-J. Models of multiple system atrophy. Exp. Mol. Med. 2019, 51, 1–10. [Google Scholar] [CrossRef]
- Schrag, A.; Ben-Shlomo, Y.; Quinn, N.P. Prevalence of progressive supranuclear palsy and multiple system atrophy: A cross-sectional study. Lancet 1999, 354, 1771–1775. [Google Scholar] [CrossRef]
- Fanciulli, A.; Wenning, G.K. Multiple-system atrophy. N Engl J. Med. 2015, 372, 249–263. [Google Scholar] [CrossRef]
- Abos, A.; Baggio, H.C.; Segura, B.; Campabadal, A.; Uribe, C.; Giraldo, D.M.; Perez-Soriano, A.; Munoz, E.; Compta, Y.; Junque, C.; et al. Differentiation of multiple system atrophy from Parkinson’s disease by structural connectivity derived from probabilistic tractography. Sci. Rep. 2019, 9, 16488. [Google Scholar] [CrossRef]
- Lipp, A.; Sandroni, P.; Ahlskog, J.E.; Fealey, R.D.; Kimpinski, K.; Iodice, V.; Gehrking, T.L.; Weigand, S.D.; Sletten, D.M.; Gehrking, J.A.; et al. Prospective differentiation of multiple system atrophy from Parkinson disease, with and without autonomic failure. Arch. Neurol. 2009, 66, 742–750. [Google Scholar] [CrossRef]
- Jan, A.; Goncalves, N.P.; Vaegter, C.B.; Jensen, P.H.; Ferreira, N. The Prion-Like Spreading of Alpha-synuclein in Parkinson’s Disease: Update on Models and Hypotheses. Int. J. Mol. Sci. 2021, 22, 8338. [Google Scholar] [CrossRef] [PubMed]
- Van Den Berge, N.; Ferreira, N.; Mikkelsen, T.W.; Alstrup, A.K.O.; Tamguney, G.; Karlsson, P.; Terkelsen, A.J.; Nyengaard, J.R.; Jensen, P.H.; Borghammer, P. Ageing promotes pathological alpha-synuclein propagation and autonomic dysfunction in wild-type rats. Brain 2021, 144, 1853–1868. [Google Scholar] [CrossRef] [PubMed]
- Schaser, A.J.; Stackhouse, T.L.; Weston, L.J.; Kerstein, P.C.; Osterberg, V.R.; Lopez, C.S.; Dickson, D.W.; Luk, K.C.; Meshul, C.K.; Woltjer, R.L.; et al. Trans-synaptic and retrograde axonal spread of Lewy pathology following pre-formed fibril injection in an in vivo A53T alpha-synuclein mouse model of synucleinopathy. Acta Neuropathol. Commun. 2020, 8, 150. [Google Scholar] [CrossRef]
- Prusiner, S.B.; Woerman, A.L.; Mordes, D.A.; Watts, J.C.; Rampersaud, R.; Berry, D.B.; Patel, S.; Oehler, A.; Lowe, J.K.; Kravitz, S.N.; et al. Evidence for α-synuclein prions causing multiple system atrophy in humans with parkinsonism. Proc. Natl. Acad. Sci. USA 2015, 112, E5308–E5317. [Google Scholar] [CrossRef] [PubMed]
- Shahnawaz, M.; Mukherjee, A.; Pritzkow, S.; Mendez, N.; Rabadia, P.; Liu, X.; Hu, B.; Schmeichel, A.; Singer, W.; Wu, G.; et al. Discriminating α-synuclein strains in Parkinson’s disease and multiple system atrophy. Nature 2020, 578, 273–277. [Google Scholar] [CrossRef]
- Mavroudis, I.; Petridis, F.; Chatzikonstantinou, S.; Kazis, D. Alpha-synuclein Levels in the Differential Diagnosis of Lewy Bodies Dementia and Other Neurodegenerative Disorders: A Meta-analysis. Alzheimer Dis. Assoc. Disord. 2020, 34, 220–224. [Google Scholar] [CrossRef]
