Effect of Aphidicolin, a Reversible Inhibitor of Eukaryotic Nuclear DNA Replication, on the Production of Genetically Modified Porcine Embryos by CRISPR/Cas9
Abstract
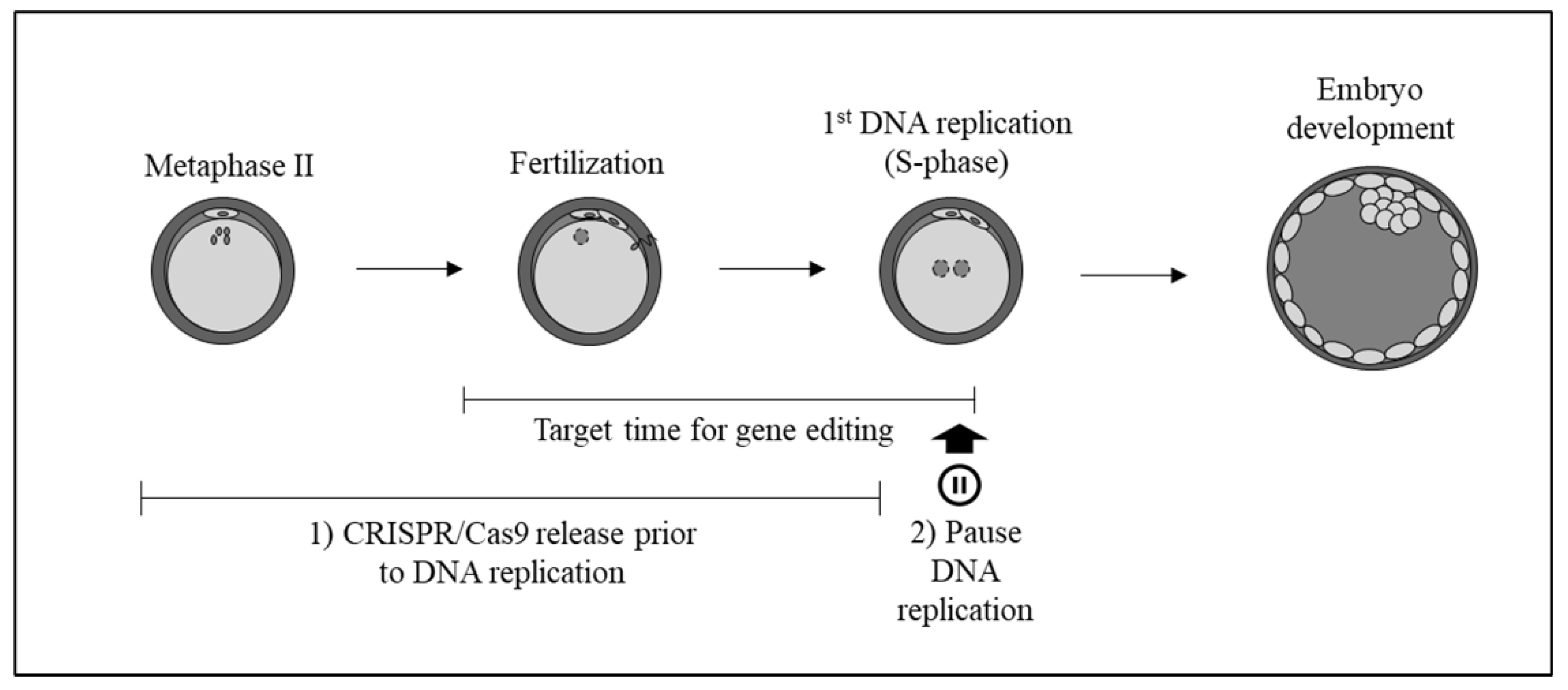
:1. Introduction
2. Results
2.1. Effect of Aphidicolin on Reversible Inhibition of DNA Replication in Porcine Zygotes
2.2. Effect of Aphidicolin on Porcine Embryo Development
2.3. Effect of Aphidicolin and RNP Concentration on Gene Editing
2.4. Effect of Aphidicolin and Methodology on Gene Editing (Electroporation vs. Microinjection)
3. Discussion
4. Materials and Methods
4.1. Ethical Issues
4.2. Culture Media Reagents
4.3. Design of Single Guide RNAs
4.4. In Vitro Maturation (IVM)
4.5. CRISPR/Cas9 Electroporation
4.6. In Vitro Fertilization (IVF)
4.7. In Vitro Embryo Culture (EC)
4.8. DNA Replication Test
4.9. Mutation Analysis
4.10. Statistical Analysis
4.11. Experimental Design
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gordon, J.W.; Scangos, G.A.; Plotkin, D.J.; Barbosa, J.A.; Ruddle, F.H. Genetic transformation of mouse embryos by microinjection of purified DNA. Proc. Natl. Acad. Sci. USA 1980, 77, 7380–7384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brem, G.; Brenig, B.; Goodman, H.M.; Selden, R.C.; Graf, F.; Kruff, B.; Springman, K.; Hondele, J.; Meyer, J.; Winnacker, H.; et al. Production Of Transgenic Mice, Rabbits And Pigs By Microinjection Into Pronuclei. Zuchthyg.-Reprod. Domest. Anim. 1985, 20, 251–252. [Google Scholar]
- Hammer, R.E.; Pursel, V.G.; Rexroad, C.E.; Wall, R.J.; Bolt, D.J.; Ebert, K.M.; Palmiter, R.D.; Brinster, R.L. Production of transgenic rabbits, sheep and pigs by microinjection. Nature 1985, 315, 680–683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bibikova, M.; Carroll, D.; Segal, D.J.; Trautman, J.K.; Smith, J.; Kim, Y.G.; Chandrasegaran, S. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol. Cell. Biol. 2001, 21, 289–297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christian, M.; Cermak, T.; Doyle, E.L.; Schmidt, C.; Zhang, F.; Hummel, A.; Bogdanove, A.J.; Voytas, D.F. Targeting DNA Double-Strand Breaks with TAL Effector Nucleases. Genetics 2010, 186, 757–761. [Google Scholar] [CrossRef] [Green Version]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [Green Version]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Yang, H.; Shivalila, C.S.; Dawlaty, M.M.; Cheng, A.W.; Zhang, F.; Jaenisch, R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 2013, 153, 910–918. [Google Scholar] [CrossRef] [Green Version]
- Whitelaw, C.B.A.; Sheets, T.P.; Lillico, S.G.; Telugu, B.P. Engineering large animal models of human disease. J. Pathol. 2016, 238, 247–256. [Google Scholar] [CrossRef]
