The Induced Expression of BPV E4 Gene in Equine Adult Dermal Fibroblast Cells as a Potential Model of Skin Sarcoid-like Neoplasia
Abstract
:1. Introduction
2. Results
2.1. Preliminary Validation of the Samples Used
2.2. Comparative Statistical Estimations Resulting from Next-Generation Sequencing (NGS) among BPV-E4 and BPV-E1^E4 Transgenic Equine ACFC-Derived Neoplastic Cells
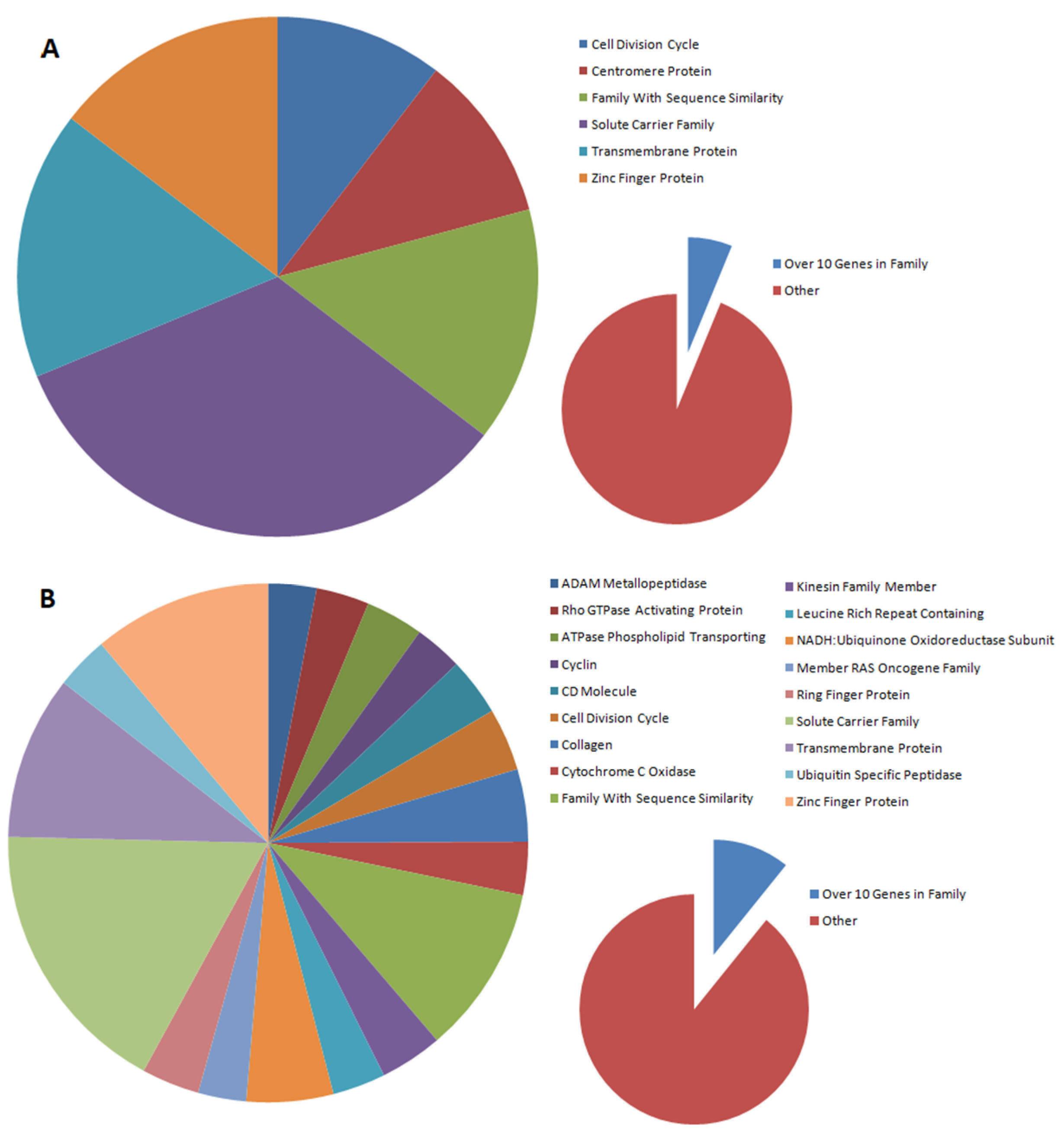
2.3. Analysis of Differentially Expressed Genes (DEGs) in Oncogenically Transformed Equine ACFCs Expressing BPV-E4 and BPV-E1^E4 Transgenes
2.4. Gene Ontology (GO) Enrichment Analysis of BPV-E4 and BPV-E1^E4 Transgenic Equine ACFC-Derived Neoplastic Cells
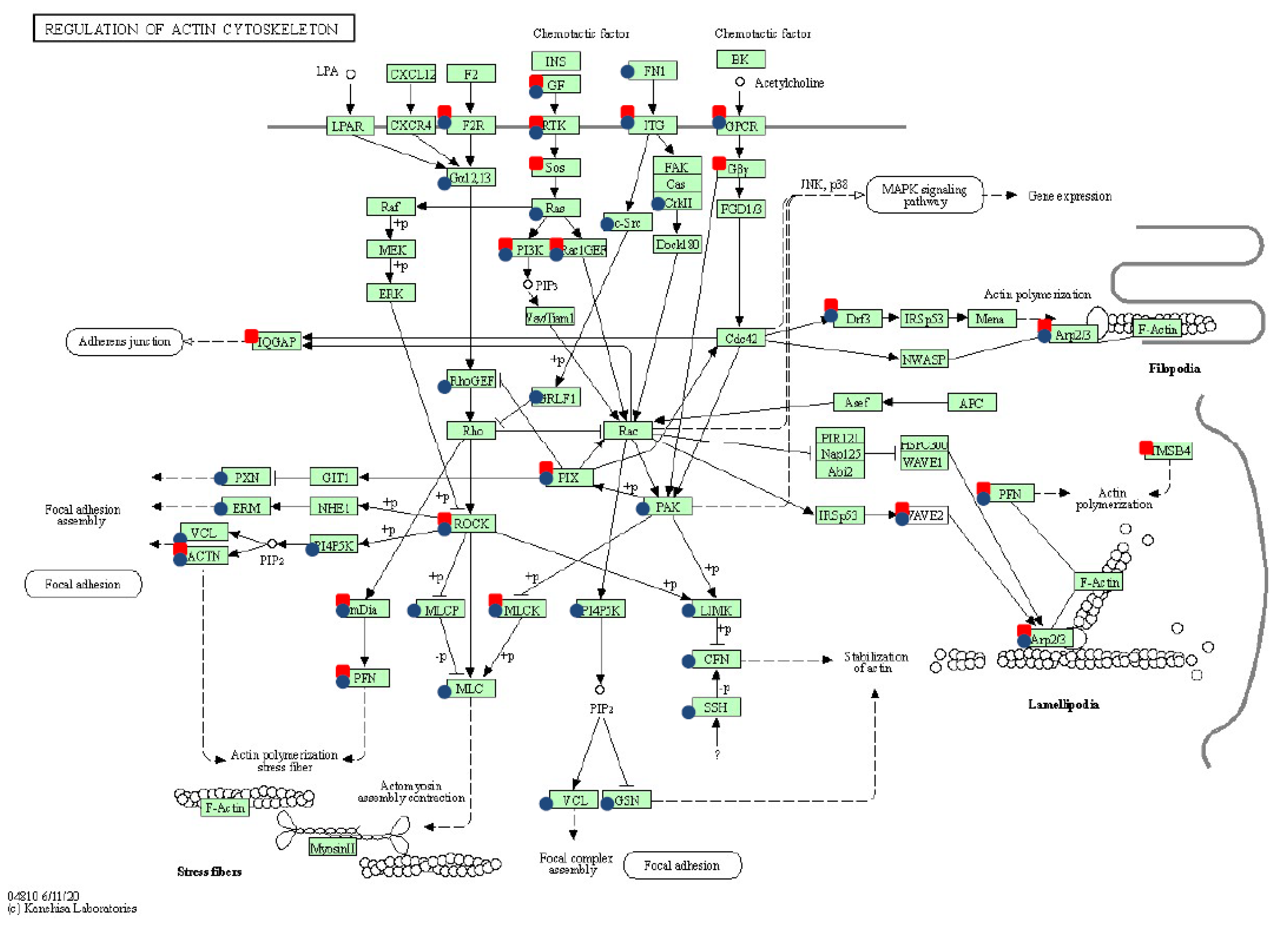
2.5. Pathway Enrichment Analysis among Oncogenically Transformed Equine ACFCs Expressing BPV-E4 and BPV-E4^E1 Transgenes
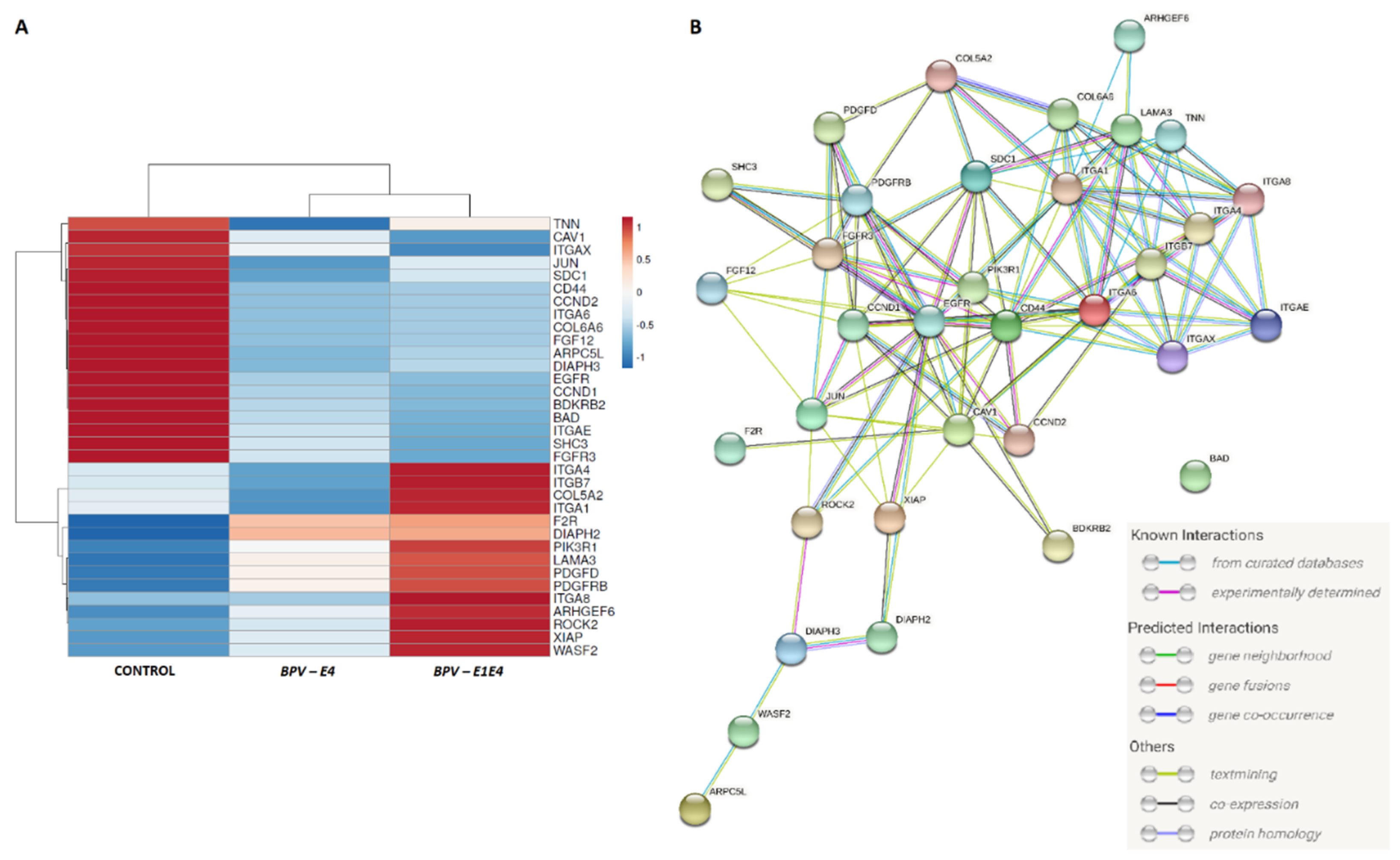
2.6. Enrichment Analysis for Identification of DEGs in BPV-E4 and BPV-E1^E4 Transgenic ACFCs Undergoing Sarcoid-Dependent Oncogenic Transformation
2.7. qPCR-Assisted Validation Accomplished for Transcriptional Activity Levels of Genes in Neoplastically Transformed Equine ACFCs Expressing BPV-E4 and BPV-E1^E4 Transgenes
3. Discussion
4. Materials and Methods
4.1. Experimental Schedule
4.2. Designing Gene Inserts for Further Experiments Aimed at Nucleofection of Equine ACFCs
4.3. The Reactions of Enzymatic Restriction and Ligation
4.4. Molecular Cloning of DNA Plasmid Constructs with Inserted BPV-E4 or BPV-E1^E4 Gene Sequences
4.5. Establishment of Primary Cultures and Mitotically Stable Lines of Equine Adult Cutaneous Fibroblast Cells (ACFCs)
4.6. Genetic Transformation of Equine ACFCs Mediated by Nucleofection
4.7. Treatment of Cell Nucleofectants Leading to Positive Antibiotic-Dependent Selection of BPV-E4 or BPV-E1^E4 Transgenic Equine ACFCs and Their Subsequent Tetracycline-Induced Neoplastic Transformation into Sarcoid-like Cells
4.8. Detection of BPV DNA in Equine ACFCs Subjected to Oncogenic Transformation with the Aid of Nucleofection
4.9. Isolation of RNA Samples from BPV-E4 and BPV-E1^E4 Transgenic Equine ACFC-Derived Neoplastic Cells
4.10. NGS Sequencing among Oncogenically Transformed Equine ACFCs Expressing BPV-E4 and BPV-E4^E1 Transgenes
4.11. qPCR-Assisted Validation Accomplished for Transcriptional Activity Levels of Genes in Neoplastically Transformed Equine ACFCs Expressing BPV-E4 and BPV-E1^E4 Transgenes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACFC | Adult cutaneous fibroblast cell |
| ACTB | Actin β |
| ADAM | Disintegrin and metalloproteinase domain |
| Akt | Serine/threonine kinase |
| ART | Assisted reproductive technology |
| BAD | Bcl2-associated agonist of cell death |
| BCG | Bacillus Calmette–Guérin |
| BMI | Polycomb ring finger |
| BPV | Bovine Papillomavirus |
| CCND | Cyclin D |
| CD | Cluster of differentiation |
| CDK | Cyclin-dependent kinase |
| COL | Collagen |
| DAVID | Database for Annotation, Visualization, and Integrated Discovery |
| DEG | Differentially expressed gene |
| DMEM | Dulbecco’s Modified Eagle’s Medium |
| DPBS | Dulbecco’s phosphate-buffered saline |
| ECM | Extracellular matrix |
| EGFR | Epidermal growth factor receptor |
| F2R | Coagulation factor II thrombin receptor |
| FBS | Fetal bovine serum |
| FDR | False discovery rate |
| FGF | Fibroblast growth factor |
| FGFR | Fibroblast growth factor receptor |
| FOS | Fos proto-oncogene |
| FoxO | Forkhead box O |
