Structural and Biochemical Analysis of the Furan Aldehyde Reductase YugJ from Bacillus subtilis
Abstract
:1. Introduction
2. Results and Discussion
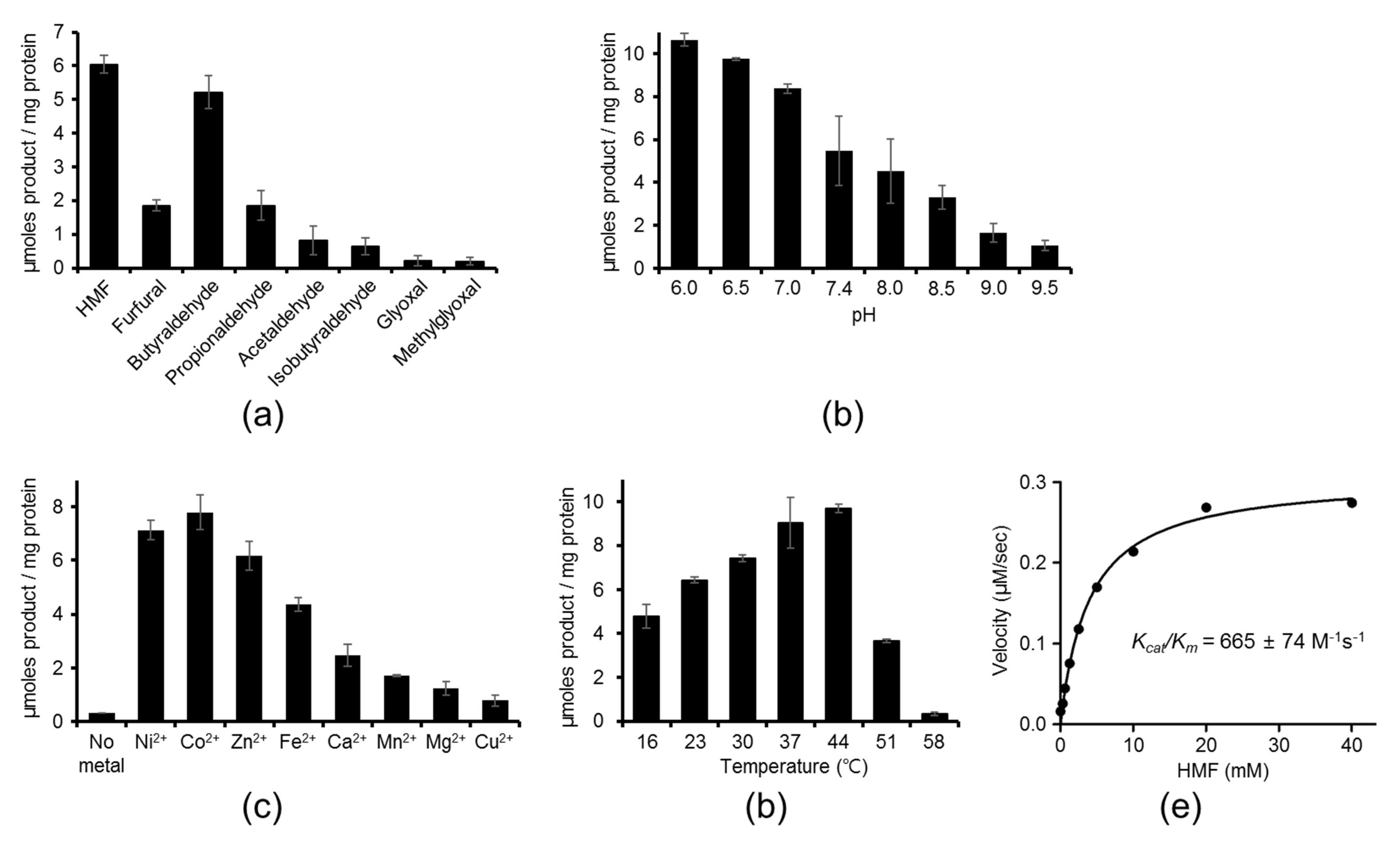
2.1. Aldehyde Reductase Activity of YugJ
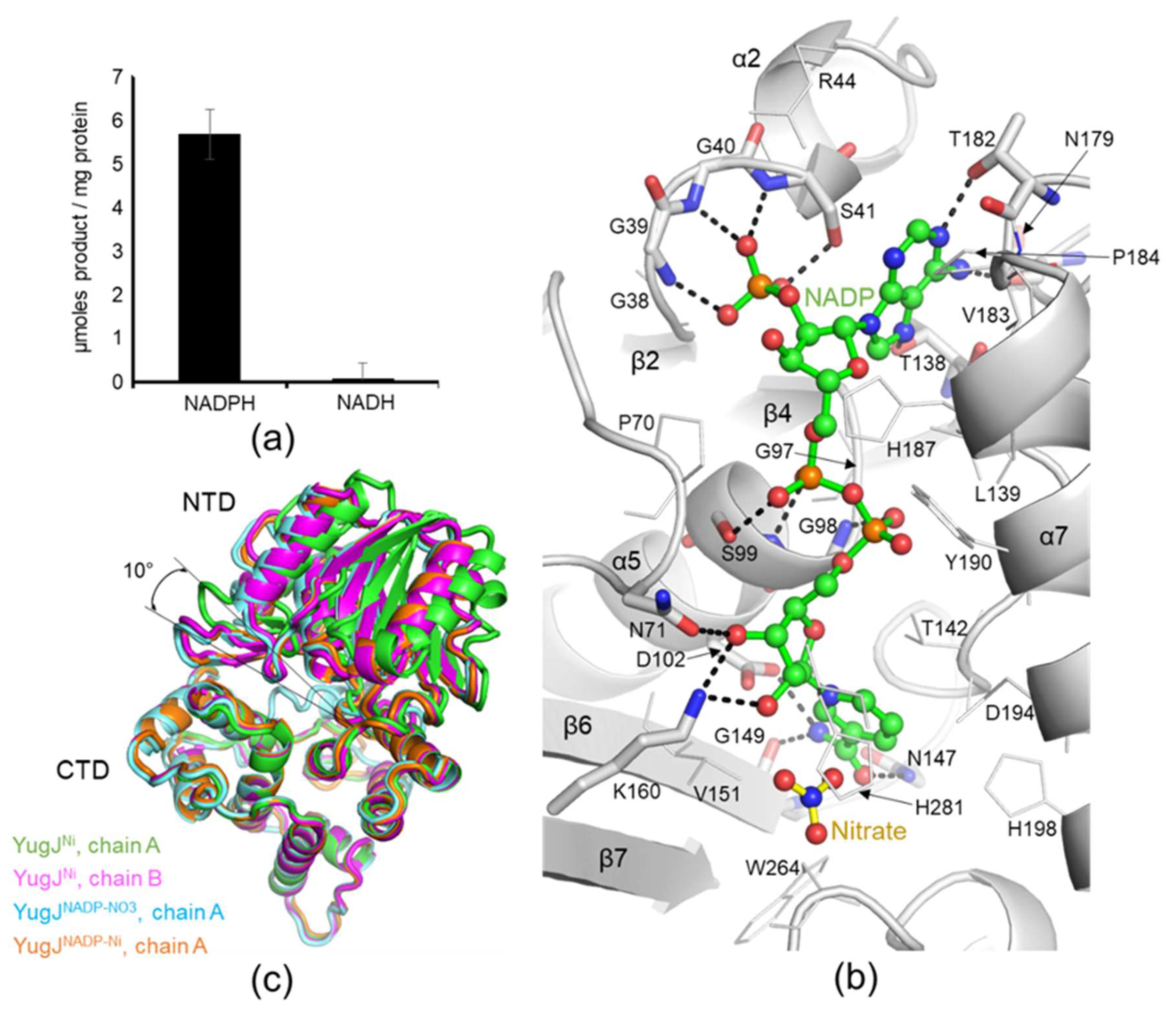
2.2. Metal Ion Recognition by YugJ
2.3. NADP(H) Recognition by YugJ
2.4. Putative Substrate-Binding Site of YugJ
3. Materials and Methods
3.1. Construction of a YugJ Expression Plasmid
3.2. Expression and Purification of the YugJ Protein
3.3. Crystallization and X-ray Diffraction
3.4. Structure Determination
3.5. Measurement of the Catalytic Activity of YugJ
3.6. In Silico Molecular Docking Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Reid, M.F.; Fewson, C.A. Molecular characterization of microbial alcohol dehydrogenases. Crit. Rev. Microbiol. 1994, 20, 13–56. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Tobias, A.; Julian-Sanchez, A.; Pina, E.; Riveros-Rosas, H. Natural alcohol exposure: Is ethanol the main substrate for alcohol dehydrogenases in animals? Chem. Biol. Interact. 2011, 191, 14–25. [Google Scholar] [CrossRef] [PubMed]
- De Smidt, O.; du Preez, J.C.; Albertyn, J. The alcohol dehydrogenases of Saccharomyces cerevisiae: A comprehensive review. FEMS Yeast. Res. 2008, 8, 967–978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, G.H.; Hsieh, M.C.; Shu, H.Y. Role of iron-containing alcohol dehydrogenases in Acinetobacter baumannii ATCC 19606 stress resistance and virulence. Int. J. Mol. Sci. 2021, 22, 9921. [Google Scholar] [CrossRef]
- Lee, C.; Kim, I.; Lee, J.; Lee, K.L.; Min, B.; Park, C. Transcriptional activation of the aldehyde reductase YqhD by YqhC and its implication in glyoxal metabolism of Escherichia coli K-12. J. Bacteriol. 2010, 192, 4205–4214. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.; Yang, X.; Yang, J.; Qiao, X.; Li, F.; Liu, X.; Wei, J.; Wang, L. Alcohol dehydrogenase modulates quorum sensing in biofilm formations of Acinetobacter baumannii. Microb. Pathog. 2020, 148, 104451. [Google Scholar] [CrossRef]
- Luong, T.T.; Kim, E.H.; Bak, J.P.; Nguyen, C.T.; Choi, S.; Briles, D.E.; Pyo, S.; Rhee, D.K. Ethanol-induced alcohol dehydrogenase E (AdhE) potentiates pneumolysin in Streptococcus pneumoniae. Infect. Immun. 2015, 83, 108–119. [Google Scholar] [CrossRef] [Green Version]