- Przedborski, S.; Jackson-Lewis, V.; Naini, A.B.; Jakowec, M.; Petzinger, G.; Miller, R.; Akram, M. The parkinsonian toxin 1-methyl-4-phenyl-1, 2, 3, 6-tetrahydropyridine (MPTP): A technical review of its utility and safety. J. Neurochem. 2001, 76, 1265–1274. [Google Scholar] [CrossRef]
- Smeyne, R.J.; Jackson-Lewis, V. The MPTP model of Parkinson’s disease. Mol. Brain Res. 2005, 134, 57–66. [Google Scholar] [CrossRef]
- Bernstein, A.I.; Garrison, S.P.; Zambetti, G.P.; O’Malley, K.L. 6-OHDA generated ROS induces DNA damage and p53-and PUMA-dependent cell death. Mol. Neurodegener. 2011, 6, 2. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Anantharam, V.; Zhang, D.; Latchoumycandane, C.; Kanthasamy, A.; Kanthasamy, A.G. Proteasome inhibitor MG-132 induces dopaminergic degeneration in cell culture and animal models. Neurotoxicology 2006, 27, 807–815. [Google Scholar] [CrossRef] [PubMed]
- Yazawa, I.; Giasson, B.I.; Sasaki, R.; Zhang, B.; Joyce, S.; Uryu, K.; Trojanowski, J.Q.; Lee, V.M.-Y. Mouse model of multiple system atrophy alpha-synuclein expression in oligodendrocytes causes glial and neuronal degeneration. Neuron 2005, 45, 847–859. [Google Scholar] [CrossRef]
- Kahle, P.J.; Neumann, M.; Ozmen, L.; Müller, V.; Jacobsen, H.; Spooren, W.; Fuss, B.; Mallon, B.; Macklin, W.B.; Fujiwara, H. Hyperphosphorylation and insolubility of alpha-synuclein in transgenic mouse oligodendrocytes. EMBO Rep. 2002, 3, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Shults, C.W.; Rockenstein, E.; Crews, L.; Adame, A.; Mante, M.; Larrea, G.; Hashimoto, M.; Song, D.; Iwatsubo, T.; Tsuboi, K. Neurological and neurodegenerative alterations in a transgenic mouse model expressing human alpha-synuclein under oligodendrocyte promoter: Implications for multiple system atrophy. J. Neurosci. 2005, 25, 10689–10699. [Google Scholar] [CrossRef] [PubMed]
- Ubhi, K.; Lee, P.H.; Adame, A.; Inglis, C.; Mante, M.; Rockenstein, E.; Stefanova, N.; Wenning, G.K.; Masliah, E. Mitochondrial inhibitor 3-nitroproprionic acid enhances oxidative modification of alpha-synuclein in a transgenic mouse model of multiple system atrophy. J. Neurosci. Res. 2009, 87, 2728–2739. [Google Scholar] [CrossRef] [PubMed]
- Fernagut, P.-O.; Tison, F. Animal models of multiple system atrophy. Neuroscience 2012, 211, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Wenning, G.; Granata, R.; Laboyrie, P.; Quinn, N.; Jenner, P.; Marsden, C. Reversal of behavioural abnormalities by fetal allografts in a novel rat model of striatonigral degeneration. Mov. Disod. Off. J. Mov. Disord. Soc. 1996, 11, 522–532. [Google Scholar] [CrossRef] [PubMed]
- Zlatkis, A.; Brazell, R.S.; Poole, C.F. The role of organic volatile profiles in clinical diagnosis. Clin. Chem. 1981, 27, 789–797. [Google Scholar] [CrossRef]
- Bizzarri, D.; Reinders, M.J.T.; Beekman, M.; Slagboom, P.E.; Bbmri, N.; van den Akker, E.B. 1H-NMR metabolomics-based surrogates to impute common clinical risk factors and endpoints. EBioMedicine 2021, 75, 103764. [Google Scholar] [CrossRef]