| GEO | Gene Expression Omnibus |
| GO | Gene Ontology |
| HEPES | 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid |
| IGF | Insulin-like growth factor |
| ISNR | Insulin receptor |
| ITG | Integrin |
| ITGA | Integrin subunit α |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| KIF | Kinesin superfamily protein |
| LAMA | Laminin subunit α |
| LB | Luria–Bertani |
| MAPK | Mitogen-activated protein kinase |
| MCS | Multiple cloning site |
| MMP | Matrix metalloproteinase |
| NANOG | Nanog homeobox |
| NCR | Noncoding region |
| NDC | Nuclear donor cell |
| NGS | Next generation sequencing |
| PCA | Principal component analysis |
| PCR | Polymerase chain reaction |
| PI3K | Phosphoinositide 3-kinase |
| PTGER | Prostaglandin E receptor |
| Rap | Ras-related protein |
| RECK | Reversion inducing cysteine rich protein with kazal motifs |
| Rho | Rhodopsin |
| ROCK | Rho-associated coiled-coil containing protein kinase |
| SCNT | Somatic cell nuclear transfer |
| SOX | SRY-box transcription |
| TBE | Tris/borate/EDTA |
| TCM | Tissue culture medium |
| TE | Tris-EDTA |
| TGFB | Transforming growth factor β |
| TGFBR | Transforming growth factor β receptor |
| TIMP | Tissue inhibitor of metalloproteinase |
| TNF | Tumor necrosis factor |
| UB | Ubiquitin |
| UBP | Ubiquitin-specific peptidase |
| URR | Upstream regulatory region |
| VEGF | Vascular endothelial growth factor |
| Wnt | Wingless-type MMTV integration site family |
| XIAP | X-linked inhibitor of apoptosis |
References
- Haralambus, R.; Burgstaller, J.; Klukowska-Rötzler, J.; Steinborn, R.; Buchinger, S.; Gerber, V.; Brandt, S. Intralesional Bovine Papillomavirus DNA Loads Reflect Severity of Equine Sarcoid Disease. Equine Vet. J. 2010, 42, 327–331. [Google Scholar] [CrossRef]
- Knottenbelt, D.C. A Suggested Clinical Classification for the Equine Sarcoid. Clin. Tech. Equine Pract. 2005, 4, 278–295. [Google Scholar] [CrossRef]
- Yuan, Z.; Gallagher, A.; Gault, E.A.; Campo, M.S.; Nasir, L. Bovine Papillomavirus Infection in Equine Sarcoids and in Bovine Bladder Cancers. Vet. J. 2007, 174, 599–604. [Google Scholar] [CrossRef] [PubMed]
- Bogaert, L.; Martens, A.; Van Poucke, M.; Ducatelle, R.; De Cock, H.; Dewulf, J.; De Baere, C.; Peelman, L.; Gasthuys, F. High Prevalence of Bovine Papillomaviral DNA in the Normal Skin of Equine Sarcoid-Affected and Healthy Horses. Vet. Microbiol. 2008, 129, 58–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nasir, L.; Reid, S.W.J. Bovine Papillomaviral Gene Expression in Equine Sarcoid Tumours. Virus Res. 1999, 61, 171–175. [Google Scholar] [CrossRef]
- Yuan, Z.Q.; Gault, E.A.; Gobeil, P.; Nixon, C.; Campo, M.S.; Nasir, L. Establishment and Characterization of Equine Fibroblast Cell Lines Transformed in Vivo and in Vitro by BPV-1: Model Systems for Equine Sarcoids. Virology 2008, 373, 352–361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bogaert, L.; Martens, A.; Kast, W.M.; Van Marck, E.; De Cock, H. Bovine Papillomavirus DNA Can Be Detected in Keratinocytes of Equine Sarcoid Tumors. Vet. Microbiol. 2010, 146, 269–275. [Google Scholar] [CrossRef]
- Bocaneti, F.; Altamura, G.; Corteggio, A.; Velescu, E.; Roperto, F.; Borzacchiello, G. Bovine Papillomavirus: New Insights into an Old Disease. Transbound. Emerg. Dis. 2016, 63, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Nasir, L.; Campo, M.S. Bovine Papillomaviruses: Their Role in the Aetiology of Cutaneous Tumours of Bovids and Equids. Vet. Dermatol. 2008, 19, 243–254. [Google Scholar] [CrossRef]