- Radianingtyas, H.; Wright, P.C. Alcohol dehydrogenases from thermophilic and hyperthermophilic archaea and bacteria. FEMS Microbiol. Rev. 2003, 27, 593–616. [Google Scholar] [CrossRef]
- Montella, C.; Bellsolell, L.; Perez-Luque, R.; Badia, J.; Baldoma, L.; Coll, M.; Aguilar, J. Crystal structure of an iron-dependent group III dehydrogenase that interconverts L-lactaldehyde and L-1,2-propanediol in Escherichia coli. J. Bacteriol. 2005, 187, 4957–4966. [Google Scholar] [CrossRef] [Green Version]
- Konig, C.; Meyer, M.; Lender, C.; Nehls, S.; Wallaschkowski, T.; Holm, T.; Matthies, T.; Lercher, D.; Matthiesen, J.; Fehling, H.; et al. An alcohol dehydrogenase 3 (ADH3) from Entamoeba histolytica is involved in the detoxification of toxic aldehydes. Microorganisms 2020, 8, 1608. [Google Scholar] [CrossRef]
- Elleuche, S.; Fodor, K.; Klippel, B.; von der Heyde, A.; Wilmanns, M.; Antranikian, G. Structural and biochemical characterisation of a NAD+-dependent alcohol dehydrogenase from Oenococcus oeni as a new model molecule for industrial biotechnology applications. Appl. Microbiol. Biotechnol. 2013, 97, 8963–8975. [Google Scholar] [CrossRef] [PubMed]
- Sulzenbacher, G.; Alvarez, K.; Van Den Heuvel, R.H.; Versluis, C.; Spinelli, S.; Campanacci, V.; Valencia, C.; Cambillau, C.; Eklund, H.; Tegoni, M. Crystal structure of E. coli alcohol dehydrogenase YqhD: Evidence of a covalently modified NADP coenzyme. J. Mol. Biol. 2004, 342, 489–502. [Google Scholar] [CrossRef] [PubMed]
- Elleuche, S.; Fodor, K.; von der Heyde, A.; Klippel, B.; Wilmanns, M.; Antranikian, G. Group III alcohol dehydrogenase from Pectobacterium atrosepticum: Insights into enzymatic activity and organization of the metal ion-containing region. Appl. Microbiol. Biotechnol. 2014, 98, 4041–4051. [Google Scholar] [CrossRef] [PubMed]
- Extance, J.; Crennell, S.J.; Eley, K.; Cripps, R.; Hough, D.W.; Danson, M.J. Structure of a bifunctional alcohol dehydrogenase involved in bioethanol generation in Geobacillus thermoglucosidasius. Acta Crystallogr. D Biol. Crystallogr. 2013, 69, 2104–2115. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.Y.; Park, S.H.; Oh, S.H.; Lee, J.J.; Kwon, K.K.; Kim, S.J.; Choi, M.; Rha, E.; Lee, H.; Lee, D.H.; et al. Discovery and biochemical characterization of a methanol dehydrogenase from Lysinibacillus xylanilyticus. Front. Bioeng. Biotechnol. 2020, 8, 67. [Google Scholar] [CrossRef]