- Collino, S.; Martin, F.P.; Rezzi, S. Clinical metabolomics paves the way towards future healthcare strategies. Br. J. Clin. Pharm. 2013, 75, 619–629. [Google Scholar] [CrossRef]
- Lu, C.; Thompson, C.B. Metabolic regulation of epigenetics. Cell Metab. 2012, 16, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Zabala-Letona, A.; Arruabarrena-Aristorena, A.; Martin-Martin, N.; Fernandez-Ruiz, S.; Sutherland, J.D.; Clasquin, M.; Tomas-Cortazar, J.; Jimenez, J.; Torres, I.; Quang, P.; et al. mTORC1-dependent AMD1 regulation sustains polyamine metabolism in prostate cancer. Nature 2017, 547, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Shin, T.H.; Lee, D.Y.; Lee, H.S.; Park, H.J.; Jin, M.S.; Paik, M.J.; Manavalan, B.; Mo, J.S.; Lee, G. Integration of metabolomics and transcriptomics in nanotoxicity studies. BMB Rep. 2018, 51, 14–20. [Google Scholar] [CrossRef]
- Shin, T.H.; Nithiyanandam, S.; Lee, D.Y.; Kwon, D.H.; Hwang, J.S.; Kim, S.G.; Jang, Y.E.; Basith, S.; Park, S.; Mo, J.-S.; et al. Analysis of Nanotoxicity with Integrated Omics and Mechanobiology. Nanomaterials 2021, 11, 2385. [Google Scholar] [CrossRef] [PubMed]
- Procaccini, C.; Santopaolo, M.; Faicchia, D.; Colamatteo, A.; Formisano, L.; de Candia, P.; Galgani, M.; De Rosa, V.; Matarese, G. Role of metabolism in neurodegenerative disorders. Metabolism 2016, 65, 1376–1390. [Google Scholar] [CrossRef]
- Emwas, A.H.; Roy, R.; McKay, R.T.; Tenori, L.; Saccenti, E.; Gowda, G.A.N.; Raftery, D.; Alahmari, F.; Jaremko, L.; Jaremko, M.; et al. NMR Spectroscopy for Metabolomics Research. Metabolites 2019, 9, 123. [Google Scholar] [CrossRef]
- Larive, C.K.; Barding, G.A., Jr.; Dinges, M.M. NMR spectroscopy for metabolomics and metabolic profiling. Anal. Chem. 2015, 87, 133–146. [Google Scholar] [CrossRef]
- Keun, H.C.; Beckonert, O.; Griffin, J.L.; Richter, C.; Moskau, D.; Lindon, J.C.; Nicholson, J.K. Cryogenic probe 13C NMR spectroscopy of urine for metabonomic studies. Anal. Chem. 2002, 74, 4588–4593. [Google Scholar] [CrossRef]
- Pan, Z.; Raftery, D. Comparing and combining NMR spectroscopy and mass spectrometry in metabolomics. Anal. Bioanal. Chem. 2007, 387, 525–527. [Google Scholar] [CrossRef]
- Lee, P.H.; Lee, G.; Paik, M.J. Polyunsaturated fatty acid levels in the cerebrospinal fluid of patients with Parkinson’s disease and multiple system atrophy. Mov Disord. 2008, 23, 309–310. [Google Scholar] [CrossRef] [PubMed]
- Plewa, S.; Poplawska-Domaszewicz, K.; Florczak-Wyspianska, J.; Klupczynska-Gabryszak, A.; Sokol, B.; Miltyk, W.; Jankowski, R.; Kozubski, W.; Kokot, Z.J.; Matysiak, J. The Metabolomic Approach Reveals the Alteration in Human Serum and Cerebrospinal Fluid Composition in Parkinson’s Disease Patients. Pharmaceuticals 2021, 14, 935. [Google Scholar] [CrossRef]
- Stoessel, D.; Schulte, C.; Teixeira dos Santos, M.C.; Scheller, D.; Rebollo-Mesa, I.; Deuschle, C.; Walther, D.; Schauer, N.; Berg, D.; Nogueira da Costa, A.; et al. Promising Metabolite Profiles in the Plasma and CSF of Early Clinical Parkinson’s Disease. Front. Aging Neurosci. 2018, 10, 51. [Google Scholar] [CrossRef] [PubMed]
