Quantitative Visualization of the Interaction between Complement Component C1 and Immunoglobulin G: The Effect of CH1 Domain Deletion
Abstract
:1. Introduction
2. Results and Discussion
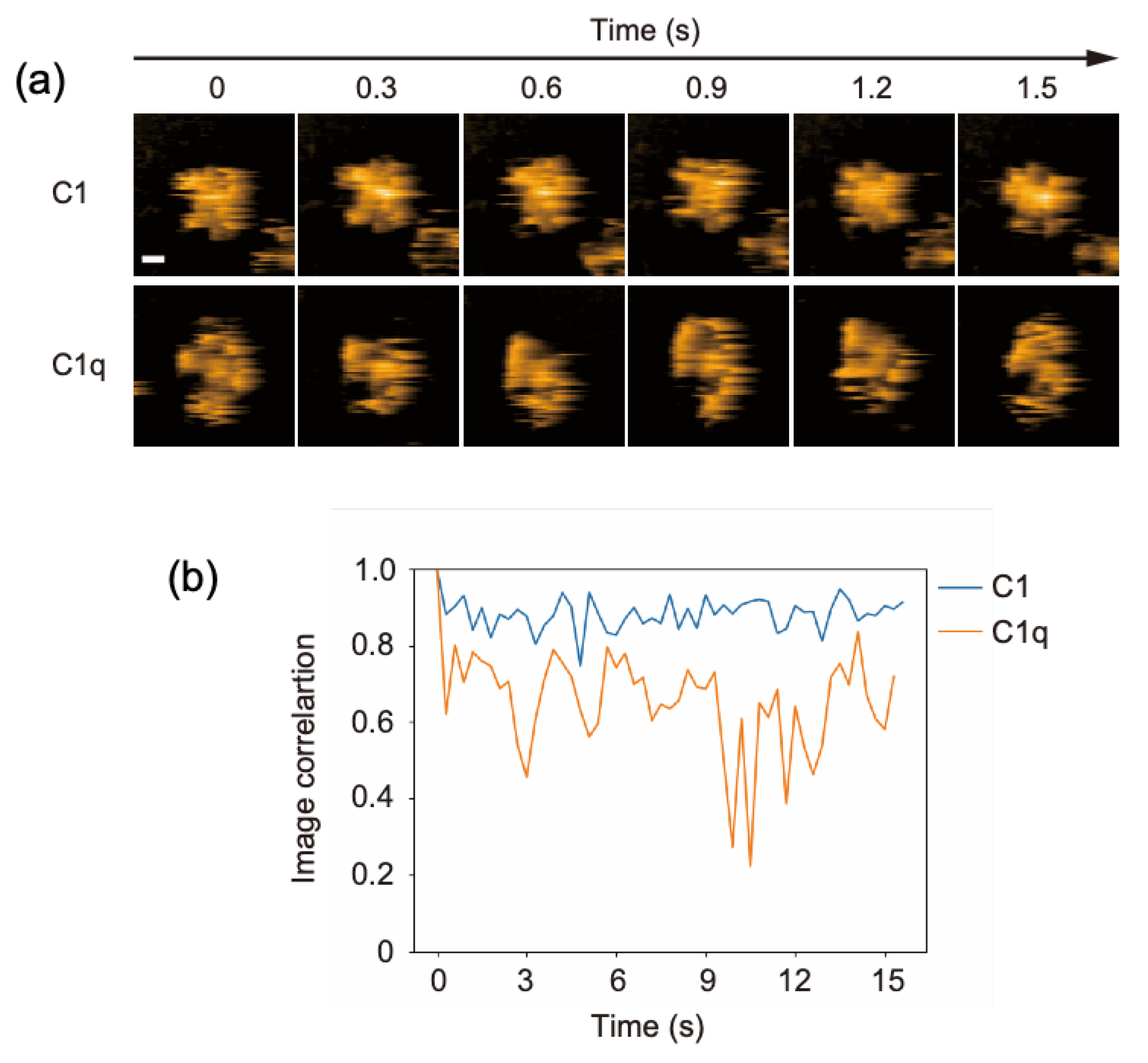
2.1. Comparing the Conformational Flexibility between C1 and C1q
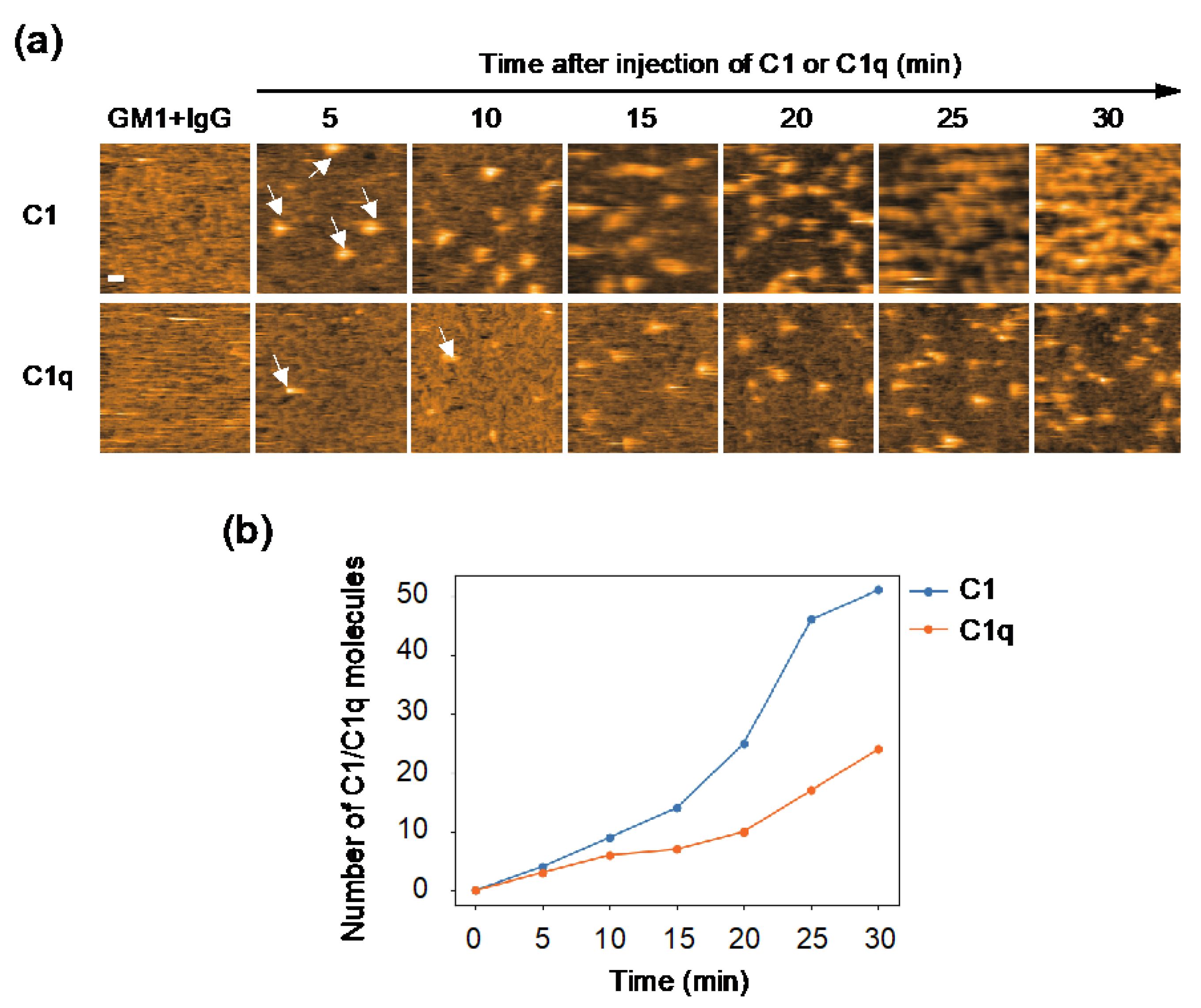
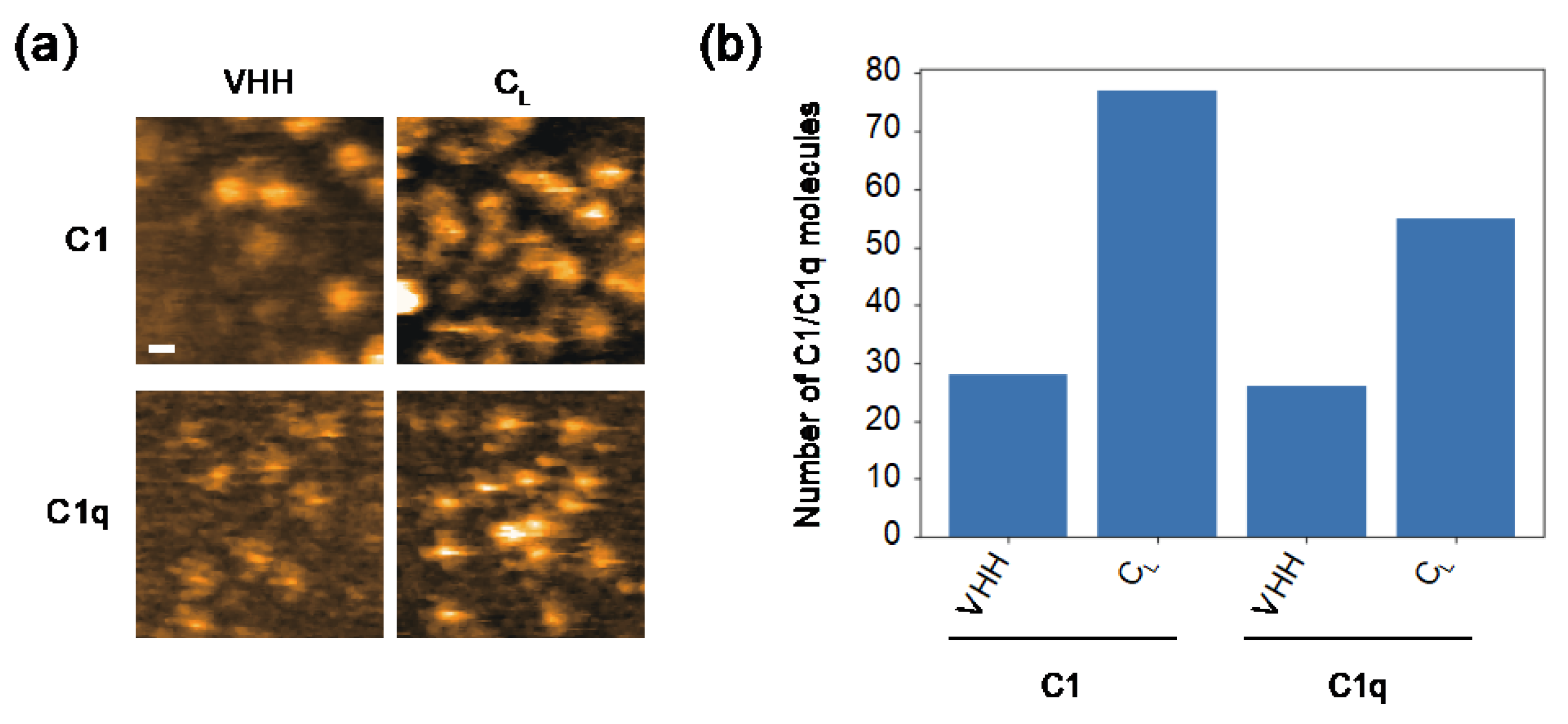
2.2. Comparing Dynamic Interactions of IgG Assemblages with C1 and C1q on Antigen-Incorporated Membranes
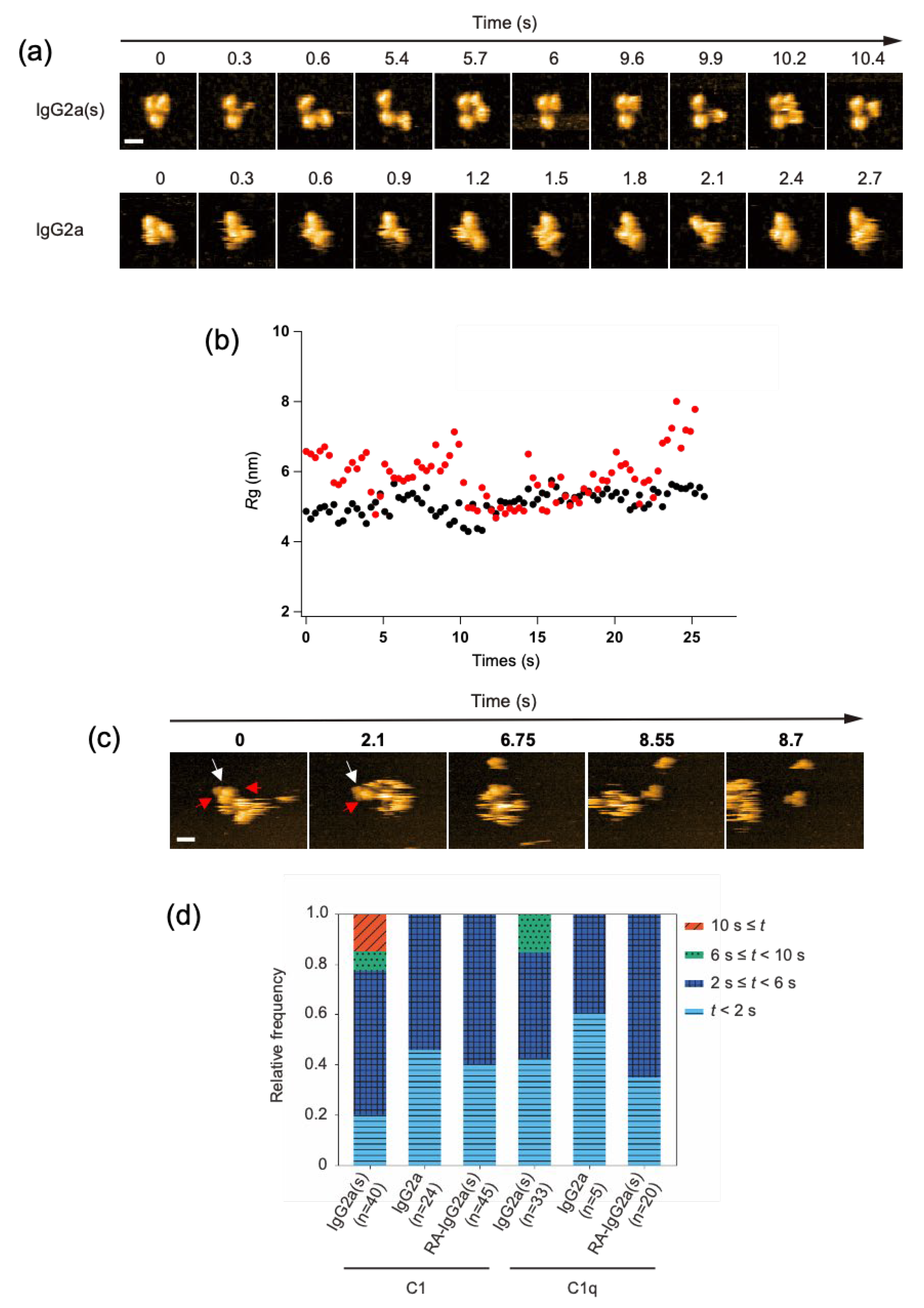
2.3. C1/C1q Interaction with IgG2a(s)
3. Materials and Methods
3.1. Chemicals
3.2. Protein Preparation
3.2.1. Antibody
3.2.2. C1q
3.3. HS-AFM Observation
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Delves, P.; Martin, S.; Burton, D.; Roitt, I. Roitt’s Essential Immunology, 13th ed.; WILEY-Blackwell: Hoboken, NJ, USA, 2016. [Google Scholar]
- Lu, R.M.; Hwang, Y.C.; Liu, I.J.; Lee, C.C.; Tsai, H.Z.; Li, H.J.; Wu, H.C. Development of therapeutic antibodies for the treatment of diseases. J. Biomed. Sci. 2020, 27, 1. [Google Scholar] [CrossRef] [PubMed]
- Kaplon, H.; Muralidharan, M.; Schneider, Z.; Reichert, J.M. Antibodies to watch in 2020. MAbs 2020, 12, 1703531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bournazos, S.; Gupta, A.; Ravetch, J.V. The role of IgG Fc receptors in antibody-dependent enhancement. Nat. Rev. Immunol. 2020, 20, 633–643. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, B.S.; Ackerman, M.E. Antibody-mediated complement activation in pathology and protection. Immunol. Cell Biol. 2020, 98, 305–317. [Google Scholar] [CrossRef] [PubMed]