- Jarboe, L.R. YqhD: A broad-substrate range aldehyde reductase with various applications in production of biorenewable fuels and chemicals. Appl. Microbiol. Biotechnol. 2011, 89, 249–257. [Google Scholar] [CrossRef] [Green Version]
- Perez, J.M.; Arenas, F.A.; Pradenas, G.A.; Sandoval, J.M.; Vasquez, C.C. Escherichia coli YqhD exhibits aldehyde reductase activity and protects from the harmful effect of lipid peroxidation-derived aldehydes. J. Biol. Chem. 2008, 283, 7346–7353. [Google Scholar] [CrossRef] [Green Version]
- Ying, X.; Wang, Y.; Badiei, H.R.; Karanassios, V.; Ma, K. Purification and characterization of an iron-containing alcohol dehydrogenase in extremely thermophilic bacterium Thermotoga hypogea. Arch. Microbiol. 2007, 187, 499–510. [Google Scholar] [CrossRef]
- Allen, S.A.; Clark, W.; McCaffery, J.M.; Cai, Z.; Lanctot, A.; Slininger, P.J.; Liu, Z.L.; Gorsich, S.W. Furfural induces reactive oxygen species accumulation and cellular damage in Saccharomyces cerevisiae. Biotechnol. Biofuels. 2010, 3, 2. [Google Scholar] [CrossRef] [Green Version]
- Khan, Q.A.; Shamsi, F.A.; Hadi, S.M. Mutagenicity of furfural in plasmid DNA. Cancer Lett. 1995, 89, 95–99. [Google Scholar] [CrossRef]
- Hristozova, T.; Angelov, A.; Tzvetkova, B.; Paskaleva, D.; Gotcheva, V.; Gargova, S.; Pavlova, K. Effect of furfural on carbon metabolism key enzymes of lactose-assimilating yeasts. Enzyme Microb. Technol. 2006, 39, 1108–1112. [Google Scholar] [CrossRef]
- Iwaki, A.; Kawai, T.; Yamamoto, Y.; Izawa, S. Biomass conversion inhibitors furfural and 5-hydroxymethylfurfural induce formation of messenger RNP granules and attenuate translation activity in Saccharomyces cerevisiae. Appl. Environ. Microb. 2013, 79, 1661–1667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishii, J.; Yoshimura, K.; Hasunuma, T.; Kondo, A. Reduction of furan derivatives by overexpressing NADH-dependent Adh1 improves ethanol fermentation using xylose as sole carbon source with Saccharomyces cerevisiae harboring XR–XDH pathway. Appl. Microbiol. Biotechnol. 2013, 97, 2597–2607. [Google Scholar] [CrossRef]