- Lancaster, W.D.; Olson, C. Animal papillomaviruses. Microbiol. Rev. 1982, 46, 191–207. [Google Scholar] [CrossRef]
- Rector, A.; Van Ranst, M. Animal Papillomaviruses. Virology 2013, 445, 213–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bravo, I.G.; Félez-Sánchez, M. Papillomaviruses. Evol. Med. Public Health 2015, 2015, 32–51. [Google Scholar] [CrossRef]
- Knottenbelt, D.C. The Equine Sarcoid: Why Are There so Many Treatment Options? Vet. Clin. N. Am. Equine Pract. 2019, 35, 243–262. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.D.; Toth, B.; Baseler, L.J.; Charney, V.A.; Miller, M.A. Lack of Correlation between Papillomaviral DNA in Surgical Margins and Recurrence of Equine Sarcoids. J. Equine Vet. Sci. 2014, 34, 722–725. [Google Scholar] [CrossRef]
- Funiciello, B.; Roccabianca, P. Equine Sarcoid; IntechOpen: London, UK, 2020; ISBN 978-1-83962-317-2. [Google Scholar]
- Semik, E.; Gurgul, A.; Ząbek, T.; Ropka-Molik, K.; Koch, C.; Mählmann, K.; Bugno-Poniewierska, M. Transcriptome Analysis of Equine Sarcoids. Vet. Comp. Oncol. 2017, 15, 1370–1381. [Google Scholar] [CrossRef] [PubMed]
- Semik-Gurgul, E. Molecular Approaches to Equine Sarcoids. Equine Vet. J. 2021, 53, 221–230. [Google Scholar] [CrossRef]
- Bogaert, L.; Woodham, A.W.; Da Silva, D.M.; Martens, A.; Meyer, E.; Kast, W.M. A Novel Murine Model for Evaluating Bovine Papillomavirus Prophylactics/Therapeutics for Equine Sarcoid-like Tumours. J. Gen. Virol. 2015, 96, 2764–2768. [Google Scholar] [CrossRef]
- Li, L.; Connelly, M.C.; Wetmore, C.; Curran, T.; Morgan, J.I. Mouse embryos cloned from brain tumors. Cancer Res. 2003, 63, 2733–2736. [Google Scholar]
- Shao, G.-B.; Ding, H.-M.; Gao, W.-L.; Li, S.-H.; Wu, C.-F.; Xu, Y.-X.; Liu, H.-L. Effect of Trychostatin A Treatment on Gene Expression in Cloned Mouse Embryos. Theriogenology 2009, 71, 1245–1252. [Google Scholar] [CrossRef]
- Skrzyszowska, M.; Samiec, M.; Słomski, R.; Lipiński, D.; Mały, E. Development of porcine transgenic nuclear-transferred embryos derived from fibroblast cells transfected by the novel technique of nucleofection or standard lipofection. Theriogenology 2008, 70, 248–259. [Google Scholar] [CrossRef]
- Samiec, M.; Skrzyszowska, M.; Lipiński, D. Pseudophysiological transcomplementary activation of reconstructed oocytes as a highly efficient method used for producing nuclear-transferred pig embryos originating from transgenic foetal fibroblast cells. Pol. J. Vet. Sci. 2012, 15, 509–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R: The R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 21 January 2022).
- STRING: Functional Protein Association Networks. Available online: https://string-db.org/ (accessed on 21 January 2022).
- Doorbar, J.; Foo, C.; Coleman, N.; Medcalf, L.; Hartley, O.; Prospero, T.; Napthine, S.; Sterling, J.; Winter, G.; Griffin, H. Characterization of Events during the Late Stages of HPV16 Infectionin VivoUsing High-Affinity Synthetic Fabs to E4. Virology 1997, 238, 40–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pray, T.R.; Laimins, L.A. Differentiation-Dependent Expression of E1^E4 Proteins in Cell Lines Maintaining Episomes of Human Papillomavirus Type 31b. Virology 1995, 206, 679–685. [Google Scholar] [CrossRef] [Green Version]
- Wilson, R.; Fehrmann, F.; Laimins, L.A. Role of the E1–E4 Protein in the Differentiation-Dependent Life Cycle of Human Papillomavirus Type 31. J. Virol. 2005, 79, 6732–6740. [Google Scholar] [CrossRef] [Green Version]