- Willkommen, D.; Lucio, M.; Moritz, F.; Forcisi, S.; Kanawati, B.; Smirnov, K.S.; Schroeter, M.; Sigaroudi, A.; Schmitt-Kopplin, P.; Michalke, B. Metabolomic investigations in cerebrospinal fluid of Parkinson’s disease. PLoS ONE 2018, 13, e0208752. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Irigoyen, J.; Cartas-Cejudo, P.; Iruarrizaga-Lejarreta, M.; Santamaría, E. Alteration in the Cerebrospinal Fluid Lipidome in Parkinson’s Disease: A Post-Mortem Pilot Study. Biomedicines 2021, 9, 491. [Google Scholar] [CrossRef]
- Molina, J.A.; Jiménez-Jiménez, F.J.; Gómez, P.; Vargas, C.; Navarro, J.A.; Ortí-Pareja, M.; Gasalla, T.; Benito-León, J.; Bermejo, F.; Arenas, J. Decreased cerebrospinal fluid levels of neutral and basic amino acids in patients with Parkinson’s disease. J. Neurol. Sci. 1997, 150, 123–127. [Google Scholar] [CrossRef]
- Kramer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- Duarte, J.M.; Schuck, P.F.; Wenk, G.L.; Ferreira, G.C. Metabolic disturbances in diseases with neurological involvement. Aging Dis. 2014, 5, 238–255. [Google Scholar] [CrossRef]
- Chitnis, T.; Weiner, H.L. CNS inflammation and neurodegeneration. J. Clin. Invest. 2017, 127, 3577–3587. [Google Scholar] [CrossRef]
- Nakano, M.; Imamura, H.; Sasaoka, N.; Yamamoto, M.; Uemura, N.; Shudo, T.; Fuchigami, T.; Takahashi, R.; Kakizuka, A. ATP Maintenance via Two Types of ATP Regulators Mitigates Pathological Phenotypes in Mouse Models of Parkinson’s Disease. EBioMedicine 2017, 22, 225–241. [Google Scholar] [CrossRef]
- Pathan, M.; Wu, J.; Lakso, H.A.; Forsgren, L.; Ohman, A. Plasma Metabolite Markers of Parkinson’s Disease and Atypical Parkinsonism. Metabolites 2021, 11, 860. [Google Scholar] [CrossRef] [PubMed]
- Kaiserova, M.; Chudackova, M.; Vranova, H.P.; Mensikova, K.; Kastelikova, A.; Stejskal, D.; Kanovsky, P. Cerebrospinal Fluid Levels of 5-Hydroxyindoleacetic Acid in Parkinson’s Disease and Atypical Parkinsonian Syndromes. Neurodegener. Dis. 2021, 21, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Abdo, W.F.; De Jong, D.; Hendriks, J.C.; Horstink, M.W.; Kremer, B.P.; Bloem, B.R.; Verbeek, M.M. Cerebrospinal fluid analysis differentiates multiple system atrophy from Parkinson’s disease. Mov. Disord. Off. J. Mov. Disord. Soc. 2004, 19, 571–579. [Google Scholar] [CrossRef] [PubMed]
- Engelborghs, S.; Marescau, B.; De Deyn, P. Amino acids and biogenic amines in cerebrospinal fluid of patients with Parkinson’s disease. Neurochem. Res. 2003, 28, 1145–1150. [Google Scholar] [CrossRef]
- Heilman, P.L.; Wang, E.W.; Lewis, M.M.; Krzyzanowski, S.; Capan, C.D.; Burmeister, A.R.; Du, G.; Escobar Galvis, M.L.; Brundin, P.; Huang, X. Tryptophan Metabolites Are Associated With Symptoms and Nigral Pathology in Parkinson’s Disease. Mov. Disord. 2020, 35, 2028–2037. [Google Scholar] [CrossRef]