- Yagi, H.; Yanaka, S.; Kato, K. Structural and Functional Roles of the N-Glycans in Therapeutic Antibodies; Elsevier: Oxford, UK, 2020. [Google Scholar] [CrossRef]
- Brekke, O.H.; Michaelsen, T.E.; Sandlie, I. The structural requirements for complement activation by IgG—Does it hinge on the hinge? Immun. Today 1995, 16, 85–90. [Google Scholar] [CrossRef]
- Yanaka, S.; Yogo, R.; Kato, K. Biophysical characterization of dynamic structures of immunoglobulin G. Biophys. Rev. 2020, 12, 637–645. [Google Scholar] [CrossRef] [PubMed]
- Chiu, M.L.; Goulet, D.R.; Teplyakov, A.; Gilliland, G.L. Antibody Structure and Function: The Basis for Engineering Therapeutics. Antibodies 2019, 8, 55. [Google Scholar] [CrossRef] [Green Version]
- Yang, D.; Kroe-Barrett, R.; Singh, S.; Roberts, C.J.; Laue, T.M. IgG cooperativity—Is there allostery? Implications for antibody functions and therapeutic antibody development. MAbs 2017, 9, 1231–1252. [Google Scholar] [CrossRef] [Green Version]
- Yogo, R.; Yamaguchi, Y.; Watanabe, H.; Yagi, H.; Satoh, T.; Nakanishi, M.; Onitsuka, M.; Omasa, T.; Shimada, M.; Maruno, T.; et al. The Fab portion of immunoglobulin G contributes to its binding to Fcγ receptor Ⅲ. Sci. Rep. 2019, 9, 11957. [Google Scholar] [CrossRef]