- Boer, J.M.; Huber, W.K.; Sültmann, H.; Wilmer, F.; von Heydebreck, A.; Haas, S.; Korn, B.; Gunawan, B.; Vente, A.; Füzesi, L.; et al. Identification and Classification of Differentially Expressed Genes in Renal Cell Carcinoma by Expression Profiling on a Global Human 31,500-Element CDNA Array. Genome Res. 2001, 11, 1861–1870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, T.; Furukawa, Y.; Nakagawa, H.; Tsunoda, T.; Ohigashi, H.; Murata, K.; Ishikawa, O.; Ohgaki, K.; Kashimura, N.; Miyamoto, M.; et al. Genome-Wide CDNA Microarray Analysis of Gene Expression Profiles in Pancreatic Cancers Using Populations of Tumor Cells and Normal Ductal Epithelial Cells Selected for Purity by Laser Microdissection. Oncogene 2004, 23, 2385–2400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, J.; Lee, J.; Namkoong, S.; Bae, S.; Kim, Y.W.; Han, S.; Cho, Y.; Nam, G.; Kim, C.-K.; Seo, J.-S.; et al. cDNA Microarray Analysis of Gene Expression Profiles Associated with Cervical Cancer. Cancer Res. Treat. 2003, 35, 451–459. [Google Scholar] [CrossRef]
- Desgrosellier, J.S.; Cheresh, D.A. Integrins in Cancer: Biological Implications and Therapeutic Opportunities. Nat. Rev. Cancer 2010, 10, 9–22. [Google Scholar] [CrossRef] [Green Version]
- Hamidi, H.; Ivaska, J. Every Step of the Way: Integrins in Cancer Progression and Metastasis. Nat. Rev. Cancer 2018, 18, 533–548. [Google Scholar] [CrossRef] [Green Version]
- Valdembri, D.; Serini, G. The Roles of Integrins in Cancer. Fac. Rev. 2021, 10, 45. [Google Scholar] [CrossRef]
- Lucanus, A.J.; Yip, G.W. Kinesin Superfamily: Roles in Breast Cancer, Patient Prognosis and Therapeutics. Oncogene 2018, 37, 833–838. [Google Scholar] [CrossRef] [PubMed]
- Koni, M.; Pinnarò, V.; Brizzi, M.F. The Wnt Signalling Pathway: A Tailored Target in Cancer. Int. J. Mol. Sci. 2020, 21, 7697. [Google Scholar] [CrossRef] [PubMed]
- Aldaz, P.; Otaegi-Ugartemendia, M.; Saenz-Antoñanzas, A.; Garcia-Puga, M.; Moreno-Valladares, M.; Flores, J.M.; Gerovska, D.; Arauzo-Bravo, M.J.; Samprón, N.; Matheu, A.; et al. SOX9 Promotes Tumor Progression through the Axis BMI1-P21CIP. Sci. Rep. 2020, 10, 357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, Y.; Lai, L.; Lian, W.; Tu, X.; Zhou, J.; Dong, P.; Su, D.; Wang, X.; Cao, X.; Chen, Y.; et al. SOX9/FXYD3/Src Axis Is Critical for ER+ Breast Cancer Stem Cell Function. Mol. Cancer Res. MCR 2019, 17, 238–249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Y.; Shepherd, J.; Zhao, D.; Bollu, L.R.; Tahaney, W.M.; Hill, J.; Zhang, Y.; Mazumdar, A.; Brown, P.H. SOX9 Is Essential for Triple-Negative Breast Cancer Cell Survival and Metastasis. Mol. Cancer Res. MCR 2020, 18, 1825–1838. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. The Hallmarks of Cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef] [Green Version]
- Jakowlew, S.B. Transforming Growth Factor-β in Cancer and Metastasis. Cancer Metastasis Rev. 2006, 25, 435. [Google Scholar] [CrossRef]
- Kim, W.; Kim, E.; Lee, S.; Kim, D.; Chun, J.; Park, K.H.; Youn, H.; Youn, B. TFAP2C-Mediated Upregulation of TGFBR1 Promotes Lung Tumorigenesis and Epithelial–Mesenchymal Transition. Exp. Mol. Med. 2016, 48, e273. [Google Scholar] [CrossRef] [Green Version]
- Zeng, Q.; Phukan, S.; Xu, Y.; Sadim, M.; Rosman, D.S.; Pennison, M.; Liao, J.; Yang, G.-Y.; Huang, C.-C.; Valle, L.; et al. Tgfbr1 Haploinsufficiency Is a Potent Modifier of Colorectal Cancer Development. Cancer Res. 2009, 69, 678–686. [Google Scholar] [CrossRef] [Green Version]