- Kuiper, M.; Teerlink, T.; Visser, J.; Bergmans, P.; Scheltens, P.; Wolters, E.C. L-glutamate, L-arginine and L-citrulline levels in cerebrospinal fluid of Parkinson’s disease, multiple system atrophy, and Alzheimer’s disease patients. J. Neural. Transm. 2000, 107, 183–189. [Google Scholar] [CrossRef]
- Compta, Y.; Giraldo, D.M.; Munoz, E.; Antonelli, F.; Fernández, M.; Bravo, P.; Soto, M.; Cámara, A.; Torres, F.; Martí, M.J. Cerebrospinal fluid levels of coenzyme Q10 are reduced in multiple system atrophy. Parkinsonism Relat. Disord. 2018, 46, 16–23. [Google Scholar] [CrossRef]
- Konings, C.; Kuiper, M.; Teerlink, T.; Mulder, C.; Scheltens, P.; Wolters, E.C. Normal cerebrospinal fluid glutathione concentrations in Parkinson’s disease, Alzheimer’s disease and multiple system atrophy. J. Neurol. Sci. 1999, 168, 112–115. [Google Scholar] [CrossRef]
- Kuiper, M.A.; Visser, J.J.; Bergmans, P.L.; Scheltens, P.; Wolters, E.C. Decreased cerebrospinal fluid nitrate levels in Parkinson’s disease, Alzheimer’s disease and multiple system atrophy patients. J. Neurol. Sci. 1994, 121, 46–49. [Google Scholar] [CrossRef]
- Li, J.; Meng, Y.; Wu, X.; Sun, Y. Polyamines and related signaling pathways in cancer. Cancer Cell Int. 2020, 20, 539. [Google Scholar] [CrossRef]
- Saiki, S.; Sasazawa, Y.; Fujimaki, M.; Kamagata, K.; Kaga, N.; Taka, H.; Li, Y.; Souma, S.; Hatano, T.; Imamichi, Y.; et al. A metabolic profile of polyamines in parkinson disease: A promising biomarker. Ann. Neurol. 2019, 86, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Tortosa, E.; Newell, K.; Irizarry, M.C.; Sanders, J.L.; Hyman, B.T. alpha-Synuclein immunoreactivity in dementia with Lewy bodies: Morphological staging and comparison with ubiquitin immunostaining. Acta Neuropathol. 2000, 99, 352–357. [Google Scholar] [CrossRef] [PubMed]
- Chung, K.K.K.; Zhang, Y.; Lim, K.L.; Tanaka, Y.; Huang, H.; Gao, J.; Ross, C.A.; Dawson, V.L.; Dawson, T.M. Parkin ubiquitinates the alpha-synuclein–interacting protein, synphilin-1: Implications for Lewy-body formation in Parkinson disease. Nat. Med. 2001, 7, 1144–1150. [Google Scholar] [CrossRef] [PubMed]
- Engelender, S.; Kaminsky, Z.; Guo, X.; Sharp, A.H.; Amaravi, R.K.; Kleiderlein, J.J.; Margolis, R.L.; Troncoso, J.C.; Lanahan, A.A.; Worley, P.F.; et al. Synphilin-1 associates with alpha-synuclein and promotes the formation of cytosolic inclusions. Nat. Genet. 1999, 22, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Phukan, G.; Shin, T.H.; Shim, J.S.; Paik, M.J.; Lee, J.-K.; Choi, S.; Kim, Y.M.; Kang, S.H.; Kim, H.S.; Kang, Y.; et al. Silica-coated magnetic nanoparticles impair proteasome activity and increase the formation of cytoplasmic inclusion bodies in vitro. Sci. Rep. 2016, 6, 29095. [Google Scholar] [CrossRef]
- Bright, G.R.; Fisher, G.W.; Rogowska, J.; Taylor, D.L. Fluorescence ratio imaging microscopy: Temporal and spatial measurements of cytoplasmic pH. J. Cell Biol. 1987, 104, 1019–1033. [Google Scholar] [CrossRef]