- Sun, Y.; Izadi, S.; Callahan, M.; Deperalta, G.; Wecksler, A.T. Antibody-receptor interactions mediate antibody-dependent cellular cytotoxicity. J. Biol. Chem. 2021, 297, 100826. [Google Scholar] [CrossRef]
- Yanaka, S.; Yogo, R.; Watanabe, H.; Taniguchi, Y.; Satoh, T.; Komura, N.; Ando, H.; Yagi, H.; Yuki, N.; Uchihashi, T.; et al. On-Membrane Dynamic Interplay between Anti-GM1 IgG Antibodies and Complement Component C1q. Int. J. Mol. Sci. 2019, 21, 147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strasser, J.; de Jong, R.N.; Beurskens, F.J.; Wang, G.B.; Heck, A.J.R.; Schuurman, J.; Parren, P.W.H.I.; Hinterdorfer, P.; Preiner, J. Unraveling the macromolecular pathways of IgG oligomerization and complement activation on antigenic surfaces. Nano Lett. 2019, 19, 4787–4796. [Google Scholar] [CrossRef] [PubMed]
- Diebolder, C.A.; Beurskens, F.J.; de Jong, R.N.; Koning, R.I.; Strumane, K.; Lindorfer, M.A.; Voorhorst, M.; Ugurlar, D.; Rosati, S.; Heck, A.J.R.; et al. Complement is activated by IgG hexamers assembled at the cell surface. Science 2014, 343, 1260–1263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ugurlar, D.; Howes, S.C.; de Kreuk, B.J.; Koning, R.I.; de Jong, R.N.; Beurskens, F.J.; Schuurman, J.; Koster, A.J.; Sharp, T.H.; Parren, P.W.H.I.; et al. Structures of C1-IgG1 provide insights into how danger pattern recognition activates complement. Science 2018, 359, 794–797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizutani, R.; Igarashi, T.; Tanaka, T.; Shimada, I.; Arata, Y. Effector functions of a mouse IgG that lacks the entire CH1 domain. C1q binding and complement fixation in the absence of antigen. J. Immunol. 1993, 150, 131–138. [Google Scholar]