- Zhou, R.; Huang, Y.; Cheng, B.; Wang, Y.; Xiong, B. TGFBR1*6A Is a Potential Modifier of Migration and Invasion in Colorectal Cancer Cells. Oncol. Lett. 2018, 15, 3971–3976. [Google Scholar] [CrossRef] [Green Version]
- Mahner, S.; Baasch, C.; Schwarz, J.; Hein, S.; Wölber, L.; Jänicke, F.; Milde-Langosch, K. C-Fos Expression Is a Molecular Predictor of Progression and Survival in Epithelial Ovarian Carcinoma. Br. J. Cancer 2008, 99, 1269–1275. [Google Scholar] [CrossRef]
- Muhammad, N.; Bhattacharya, S.; Steele, R.; Phillips, N.; Ray, R.B. Involvement of C-Fos in the Promotion of Cancer Stem-like Cell Properties in Head and Neck Squamous Cell Carcinoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2017, 23, 3120–3128. [Google Scholar] [CrossRef] [Green Version]
- Gao, F.; Zhou, L.; Li, M.; Liu, W.; Yang, S.; Li, W. Inhibition of ERKs/Akt-Mediated c-Fos Expression Is Required for Piperlongumine-Induced Cyclin D1 Downregulation and Tumor Suppression in Colorectal Cancer Cells. OncoTargets Ther. 2020, 13, 5591–5603. [Google Scholar] [CrossRef]
- Nallanthighal, S.; Heiserman, J.P.; Cheon, D.-J. The Role of the Extracellular Matrix in Cancer Stemness. Front. Cell Dev. Biol. 2019, 7, 86. [Google Scholar] [CrossRef] [PubMed]
- Winkler, J.; Abisoye-Ogunniyan, A.; Metcalf, K.J.; Werb, Z. Concepts of Extracellular Matrix Remodelling in Tumour Progression and Metastasis. Nat. Commun. 2020, 11, 5120. [Google Scholar] [CrossRef] [PubMed]
- Walker, C.; Mojares, E.; del Río Hernández, A. Role of Extracellular Matrix in Development and Cancer Progression. Int. J. Mol. Sci. 2018, 19, 3028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, S.; Xu, H.; Wang, W.; Li, S.; Li, H.; Li, T.; Zhang, W.; Yu, X.; Liu, L. The Role of Collagen in Cancer: From Bench to Bedside. J. Transl. Med. 2019, 17, 309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Zhang, S.; Li, H.; Chou, H. FGFR3 Promotes the Growth and Malignancy of Melanoma by Influencing EMT and the Phosphorylation of ERK, AKT, and EGFR. BMC Cancer 2019, 19, 963. [Google Scholar] [CrossRef]
- Gao, G.; Yang, M.; Wang, F.; Dang, G.; Zhang, X.; Zhao, J.; Wang, X.; Jin, B. Coagulation Factor 2 Thrombin Receptor Promotes Malignancy in Glioma under SOX2 Regulation. Aging 2020, 12, 10594–10613. [Google Scholar] [CrossRef] [PubMed]
- Kaczorowski, M.; Biecek, P.; Donizy, P.; Pieniążek, M.; Matkowski, R.; Hałoń, A. ROCK1 and ROCK2 Are Down-Regulated in Aggressive and Advanced Skin Melanomas—A Clinicopathological Perspective. Anticancer Res. 2020, 40, 1931–1942. [Google Scholar] [CrossRef]
- Zucchini, C.; Manara, M.C.; Cristalli, C.; Carrabotta, M.; Greco, S.; Pinca, R.S.; Ferrari, C.; Landuzzi, L.; Pasello, M.; Lollini, P.-L.; et al. ROCK2 Deprivation Leads to the Inhibition of Tumor Growth and Metastatic Potential in Osteosarcoma Cells through the Modulation of YAP Activity. J. Exp. Clin. Cancer Res. 2019, 38, 503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, Y.; Li, J.; Han, F.; Chen, H.; Zhao, X.; Qin, Q.; Shi, R.; Liu, J. High IGF2 Expression Is Associated with Poor Clinical Outcome in Human Ovarian Cancer. Oncol. Rep. 2015, 34, 936–942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spano, J.-P.; Lagorce, C.; Atlan, D.; Milano, G.; Domont, J.; Benamouzig, R.; Attar, A.; Benichou, J.; Martin, A.; Morere, J.