- Kim, Y.-S.; Lee, D.; Lee, E.-K.; Sung, J.Y.; Chung, K.C.; Kim, J.; Paik, S.R. Multiple ligand interaction of alpha-synuclein produced various forms of protein aggregates in the presence of Abeta25-35, copper, and eosin. Brain Res. 2001, 908, 93–98. [Google Scholar] [CrossRef]
- Jellinger, K.A. Recent developments in the pathology of Parkinson’s disease. J. Neural Transm. Suppl. 2002, 62, 347–376. [Google Scholar]
- Antony, T.; Hoyer, W.; Cherny, D.; Heim, G.; Jovin, T.M.; Subramaniam, V. Cellular polyamines promote the aggregation of alpha-synuclein. J. Biol. Chem. 2003, 278, 3235–3240. [Google Scholar] [CrossRef]
- Holmes, E.; Wilson, I.D.; Nicholson, J.K. Metabolic phenotyping in health and disease. Cell 2008, 134, 714–717. [Google Scholar] [CrossRef]
- Szeto, S.S.; Reinke, S.N.; Sykes, B.D.; Lemire, B.D. Mutations in the Saccharomyces cerevisiae succinate dehydrogenase result in distinct metabolic phenotypes revealed through (1)H NMR-based metabolic footprinting. J. Proteome Res. 2010, 9, 6729–6739. [Google Scholar] [CrossRef] [PubMed]
- Everett, J.R. Pharmacometabonomics: The prediction of drug effects using metabolic profiling. Concepts Princ. Pharmacol. 2019, 260, 263–299. [Google Scholar]
- Li, K.W.; Ganz, A.B.; Smit, A.B. Proteomics of neurodegenerative diseases: Analysis of human post-mortem brain. J. Neurochem. 2019, 151, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Lehnert, S.; Jesse, S.; Rist, W.; Steinacker, P.; Soininen, H.; Herukka, S.-K.; Tumani, H.; Lenter, M.; Oeckl, P.; Ferger, B. iTRAQ and multiple reaction monitoring as proteomic tools for biomarker search in cerebrospinal fluid of patients with Parkinson’s disease dementia. Exp. Neurol. 2012, 234, 499–505. [Google Scholar] [CrossRef]
- Compta, Y.; Dias, S.P.; Giraldo, D.M.; Pérez-Soriano, A.; Muñoz, E.; Saura, J.; Fernández, M.; Bravo, P.; Cámara, A.; Pulido-Salgado, M. Cerebrospinal fluid cytokines in multiple system atrophy: A cross-sectional Catalan MSA registry study. Parkinsonism Relat. Disord. 2019, 65, 3–12. [Google Scholar] [CrossRef]
- Wang, J.; Chen, J.; Sen, S. MicroRNA as Biomarkers and Diagnostics. J. Cell. Physiol. 2016, 231, 25–30. [Google Scholar] [CrossRef]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef]
- Lau, N.C.; Lim, L.P.; Weinstein, E.G.; Bartel, D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 2001, 294, 858–862. [Google Scholar] [CrossRef]
- Hydbring, P.; Badalian-Very, G. Clinical applications of microRNAs. F1000Res 2013, 2, 136. [Google Scholar] [CrossRef]
- Maciotta, S.; Meregalli, M.; Torrente, Y. The involvement of microRNAs in neurodegenerative diseases. Front. Cell Neurosci. 2013, 7, 265. [Google Scholar] [CrossRef]
- Van Assche, R.; Broeckx, V.; Boonen, K.; Maes, E.; De Haes, W.; Schoofs, L.; Temmerman, L. Integrating -Omics: Systems Biology as Explored Through C. elegans Research. J. Mol. Biol. 2015, 427, 3441–3451. [Google Scholar] [CrossRef]