- Poon, P.H.; Schumaker, V.N.; Phillips, M.L.; Strang, C.J. Conformation and restricted segmental flexibility of C1, the first component of human complement. J. Mol. Biol. 1983, 168, 563–577. [Google Scholar] [CrossRef]
- Uchihashi, T.; Iino, R.; Ando, T.; Noji, H. High-speed atomic force microscopy reveals rotary catalysis of rotorless F(1)-ATPase. Science 2011, 333, 755–758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuki, N.; Susuki, K.; Koga, M.; Nishimoto, Y.; Odaka, M.; Hirata, K.; Taguchi, K.; Miyatake, T.; Furukawa, K.; Kobata, T.; et al. Carbohydrate mimicry between human ganglioside GM1 and Campylobacter jejuni lipooligosaccharide causes Guillain-Barré syndrome. Proc. Natl. Acad. Sci. USA 2004, 101, 11404–11409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zwarthoff, S.A.; Widmer, K.; Kuipers, A.; Strasser, J.; Ruyken, M.; Aerts, P.C.; de Haas, C.J.C.; Ugurlar, D.; den Boer, M.A.; Vidarsson, G.; et al. C1q binding to surface-bound IgG is stabilized by C1r2s2 proteases. Proc. Natl. Acad. Sci. USA 2021, 118, e2102787118. [Google Scholar] [CrossRef]
- Igarashi, T.; Sato, M.; Katsube, Y.; Takio, K.; Tanaka, T.; Nakanishi, M.; Arata, Y. Structure of a mouse immunoglobulin G that lacks the entire CH1 domain: Protein sequencing and small-angle X-ray scattering studies. Biochemistry 1990, 29, 5727–5733. [Google Scholar] [CrossRef]