- Navarro-Serna, S.; Vilarino, M.; Park, I.; Gadea, J.; Ross, P.J. Livestock Gene Editing by One-step Embryo Manipulation. J. Equine Vet. Sci. 2020, 89, 103025. [Google Scholar] [CrossRef]
- Whitworth, K.M.; Lee, K.; Benne, J.A.; Beaton, B.P.; Spate, L.D.; Murphy, S.L.; Samuel, M.S.; Mao, J.; O’Gorman, C.; Walters, E.M.; et al. Use of the CRISPR/Cas9 system to produce genetically engineered pigs from in vitro-derived oocytes and embryos. Biol. Reprod. 2014, 91, 78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hai, T.; Teng, F.; Guo, R.; Li, W.; Zhou, Q. One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Res. 2014, 24, 372–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Zheng, Y.; Kang, Y.; Yang, W.; Niu, Y.; Guo, X.; Tu, Z.; Si, C.; Wang, H.; Xing, R.; et al. Functional disruption of the dystrophin gene in rhesus monkey using CRISPR/Cas9. Hum. Mol. Genet. 2015, 24, 3764–3774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navarro-Serna, S.; Hachem, A.; Canha-Gouveia, A.; Hanbashi, A.; Garrappa, G.; Lopes, J.S.; Paris-Oller, E.; Bermejo-Álvarez, P.; Matas, C.; Romar, R.; et al. Generation of Nonmosaic, Two-Pore Channel 2 Biallelic Knockout Pigs in One Generation by CRISPR-Cas9 Microinjection Before Oocyte Insemination. Cris. J. 2021, 4, 132–146. [Google Scholar] [CrossRef]
- Sheets, T.P.; Park, C.H.; Park, K.E.; Powell, A.; Donovan, D.M.; Telugu, B.P. Somatic cell nuclear transfer followed by CRIPSR/Cas9 microinjection results in highly efficient genome editing in cloned pigs. Int. J. Mol. Sci. 2016, 17, 2031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whitworth, K.M.; Benne, J.A.; Spate, L.D.; Murphy, S.L.; Samuel, M.S.; Murphy, C.N.; Richt, J.A.; Walters, E.; Prather, R.S.; Wells, K.D. Zygote injection of CRISPR/Cas9 RNA successfully modifies the target gene without delaying blastocyst development or altering the sex ratio in pigs. Transgenic Res. 2017, 26, 97–107. [Google Scholar] [CrossRef]
- Burkard, C.; Lillico, S.G.; Reid, E.; Jackson, B.; Mileham, A.J.; Ait-Ali, T.; Whitelaw, C.B.A.; Archibald, A.L. Precision engineering for PRRSV resistance in pigs: Macrophages from genome edited pigs lacking CD163 SRCR5 domain are fully resistant to both PRRSV genotypes while maintaining biological function. PLoS Pathog. 2017, 13, e1006206. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Tang, X.; Xie, Z.; Zou, X.; Li, M.; Yuan, H.; Guo, N.; Ouyang, H.; Jiao, H.; Pang, D. CRISPR/Cas9-mediated knockout of myostatin in Chinese indigenous Erhualian pigs. Transgenic Res. 2017, 26, 799–805. [Google Scholar] [CrossRef]
- Hirata, M.; Tanihara, F.; Wittayarat, M.; Hirano, T.; Nguyen, N.T.; Le, Q.A. Genome mutation after introduction of the gene editing by electroporation of Cas9 protein ( GEEP ) system in matured oocytes and putative zygotes. In Vitro Cell. Dev. Biol. Anim. 2019, 55, 237–242. [Google Scholar] [CrossRef]