- Misra, B.B.; Langefeld, C.; Olivier, M.; Cox, L.A. Integrated omics: Tools, advances and future approaches. J. Mol. Endocrinol. 2019, 62, R21–R45. [Google Scholar] [CrossRef] [PubMed]
- Shin, T.H.; Lee, S.; Choi, K.R.; Lee, D.Y.; Kim, Y.; Paik, M.J.; Seo, C.; Kang, S.; Jin, M.S.; Yoo, T.H.; et al. Quality and freshness of human bone marrow-derived mesenchymal stem cells decrease over time after trypsinization and storage in phosphate-buffered saline. Sci. Rep. 2017, 7, 1106. [Google Scholar] [CrossRef]
- Shin, T.H.; Ketebo, A.A.; Lee, D.Y.; Lee, S.; Kang, S.H.; Basith, S.; Manavalan, B.; Kwon, D.H.; Park, S.; Lee, G. Decrease in membrane fluidity and traction force induced by silica-coated magnetic nanoparticles. J. Nanobiotechnol. 2021, 19, 21. [Google Scholar] [CrossRef] [PubMed]
- Shin, T.H.; Manavalan, B.; Lee, D.Y.; Basith, S.; Seo, C.; Paik, M.J.; Kim, S.-W.; Seo, H.; Lee, J.Y.; Kim, J.Y.; et al. Silica-coated magnetic-nanoparticle-induced cytotoxicity is reduced in microglia by glutathione and citrate identified using integrated omics. Part. Fibre Toxicol. 2021, 18, 42. [Google Scholar] [CrossRef] [PubMed]
- Shin, T.H.; Seo, C.; Lee, D.Y.; Ji, M.; Manavalan, B.; Basith, S.; Chakkarapani, S.K.; Kang, S.H.; Lee, G.; Paik, M.J.; et al. Silica-coated magnetic nanoparticles induce glucose metabolic dysfunction in vitro via the generation of reactive oxygen species. Arch. Toxicol. 2019, 93, 1201–1212. [Google Scholar] [CrossRef]
- Shin, T.H.; Lee, D.Y.; Manavalan, B.; Basith, S.; Na, Y.-C.; Yoon, C.; Lee, H.-S.; Paik, M.J.; Lee, G. Silica-coated magnetic nanoparticles activate microglia and induce neurotoxic d-serine secretion. Part. Fibre Toxicol. 2021, 18, 30. [Google Scholar] [CrossRef]
- Rotunno, M.S.; Lane, M.; Zhang, W.; Wolf, P.; Oliva, P.; Viel, C.; Wills, A.-M.; Alcalay, R.N.; Scherzer, C.R.; Shihabuddin, L.S.; et al. Cerebrospinal fluid proteomics implicates the granin family in Parkinson’s disease. Sci. Rep. 2020, 10, 2479. [Google Scholar] [CrossRef]
- Markaki, I.; Bergström, S.; Tsitsi, P.; Remnestål, J.; Månberg, A.; Hertz, E.; Paslawski, W.; Sorjonen, K.; Uhlén, M.; Mangone, G.; et al. Cerebrospinal Fluid Levels of Kininogen-1 Indicate Early Cognitive Impairment in Parkinson’s Disease. Mov. Disord. 2020, 35, 2101–2106. [Google Scholar] [CrossRef]
- Lee, D.Y.; Lee, S.E.; Kwon, D.H.; Nithiyanandam, S.; Lee, M.H.; Hwang, J.S.; Basith, S.; Ahn, J.H.; Shin, T.H.; Lee, G. Strategies to Improve the Quality and Freshness of Human Bone Marrow-Derived Mesenchymal Stem Cells for Neurological Diseases. Stem Cells Int. 2021, 2021, 8444599. [Google Scholar] [CrossRef]
- Kim, S.; Jhong, J.H.; Lee, J.; Koo, J.Y. Meta-analytic support vector machine for integrating multiple omics data. BioData Min. 2017, 10, 2. [Google Scholar] [CrossRef] [PubMed]
- Basith, S.; Manavalan, B.; Hwan Shin, T.; Lee, G. Machine intelligence in peptide therapeutics: A next-generation tool for rapid disease screening. Med. Res. Rev. 2020, 40, 1276–1314. [Google Scholar] [CrossRef] [PubMed]