- Matsunaga, C.; Kato, K.; Arata, Y. A 13C NMR study of the hinge region of a mouse monoclonal antibody. J. Biomol. NMR 1991, 1, 379–390. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Matsunaga, C.; Yoshino, A.; Kato, K.; Arata, Y. Dynamical structure of the hinge region of immunoglobulin G as studied by 13C nuclear magnetic resonance spectroscopy. J. Mol. Biol. 1994, 236, 300–309. [Google Scholar] [CrossRef] [PubMed]
- Kato, K.; Matsunaga, C.; Igarashi, T.; Kim, H.; Odaka, A.; Shimada, I.; Arata, Y. Complete assignment of the methionyl carbonyl carbon resonances in switch variant anti-dansyl antibodies labeled with [1-13C]methionine. Biochemistry 1991, 30, 270–278. [Google Scholar] [CrossRef] [PubMed]
- Hamad, N.; Watanabe, H.; Uchihashi, T.; Kurokawa, R.; Nagata, T.; Katahira, M. Direct visualization of the conformational change of FUS/TLS upon binding to promoter-associated non-coding RNA. Chem. Commun. 2020, 56, 9134–9137. [Google Scholar] [CrossRef] [PubMed]
- Kato, K.; Matsunaga, C.; Odaka, A.; Yamato, S.; Takaha, W.; Shimada, I.; Arata, Y. Carbon-13 NMR study of switch variant anti-dansyl antibodies: Antigen binding and domain-domain interactions. Biochemistry 1991, 30, 6604–6610. [Google Scholar] [CrossRef]
- Davies, J.; Riechmann, L. ‘Camelising’ human antibody fragments: NMR studies on VH domains. FEBS Lett. 1994, 339, 285–290. [Google Scholar] [CrossRef] [Green Version]