- Li, M.; Ouyang, H.; Yuan, H.; Li, J.; Xie, Z.; Wang, K.; Yu, T.; Liu, M.; Chen, X.; Tang, X.; et al. Site-Specific Fat-1 Knock-In Enables Significant Decrease of n-6PUFAs/n-3PUFAs Ratio in Pigs. G3 Genes Genomes Genet. 2018, 8, 1747–1754. [Google Scholar] [CrossRef] [Green Version]
- Dorado, B.; Pløen, G.G.; Barettino, A.; Macías, A.; Gonzalo, P.; Andrés-Manzano, M.J.; González-Gómez, C.; Galán-Arriola, C.; Alfonso, J.M.; Lobo, M.; et al. Generation and characterization of a novel knockin minipig model of Hutchinson-Gilford progeria syndrome. Cell Discov. 2019, 5, 16. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Zhou, X.; Lin, T.; Wang, L.; Liu, C.; Luo, X.; Luo, L.; Chen, H.; Guo, K.; Wei, H.; et al. Production of gene-edited pigs harboring orthologous human mutations via double cutting by CRISPR/Cas9 with long single-stranded DNAs as homology-directed repair templates by zygote injection. Transgenic Res. 2020, 29, 587–598. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhou, J.; Cao, C.; Huang, J.; Hai, T.; Wang, Y.; Zheng, Q.; Zhang, H.; Qin, G.; Miao, X.; et al. Efficient CRISPR/Cas9-mediated biallelic gene disruption and site-specific knockin after rapid selection of highly active sgRNAs in pigs. Sci. Rep. 2015, 5, 13348. [Google Scholar] [CrossRef] [PubMed]
- Petersen, B.; Frenzel, A.; Lucas-Hahn, A.; Herrmann, D.; Hassel, P.; Klein, S.; Ziegler, M.; Hadeler, K.-G.; Niemann, H. Efficient production of biallelic GGTA1 knockout pigs by cytoplasmic microinjection of CRISPR/Cas9 into zygotes. Xenotransplantation 2016, 23, 338–346. [Google Scholar] [CrossRef]
- Chuang, C.-K.; Chen, C.-H.; Huang, C.-L.; Su, Y.-H.; Peng, S.-H.; Lin, T.-Y.; Tai, H.-C.; Yang, T.-S.; Tu, C.-F. Generation of GGTA1 Mutant Pigs by Direct Pronuclear Microinjection of CRISPR/Cas9 Plasmid Vectors. Anim. Biotechnol. 2017, 28, 174–181. [Google Scholar] [CrossRef]
- Zhang, T.Y.; Dai, J.J.; Wu, C.F.; Gu, X.L.; Liu, L.; Wu, Z.Q.; Xie, Y.N.; Wu, B.; Chen, H.L.; Li, Y.; et al. Positive effects of treatment of donor cells with aphidicolin on the preimplantation development of somatic cell nuclear transfer embryos in Chinese Bama mini-pig (Sus Scrofa). Anim. Sci. J. 2012, 83, 103–110. [Google Scholar] [CrossRef]
- Miyamoto, K.; Hoshino, Y.; Minami, N.; Yamada, M.; Imai, H. Effects of synchronization of donor cell cycle on embryonic development and DNA synthesis in porcine nuclear transfer embryos. J. Reprod. Dev. 2007, 53, 237–246. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Y.; Lee, K.; Park, K.W.; Spate, L.D.; Prather, R.S.; Wells, K.D.; Roberts, R.M. Cell cycle synchronization of leukemia inhibitory factor (LIF)-dependent porcine-induced pluripotent stem cells and the generation of cloned embryos. Cell Cycle 2014, 13, 1265–1276. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.; Zheng, Z.; Jeon, Y.; Jin, Y.X.; Hwang, S.U.; Cai, L.; Lee, C.K.; Kim, N.H.; Hyun, S.H. An improved system for generation of diploid cloned porcine embryos using induced pluripotent stem cells synchronized to metaphase. PLoS ONE 2016, 11, e0160289. [Google Scholar] [CrossRef]