-F.; et al. Impact of EGFR Expression on Colorectal Cancer Patient Prognosis and Survival. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2005, 16, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Mitsudomi, T.; Yatabe, Y. Epidermal Growth Factor Receptor in Relation to Tumor Development: EGFR Gene and Cancer. FEBS J. 2010, 277, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Csordás, G.; Santra, M.; Reed, C.C.; Eichstetter, I.; McQuillan, D.J.; Gross, D.; Nugent, M.A.; Hajnóczky, G.; Iozzo, R.V. Sustained Down-Regulation of the Epidermal Growth Factor Receptor by Decorin: A Mechanism for Controlling Tumor Growth In Vivo. J. Biol. Chem. 2000, 275, 32879–32887. [Google Scholar] [CrossRef] [Green Version]
- Mizuno, E.; Iura, T.; Mukai, A.; Yoshimori, T.; Kitamura, N.; Komada, M. Regulation of Epidermal Growth Factor Receptor Down-Regulation by UBPY-Mediated Deubiquitination at Endosomes. Mol. Biol. Cell 2005, 16, 5163–5174. [Google Scholar] [CrossRef] [Green Version]
- Heidegger, I.; Kern, J.; Ofer, P.; Klocker, H.; Massoner, P. Oncogenic functions of IGF1R and INSR in prostate cancer include enhanced tumor growth, cell migration and angiogenesis. Oncotarget 2014, 5, 2723–2735. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Tang, N.; Thompson, R.C.; Mobley, B.C.; Clark, S.W.; Sarkaria, J.N.; Wang, J. InsR/IGF1R Pathway Mediates Resistance to EGFR Inhibitors in Glioblastoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2016, 22, 1767–1776. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Lu, Z.; Deng, Y.; Wang, W.; He, Q.; Yan, W.; Wang, A. Up-Regulation of INSR/IGF1R by C-Myc Promotes TSCC Tumorigenesis and Metastasis through the NF-ΚB Pathway. Biochim. Biophys. Acta BBA—Mol. Basis Dis. 2018, 1864, 1873–1882. [Google Scholar] [CrossRef]
- Roudnicky, F.; Dieterich, L.C.; Poyet, C.; Buser, L.; Wild, P.; Tang, D.; Camenzind, P.; Ho, C.H.; Otto, V.I.; Detmar, M. High Expression of Insulin Receptor on Tumour-Associated Blood Vessels in Invasive Bladder Cancer Predicts Poor Overall and Progression-Free Survival. J. Pathol. 2017, 242, 193–205. [Google Scholar] [CrossRef] [Green Version]
- BioRender. Available online: https://biorender.com/ (accessed on 25 January 2022).
- PaVE: Papilloma Virus Genome Database. Available online: https://pave.niaid.nih.gov/ (accessed on 21 January 2022).
- NEBioCalculator. Available online: https://nebiocalculator.neb.com/#!/ligation (accessed on 21 January 2022).
- Tomasek, J.J.; Haaksma, C.J.; Eddy, R.J.; Vaughan, M.B. Fibroblast Contraction Occurs on Release of Tension in Attached Collagen Lattices: Dependency on an Organized Actin Cytoskeleton and Serum. Anat. Rec. 1992, 232, 359–368. [Google Scholar] [CrossRef]
- Sharma, R.; Barakzai, S.Z.; Taylor, S.E.; Donadeu, F.X. Epidermal-like Architecture Obtained from Equine Keratinocytes in Three-Dimensional Cultures. J. Tissue Eng. Regen. Med. 2016, 10, 627–636. [Google Scholar] [CrossRef]
- Teifke, J.P.; Kidney, B.A.; Löhr, C.V.; Yager, J.A. Detection of Papillomavirus-DNA in Mesenchymal Tumour Cells and Not in the Hyperplastic Epithelium of Feline Sarcoids. Vet. Dermatol. 2003, 14, 47–56. [Google Scholar] [CrossRef] [PubMed]
- DAVID Functional Annotation Bioinformatics Microarray Analysis. Available online: https://david.ncifcrf.gov/ (accessed on 21 January 2022).
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Primer3 Input (Version 0.4.0). Available online: https://bioinfo.ut.ee/primer3-0.4.0/ (accessed on 21 January 2022).