- Laadan, B.; Almeida, J.R.; Radstrom, P.; Hahn-Hagerdal, B.; Gorwa-Grauslund, M. Identification of an NADH-dependent 5-hydroxymethylfurfural-reducing alcohol dehydrogenase in Saccharomyces cerevisiae. Yeast 2008, 25, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ujor, V.; Wick, M.; Ezeji, T.C. Identification, purification and characterization of furfural transforming enzymes from Clostridium beijerinckii NCIMB 8052. Anaerobe 2015, 33, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ezeji, T.C. Transcriptional analysis of Clostridium beijerinckii NCIMB 8052 to elucidate role of furfural stress during acetone butanol ethanol fermentation. Biotechnol. Biofuels 2013, 6, 66. [Google Scholar] [CrossRef]
- Wang, X.; Miller, E.N.; Yomano, L.P.; Zhang, X.; Shanmugam, K.T.; Ingram, L.O. Increased furfural tolerance due to overexpression of NADH-dependent oxidoreductase fuco in Escherichia coli Strains Engineered for the Production of ethanol and lactate. Appl. Environ. Microb. 2011, 77, 5132–5140. [Google Scholar] [CrossRef] [Green Version]
- Hahn-Hagerdal, B.; Galbe, M.; Gorwa-Grauslund, M.F.; Liden, G.; Zacchi, G. Bio-ethanol—The fuel of tomorrow from the residues of today. Trends Biotechnol. 2006, 24, 549–556. [Google Scholar] [CrossRef]
- Katahira, S.; Mizuike, A.; Fukuda, H.; Kondo, A. Ethanol fermentation from lignocellulosic hydrolysate by a recombinant xylose- and cellooligosaccharide-assimilating yeast strain. Appl. Microbiol. Biotechnol. 2006, 72, 1136–1143. [Google Scholar] [CrossRef]
- Agbor, V.B.; Cicek, N.; Sparling, R.; Berlin, A.; Levin, D.B. Biomass pretreatment: Fundamentals toward application. Biotechnol. Adv. 2011, 29, 675–685. [Google Scholar] [CrossRef]
- Stulke, J.; Hillen, W. Regulation of carbon catabolism in Bacillus species. Annu. Rev. Microbiol. 2000, 54, 849–880. [Google Scholar] [CrossRef] [PubMed]
- Schumann, W. Production of recombinant proteins in Bacillus subtilis. Adv. Appl. Microbiol. 2007, 62, 137–189. [Google Scholar] [PubMed]
- Westers, L.; Westers, H.; Quax, W.J. Bacillus subtilis as cell factory for pharmaceutical proteins: A biotechnological approach to optimize the host organism. Biochim. Biophys. Acta BBA-Mol. Cell Res. 2004, 1694, 299–310. [Google Scholar] [CrossRef]