- Natsume, A.; In, M.; Takamura, H.; Nakagawa, T.; Shimizu, Y.; Kitajima, K.; Wakitani, M.; Ohta, S.; Satoh, M.; Shitara, K.; et al. Engineered antibodies of IgG1/IgG3 mixed isotype with enhanced cytotoxic activities. Cancer Res. 2008, 68, 3863–3872. [Google Scholar] [CrossRef] [Green Version]
- Dangl, J.L.; Parks, D.R.; Oi, V.T.; Herzenberg, L.A. Rapid isolation of cloned isotype switch variants using fluorescence activated cell sorting. Cytometry 1982, 2, 395–401. [Google Scholar] [CrossRef]
- Akiba, H.; Tamura, H.; Kiyoshi, M.; Yanaka, S.; Sugase, K.; Caaveiro, J.M.M.; Tsumoto, K. Structural and thermodynamic basis for the recognition of the substrate-binding cleft on hen egg lysozyme by a single-domain antibody. Sci. Rep. 2019, 9, 15481. [Google Scholar] [CrossRef]
- Ando, T.; Kodera, N.; Takai, E.; Maruyama, D.; Saito, K.; Toda, A. A high-speed atomic force microscope for studying biological macromolecules. Proc. Natl. Acad. Sci. USA 2001, 98, 12468–12472. [Google Scholar] [CrossRef] [Green Version]
- Uchihashi, T.; Ganser, C. Recent advances in bioimaging with high-speed atomic force microscopy. Biophys. Rev. 2020, 12, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Wendel, M.; Lorenz, H.; Kotthaus, J.P. Sharpened electron beam deposited tips for high resolution atomic force microscope lithography and imaging. Appl. Phys. Lett. 1995, 67, 3732. [Google Scholar] [CrossRef]