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of Stable Housekeeping Genes, Differentially Regulated Target Genes and Sample Integrity: BestKeeper—Excel-Based Tool Using Pair-Wise Correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Bogaert, L.; Van Poucke, M.; De Baere, C.; Peelman, L.; Gasthuys, F.; Martens, A. Selection of a Set of Reliable Reference Genes for Quantitative Real-Time PCR in Normal Equine Skin and in Equine Sarcoids. BMC Biotechnol. 2006, 6, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Gene Ontology | Nall | Nu | Upregulated Genes | Nd | Downregulated Genes | FDR |
|---|---|---|---|---|---|---|
| positive regulation of cell migration | 24 | 14 | SEMA4D, CORO1A, INSR, F2R, DAB2, PDGFD, MMP14, SDCBP, CDH13, PIK3R1 | 10 | ATP8A1, COL18A1, ARHGEF39, CCL26, HAS2, SEMA7A, BMP2, SNAI1, EGFR, TRIP6 | <0.001 |
| negative regulation of cell proliferation | 34 | 17 | IFIT3, SKAP2, IRF1, F2R, SMAD1, KAT2B, ZEB1, TESC, GLI3, CDH13 | 17 | EREG, CER1, FEZF2, WNK2, AXIN2, HMGA1, RBPJ, PTPN14, SPRY2, BMP2 | <0.001 |
| cell-matrix adhesion | 15 | 5 | VCAM1, SNED1, ITGB4, CL2L11, ITGA8 | 10 | EPDR1, ITGB6, TECTA, OTOA, FREM1, TNN, STRC, ITGB3, ITGA6, ITGA1 | <0.001 |
| cell migration | 21 | 9 | SDC4, ASAP3, RASGEF1A, JAK2, LIMD1, MMP14, CLN3, JAK1, NDE1 | 12 | TNS3, DEPDC1B, TNN, HES1, CSPG4, ELMO1, SDC1, SNAI1, ABL2, FSCN1 | <0.001 |
| mitotic spindle assembly | 9 | 3 | KIF3B, WRAP73, ARHGEF10, | 6 | BIRC5, NEK2, MYBL2, KIFC1, KIF11, RAB11A | 0.002 |
| mitotic cytokinesis | 8 | 0 | - | 8 | NUSAP1, KIF20A, CEP55, KIF23, RACGAP1, ANLN, CKAP2, PLK1, | 0.002 |
| chromosome segregation | 11 | 2 | NDE1, NEK3 | 9 | NEK2, HJURP, CENPT, SPC25, CENPN, KIF11, CENPW, CDCA2, RCC1 | 0.002 |
| actin cytoskeleton organization | 15 | 8 | CDC42, EP2, CORO1A, RHOJ, SDCBP, NISCH, CLN3, WASF2, BCL6 | 7 | ARHGAP26, ELMO1, DIAPH3, NUAK2, ABL2, PFN1, TMSB4X | 0.002 |
| cell adhesion | 21 | 5 | GPNMB, ITGA8, CERCAM, EPHB4, TNFAIP6 | 16 | POSTN, TNC, TGFBI, COL18A1, NINJ1, HES1, SUSD5, HAS2, ITGA6, COL15A1 | 0.003 |
| Gene Ontology | Nall | Nu | Upregulated Genes | Nd | Downregulated Genes | FDR |
|---|---|---|---|---|---|---|
| negative regulation of canonical Wnt signaling pathway | 33 | 21 | EGR1, WNT5A, DKK2, SOX9, LIMD1, GREM1, BICC1, GLI3, STK4, LATS1 | 12 | NOTUM, WNT11, GPC3, DRAXIN, AXIN2, CAV1, LRP4, NPHP4, MLLT3, MAD2L2 | 0.009 |
| focal adhesion | 99 | 65 | ITGA8, SORBS1, CNN1, MCAM, ITGA11, SYNPO2, FBLN7, NEXN, LPP, PHLDB2 | 34 | CSPG4, PROCR, FLRT2, HMGA1, TNS4, TSPAN4, CAV1, FHL1, KIF22, PLAU | |
| negative regulation of extrinsic apoptotic signaling pathway | 15 | 13 | TGFBR1, COL11A1, COL1A1, LOX, COL5A1, GREM1, P4HA1, LOXL2, COL1A2, NF1 | 2 | FMOD, ANXA2 | <0.001 |
| transforming growth factor beta receptor signaling pathway | 19 | 12 | TGFBR1, FOS, SKIL, SMAD4, SMURF1, COL1A2, SMAD9 FERMT2, TGFBR3, MTMR4, | 7 | HPGD, SMAD6, PTPRK, SMURF2, TAB1, PXN, TGFB3 | <0.001 |
| collagen fibril organization | 15 | 13 | TGFBR1, COL11A1, COL1A1, LOX, COL5A1, GREM1, P4HA1, LOXL2, COL1A2, NF1 | 2 | FMOD, ANXA2 | <0.001 |
| KEGG Pathways | Nall | Nu | Nd | FDR | Most Deregulated Genes | |
|---|---|---|---|---|---|---|
| Up | Down | |||||
| Focal adhesion (ecb04510) | 31 | 11 | 20 | 0.051 | ITGB4, LAMA3, XIAP, ITGA8, PDGFD, PIK3R1, SOS2, ROCK2, LAMB2, ERBB2 | TNC, ITGB6, CCND2, TNN, CCND1, COL6A6, SHC3, ACTN3, ITGB3, BAD |
| Regulation of actin cytoskeleton (ecb04810) | 34 | 13 | 21 | 0.008 | FGF18, ITGB4, F2R, ITGA8, ARHGEF6, PDGFD, DIAPH2, PIK3R1, SOS2, ROCK2 | ITGB6, FGF12, BDKRB2, IQGAP3, FGFR3, ACTN3, ITGB3, DIAPH3, ITGB7, ITGA6 |
| ECM-receptor interaction (ecb04512) | 19 | 5 | 14 | 0.010 | ITGB4, LAMA3, SDC4, ITGA8, LAMB2 | TNC, ITGB6, TNN, COL6A6, HMMR, ITGB3, ITGB7, ITGA6, SDC1, ITGA1 |
| PI3K-Akt signaling pathway (ecb04151) | 44 | 20 | 24 | 0.051 | FGF18, ITGB4, LAMA3, INSR, CREB3L1, BCL2L11, F2R, TGA8, NR4A1, JAK2 | TNC, ITGB6, FGF12, CCND2, TNN, CCND1, ANGPT1, COL6A6, FGFR3, ITGB3 |
| Cell cycle (ecb04110) | 28 | 9 | 18 | 0.001 | CDC14A, RB1, STAG1, CDC25B, E2F5, SMC3, RBL1, RBL2, RAD21 | CCND2, CDC45, CCND1, MCM5, CCNB2, CCNB1, CDC20, E2F1, CDK1, BUB1 |