- Schallmey, M.; Singh, A.; Ward, O.P. Developments in the use of Bacillus species for industrial production. Can. J. Microbiol. 2004, 50, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Romero, S.; Merino, E.; Bolivar, F.; Gosset, G.; Martinez, A. Metabolic engineering of Bacillus subtilis for ethanol production: Lactate dehydrogenase plays a key role in fermentative metabolism. Appl. Environ. Microbiol. 2007, 73, 5190–5198. [Google Scholar] [CrossRef] [Green Version]
- Maleki, F.; Changizian, M.; Zolfaghari, N.; Rajaei, S.; Noghabi, K.A.; Zahiri, H.S. Consolidated bioprocessing for bioethanol production by metabolically engineered Bacillus subtilis strains. Sci. Rep. 2021, 11, 13731. [Google Scholar] [CrossRef]
- Miller, E.N.; Turner, P.C.; Jarboe, L.R.; Ingram, L.O. Genetic changes that increase 5-hydroxymethyl furfural resistance in ethanol-producing Escherichia coli LY180. Biotechnol. Lett. 2010, 32, 661–667. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.T.; Wu, L.H.; Knight, J.A. Stability of NADPH: Effect of various factors on the kinetics of degradation. Clin. Chem. 1986, 32, 314–319. [Google Scholar] [CrossRef]
- Iwig, J.S.; Leitch, S.; Herbst, R.W.; Maroney, M.J.; Chivers, P.T. Ni(II) and Co(II) sensing by Escherichia coli RcnR. J. Am. Chem. Soc. 2008, 130, 7592–7606. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Yu, G.; Xu, Q.; Li, N.; Xiao, C.; Yin, X.; Cao, K.; Han, J.; He, Q.Y. Putative cobalt- and nickel-binding proteins and motifs in Streptococcus pneumoniae. Metallomics 2013, 5, 928–935. [Google Scholar] [CrossRef]
- Van Beilen, J.W.A.; Brul, S. Compartment-specific pH monitoring in Bacillus subtilis using fluorescent sensor proteins: A tool to analyze the antibacterial effect of weak organic acids. Front. Microbiol. 2013, 4, 157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moon, J.H.; Lee, H.J.; Park, S.Y.; Song, J.M.; Park, M.Y.; Park, H.M.; Sun, J.; Park, J.H.; Kim, B.Y.; Kim, J.S. Structures of iron-dependent alcohol dehydrogenase 2 from Zymomonas mobilis ZM4 with and without NAD+ cofactor. J. Mol. Biol. 2011, 407, 413–424. [Google Scholar] [CrossRef] [PubMed]
- Marcal, D.; Rego, A.T.; Carrondo, M.A.; Enguita, F.J. 1,3-Propanediol dehydrogenase from Klebsiella pneumoniae: Decameric quaternary structure and possible subunit cooperativity. J. Bacteriol. 2009, 191, 1143–1151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Handing, K.B.; Niedzialkowska, E.; Shabalin, I.G.; Kuhn, M.L.; Zheng, H.; Minor, W. Characterizing metal-binding sites in proteins with X-ray crystallography. Nat. Protoc. 2018, 13, 1062–1090. [Google Scholar] [CrossRef] [PubMed]