- Dien, J. Evaluating two-step PCA of ERP data with Geomin, Infomax, Oblimin, Promax, and Varimax rotations. Psychophysiology 2010, 47, 170–183. [Google Scholar] [CrossRef] [PubMed]

| PD vs. Control | MSA vs. Control | ||||||
|---|---|---|---|---|---|---|---|
| Metabolite | Alteration | Analysis Equipment | Reference | Metabolite | Alteration | Analysis Equipment | Reference |
| EPA | ↑ | GC-MS | [61] | EPA | ↑ | GC-MS | [61] |
| 5-HIAA | ↓ | Sandwich ELISA | [72] | 5-HIAA | ↓ | Sandwich ELISA | [72] |
| HVA | ↓ | HPLC | [73] | HVA | ↓ | HPLC | [73] |
| DOPA | ↓ | LC | [9] | DOPA | ↓ | LC | [9] |
| DHPG | ↓ | LC | [9] | DA | ↓ | LC | [9] |
| DOPAC | ↓ | LC | [9] | DOPAC | ↓ | LC | [9] |
| Tyrosine | ↑ | HPLC | [62] | NE | N.S | LC | [9] |
| Taurine | ↓ | HPLC | [74] | DHPG | ↓ | LC | [9] |
| KYNA | ↓ | UPLC | [75] | MHPG | ↓ | HPLC | [73] |
| QNA | ↑ | FT-ICR-MS | [64] | N1-Acetylputrescine | ↓ | GC-MS | [8] |
| N1-Acetylcadaverine | ↑ | GC-MS | [8] | N1-Acetylcadaverine | ↓ | GC-MS | [8] |
| Putrescine | ↑ | GC-MS | [8] | Putrescine | N.S | GC-MS | [8] |
| Cadaverine | ↑ | GC-MS | [8] | Cadaverine | ↑ | GC-MS | [8] |
| N1-Acetylspermidine | ↑ | GC-MS | [8] | N1-Acetylspermidine | ↓ | GC-MS | [8] |
| N8-Acetylspermidine | ↑ | GC-MS | [8] | N8-Acetylspermidine | ↑ | GC-MS | [8] |
| Spermidine | ↓ | GC-MS | [8] | Spermidine | N.S | GC-MS | [8] |
| Arachidonic acid | ↑ | FT-ICR-MS | [64] | Citrulline | ↑ | HPLC | [76] |
| Alanine | ↓ | Amino acid analyzer | [66] | Arginine | N.S | HPLC | [76] |
| Valine | ↓ | Amino acid analyzer | [66] | Glutamate | N.S | HPLC | [76] |
| Isoleucine | ↓ | Amino acid analyzer | [66] | Coenzyme Q10 | ↓ | ELISA | [77] |
| Leucine | ↓ | Amino acid analyzer | [66] | Glutathione | N.S | HPLC | [78] |
| Ethanolamine | ↓ | Amino acid analyzer | [66] | Nitrite | N.S | ELISA | [79] |
| Nitrate | ↓ | ELISA | [79] | Nitrate | ↓ | ELISA | [79] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kwon, D.H.; Hwang, J.S.; Kim, S.G.; Jang, Y.E.; Shin, T.H.; Lee, G. Cerebrospinal Fluid Metabolome in Parkinson’s Disease and Multiple System Atrophy. Int. J. Mol. Sci. 2022, 23, 1879. https://doi.org/10.3390/ijms23031879
Kwon DH, Hwang JS, Kim SG, Jang YE, Shin TH, Lee G. Cerebrospinal Fluid Metabolome in Parkinson’s Disease and Multiple System Atrophy. International Journal of Molecular Sciences. 2022; 23(3):1879. https://doi.org/10.3390/ijms23031879
Chicago/Turabian StyleKwon, Do Hyeon, Ji Su Hwang, Seok Gi Kim, Yong Eun Jang, Tae Hwan Shin, and Gwang Lee. 2022. "Cerebrospinal Fluid Metabolome in Parkinson’s Disease and Multiple System Atrophy" International Journal of Molecular Sciences 23, no. 3: 1879. https://doi.org/10.3390/ijms23031879
APA StyleKwon, D. H., Hwang, J. S., Kim, S. G., Jang, Y. E., Shin, T. H., & Lee, G. (2022). Cerebrospinal Fluid Metabolome in Parkinson’s Disease and Multiple System Atrophy. International Journal of Molecular Sciences, 23(3), 1879. https://doi.org/10.3390/ijms23031879