| Steroid biosynthesis (ecb00100) | 9 | 1 | 8 | 0.008 | SOAT1 | HSD17B7, TM7SF2, LSS, SQLE, FDFT1, SQLE, FDFT1, FAXDC2, EBP, SC5D |
| Pathways in cancer (ecb05200) | 52 | 23 | 29 | 0.010 | FGF18, LAMA3, FOS, XIAP, F2R, TGFBR2, ADCY9, MITF, RB1, ADCY3 | CTNNA2, WNT7B, FGF12, MMP1, CXCL8, BDKRB2, TCF7, BIRC5, CCND1, AXIN2 |
| KEGG Pathways | Nall | Nu | Nd | FDR | Most Deregulated Genes | |
|---|---|---|---|---|---|---|
| Up | Down | |||||
| Focal adhesion (ecb04510) | 63 | 42 | 21 | <0.001 | ITGA8, THBS1, ITGA11, OL11A1, XIAP, PDPK1, LAMA3, MYLK3, ROCK2, PP1R12A | LAMC3, SHC3, COL5A3, COL4A1, CCND1, CCND2, COL6A6, BAD, VEGFD, COL6A3 |
| Regulation of actin cytoskeleton (ecb04810) | 57 | 43 | 14 | <0.001 | FGF21, ITGA8, FGF5, ITGA11, MYLK3, ROCK2, PPP1R12A, PFN2, FGFR2, ARHGEF6 | BDKRB2, FGFR3, ITGAX, FGF12, IQGAP2, DIAPH3, ITGA6, GSN, ITGAE, EGFR |
| ECM-receptor interaction (ecb04512) | 30 | 18 | 12 | 0.001 | ITGA8, THBS1, ITGA11, COL11A1, LAMA3, LAMA5, COL1A1, COL5A, ITGB7, FN1, | LAMC3, COL5A3, COL4A1, COL6A6, COL6A3, ITGA6, SDC1 CD44, AGRN, TNN, |
| PI3K-Akt signaling pathway (ecb04151) | 73 | 47 | 26 | 0.019 | FGF21, ITGA8, FGF5, THBS1 ITGA11, COL11A1, CREB3L1, INSR, EFNA1, DPK1 | IL6LAMC3, FGFR3, COL5A3, COL4A1, CCND1, FGF12, CCND2 IL7, COL6A6, |
| Cell cycle (ecb04110) | 32 | 10 | 22 | 0.047 | GADD45B, RBL1, SMAD4, STAG1, EP300, CDC27, AD21, YWHAG, STAG2, E2F5 | CCND1, CCND2, CCNB2, CDC20, MCM5, CCNB1, CDK1, CDC45 PKMYT1, PLK1, |
| FoxO signaling pathway (ecb04068) | 38 | 22 | 16 | 0.003 | TGFBR1, INSR, PDPK1, PRKAB2, AKT3, FBXO32, IRS2, GADD45B, SMAD4, PRKAG3 | IL6, CCND1, CCND2, CCNB2, CCNB1, TNFSF10, S1PR1, PLK1, CDKN2B, G6PC3, |
| Proteoglycans in cancer (ecb05205) | 48 | 27 | 21 | 0.019 | ITPR1, THBS1, WNT5A, PDPK1, ROCK2, PPP1R12A, AKT3, FN1, CAMK2D, PIK3R1 | WNT11, ERBB3, GPC3, CCND1, WNT7B, HPSE, MMP9, TIMP3 CAV1, IGF2, |
| Rap1 signaling pathway (ecb04015) | 49 | 34 | 15 | 0.035 | FGF21, FGF5, THBS1, INSR, ADCY5, EFNA1, AKT3, SIPA1L2, PFN2, ADCY9 | RAP1GAP, FGFR3, FGF12, ID1, ADORA2A, VEGFD, ANGPT1, ANGPT4, HGF, MAP2K3, |
| TNF signaling pathway (ecb04668) | 29 | 16 | 13 | 0.047 | MAP3K8, EDN1, CREB3L1, FOS, AKT3, CREB3L2, TAB3, PIK3R1, ITCH, MAP3K5 | CSF2, IL6, CXCL1, VCAM1, MMP9, IL15, CREB3L4, MAP2K3, CCL2, TRADD |
| Gene | Accession Number | Correlation Coefficient |
|---|---|---|
| MMP2 | ENSECAG00000000953 | 0.839 *** |
| MMP14 | ENSECAG00000008351 | 0.887 * |
| MMP9 | ENSECAG00000013081 | 0.662 * |
| MMP15 | ENSECAG00000000196 | 0.897 ** |
| MMP17 | ENSECAG00000013201 | 0.440 ns |
| MMP24 | ENSECAG00000024778 | 0.814 * |
| PTGER2 | ENSECAG00000009713 | 0.686 * |
| TIMP1 | ENSECAG00000014259 | 0.989 *** |
| FGF10 | ENSECAG00000014361 | 0.748 * |
| RECK | ENSECAG00000010426 | 0.688 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Podstawski, P.; Samiec, M.; Skrzyszowska, M.; Szmatoła, T.; Semik-Gurgul, E.; Ropka-Molik, K. The Induced Expression of BPV E4 Gene in Equine Adult Dermal Fibroblast Cells as a Potential Model of Skin Sarcoid-like Neoplasia. Int. J. Mol. Sci. 2022, 23, 1970. https://doi.org/10.3390/ijms23041970
Podstawski P, Samiec M, Skrzyszowska M, Szmatoła T, Semik-Gurgul E, Ropka-Molik K. The Induced Expression of BPV E4 Gene in Equine Adult Dermal Fibroblast Cells as a Potential Model of Skin Sarcoid-like Neoplasia. International Journal of Molecular Sciences. 2022; 23(4):1970. https://doi.org/10.3390/ijms23041970
Chicago/Turabian StylePodstawski, Przemysław, Marcin Samiec, Maria Skrzyszowska, Tomasz Szmatoła, Ewelina Semik-Gurgul, and Katarzyna Ropka-Molik. 2022. "The Induced Expression of BPV E4 Gene in Equine Adult Dermal Fibroblast Cells as a Potential Model of Skin Sarcoid-like Neoplasia" International Journal of Molecular Sciences 23, no. 4: 1970. https://doi.org/10.3390/ijms23041970
APA StylePodstawski, P., Samiec, M., Skrzyszowska, M., Szmatoła, T., Semik-Gurgul, E., & Ropka-Molik, K. (2022). The Induced Expression of BPV E4 Gene in Equine Adult Dermal Fibroblast Cells as a Potential Model of Skin Sarcoid-like Neoplasia. International Journal of Molecular Sciences, 23(4), 1970. https://doi.org/10.3390/ijms23041970