- Cahn, J.K.; Baumschlager, A.; Brinkmann-Chen, S.; Arnold, F.H. Mutations in adenine-binding pockets enhance catalytic properties of NAD(P)H-dependent enzymes. Protein Eng. Des. Sel. 2016, 29, 31–38. [Google Scholar] [CrossRef] [Green Version]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Obradors, N.; Cabiscol, E.; Aguilar, J.; Ros, J. Site-directed mutagenesis studies of the metal-binding center of the iron-dependent propanediol oxidoreductase from Escherichia coli. Eur. J. Biochem. 1998, 258, 207–213. [Google Scholar] [CrossRef] [Green Version]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [Green Version]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [Green Version]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cafardi, V.; Biagini, M.; Martinelli, M.; Leuzzi, R.; Rubino, J.T.; Cantini, F.; Norais, N.; Scarselli, M.; Serruto, D.; Unnikrishnan, M. Identification of a novel zinc metalloprotease through a global analysis of Clostridium difficile extracellular proteins. PLoS ONE 2013, 8, e81306. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Chen, J.; Fu, G.; Gindulyte, A.; Han, L.; He, J.; He, S.; Shoemaker, B.A.; et al. PubChem substance and compound databases. Nucleic Acids Res. 2016, 44, D1202–D1213. [Google Scholar] [CrossRef] [PubMed]
- Pei, J.; Kim, B.H.; Grishin, N.V. PROMALS3D: A tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008, 36, 2295–2300. [Google Scholar] [CrossRef]




| YugJNi | YugJNADP-Ni | YugJNADP-NO3 | |
|---|---|---|---|
| Data collection | |||
| Space group | P212121 | P1 | P21 |
| Cell parameters | |||
| a (Å) | 62.66 | 66.04 | 56.51 |
| b (Å) | 114.52 | 69.64 | 67.69 |
| c (Å) | 118.74 | 87.61 | 102.37 |
| α (°) | 90 | 92.29 | 90 |
| β (°) | 90 | 91.03 | 100.15 |
| γ (°) | 90 | 90.37 | 90 |
| Wavelength (Å) | 1.0000 | 0.9793 | 1.0000 |
| Resolution (Å) | 30.00–2.15 | 30.00–1.93 | 30.00–1.65 |
| Highest resolution (Å) | 2.19–2.15 | 1.96–1.93 | 1.68–1.65 |
| No. uniquere flections | 47,100 (2297) a | 112,718 (5537) a | 91,189 (4474) a |
| Rmerge (%) b | 6.8 (50.0) a | 5.5 (35.1) a | 7.7 (44.1) a |