- Shibata, M.; Nishimasu, H.; Kodera, N.; Hirano, S.; Ando, T.; Uchihashi, T.; Nureki, O. Real-space and real-time dynamics of CRISPR-Cas9 visualized by high-speed atomic force microscopy. Nat. Commun. 2017, 8, 1430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yanaka, S.; Nishiguchi, S.; Yogo, R.; Watanabe, H.; Shen, J.; Yagi, H.; Uchihashi, T.; Kato, K. Quantitative Visualization of the Interaction between Complement Component C1 and Immunoglobulin G: The Effect of CH1 Domain Deletion. Int. J. Mol. Sci. 2022, 23, 2090. https://doi.org/10.3390/ijms23042090
Yanaka S, Nishiguchi S, Yogo R, Watanabe H, Shen J, Yagi H, Uchihashi T, Kato K. Quantitative Visualization of the Interaction between Complement Component C1 and Immunoglobulin G: The Effect of CH1 Domain Deletion. International Journal of Molecular Sciences. 2022; 23(4):2090. https://doi.org/10.3390/ijms23042090
Chicago/Turabian StyleYanaka, Saeko, Shigetaka Nishiguchi, Rina Yogo, Hiroki Watanabe, Jiana Shen, Hirokazu Yagi, Takayuki Uchihashi, and Koichi Kato. 2022. "Quantitative Visualization of the Interaction between Complement Component C1 and Immunoglobulin G: The Effect of CH1 Domain Deletion" International Journal of Molecular Sciences 23, no. 4: 2090. https://doi.org/10.3390/ijms23042090
APA StyleYanaka, S., Nishiguchi, S., Yogo, R., Watanabe, H., Shen, J., Yagi, H., Uchihashi, T., & Kato, K. (2022). Quantitative Visualization of the Interaction between Complement Component C1 and Immunoglobulin G: The Effect of CH1 Domain Deletion. International Journal of Molecular Sciences, 23(4), 2090. https://doi.org/10.3390/ijms23042090