| Rmeas (%) c | 7.6 (55.8) a | 7.8 (49.7) a | 9.0 (51.7) a |
| CC1/2 d | 0.997 (0.860) a | 0.997 (0.740) a | 0.994 (0.879) a |
| I/sigma(I) | 32.6 (5.0) a | 19.1 (2.8) a | 27.4 (4.2) a |
| Completeness (%) | 99.9 (100.0) a | 96.7 (95.3) a | 99.6 (99.0) a |
| Redundancy | 4.9 (5.0) a | 2.0 (1.9) a | 3.7 (3.7) a |
| Wilson B-factor (Å2) | 33.4 | 22.1 | 16.4 |
| Refinement | |||
| Resolution (Å) | 30.00–2.15 | 30.00–1.93 | 30.00–1.65 |
| No. of reflections (work) | 44,658 | 106,897 | 86,549 |
| No. of reflections (test) | 2376 | 5775 | 4572 |
| Rwork (%) e | 20.3 | 17.0 | 17.4 |
| Rfree (%) f | 24.1 | 20.8 | 19.7 |
| Estimated coordinate error (Å) | 0.25 | 0.19 | 0.16 |
| No. atoms | |||
| Protein | 5717 | 11,878 | 5897 |
| Metals | 2 | 4 | - |
| NADP | - | 160 | 96 |
| Nitrate | - | - | 16 |
| Water | 116 | 792 | 605 |
| Average B-value (Å2) | |||
| Protein | 49.0 | 29.2 | 21.6 |
| Metals | 51.1 | 26.8 | - |
| NADP | - | 22.8 | 13.8 |
| Nitrate | - | - | 23.3 |
| Water | 37.0 | 32.5 | 29.7 |
| RMSD bonds (Å) | 0.008 | 0.007 | 0.006 |
| RMSD angles (°) | 0.864 | 0.851 | 0.899 |
| Ramachandran g (favored) | 97.4% | 97.5% | 96.8% |
| Ramachandran g (outliers) | 0.0% | 0.1% | 0.0% |
| PDB ID | 7W9X | 7W9Y | 7W9Z |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cho, H.Y.; Nam, M.S.; Hong, H.J.; Song, W.S.; Yoon, S.-i. Structural and Biochemical Analysis of the Furan Aldehyde Reductase YugJ from Bacillus subtilis. Int. J. Mol. Sci. 2022, 23, 1882. https://doi.org/10.3390/ijms23031882
Cho HY, Nam MS, Hong HJ, Song WS, Yoon S-i. Structural and Biochemical Analysis of the Furan Aldehyde Reductase YugJ from Bacillus subtilis. International Journal of Molecular Sciences. 2022; 23(3):1882. https://doi.org/10.3390/ijms23031882
Chicago/Turabian StyleCho, Hye Yeon, Mi Sun Nam, Ho Jeong Hong, Wan Seok Song, and Sung-il Yoon. 2022. "Structural and Biochemical Analysis of the Furan Aldehyde Reductase YugJ from Bacillus subtilis" International Journal of Molecular Sciences 23, no. 3: 1882. https://doi.org/10.3390/ijms23031882
APA StyleCho, H. Y., Nam, M. S., Hong, H. J., Song, W. S., & Yoon, S.-i. (2022). Structural and Biochemical Analysis of the Furan Aldehyde Reductase YugJ from Bacillus subtilis. International Journal of Molecular Sciences, 23(3), 1882. https://doi.org/10.3390/ijms23031882