- Stephens, L.; Hardin, J.; Keller, R.; Wilt, F. The effects of aphidicolin on morphogenesis and differentiation in the sea urchin embryo. Dev. Biol. 1986, 118, 64–69. [Google Scholar] [CrossRef]
- Smith, R.K.; Johnson, M. DNA replication and compaction in the cleaving embryo of the mouse. J. Embryol Exp. Morphol. 1985, 89, 133–148. [Google Scholar] [CrossRef] [PubMed]
- Dean, W.L.; Rossant, J. Effect of delaying DNA replication on blastocyst formation in the mouse. Differentiation 1984, 26, 134–137. [Google Scholar] [CrossRef] [PubMed]
- Samaké, S.; Smith, L.C. Effects of cell-cycle-arrest agents on cleavage and development of mouse embryos. J. Exp. Zool. 1996, 274, 111–120. [Google Scholar] [CrossRef]
- Wang, B.; Pfeiffer, M.J.; Schwarzer, C.; Araúzo-Bravo, M.J.; Boiani, M. DNA replication is an integral part of the mouse oocyte’s reprogramming machinery. PLoS ONE 2014, 9, e97199. [Google Scholar] [CrossRef] [Green Version]
- Memili, E.; First, N.L. Control of gene expression at the onset of bovine embryonic development. Biol. Reprod. 1999, 61, 1198–1207. [Google Scholar] [CrossRef] [Green Version]
- Gagné, M.; Pothier, F.; Sirard, M.-A. Effect of microinjection time during postfertilization S-phase on bovine embryonic development. Mol. Reprod. Dev. 1995, 41, 184–194. [Google Scholar] [CrossRef]
- Mehravar, M.; Shirazi, A.; Nazari, M.; Banan, M. Mosaicism in CRISPR/Cas9-mediated genome editing. Dev. Biol. 2019, 445, 156–162. [Google Scholar] [CrossRef]
- Yamashita, S.; Kogasaka, Y.; Hiradate, Y.; Tanemura, K.; Sendai, Y. Suppression of mosaic mutation by co-delivery of crispr associated protein 9 and three-prime repair exonuclease 2 into porcine zygotes via electroporation. J. Reprod. Dev. 2020, 66, 41–48. [Google Scholar] [CrossRef] [Green Version]
- Tu, Z.; Yang, W.; Yan, S.; Yin, A.; Gao, J.; Liu, X.; Zheng, Y.; Zheng, J.; Li, Z.; Yang, S.; et al. Promoting Cas9 degradation reduces mosaic mutations in non-human primate embryos. Sci. Rep. 2017, 7, 42081. [Google Scholar] [CrossRef] [Green Version]
- Tanihara, F.; Hirata, M.; Nguyen, N.T.; Sawamoto, O.; Kikuchi, T.; Otoi, T. One-step generation of multiple gene-edited pigs by electroporation of the crispr/cas9 system into zygotes to reduce xenoantigen biosynthesis. Int. J. Mol. Sci. 2021, 22, 2249. [Google Scholar] [CrossRef]
- Tao, L.; Yang, M.; Wang, X.; Zhang, Z.; Wu, Z.; Tian, J.; An, L.; Wang, S. Efficient biallelic mutation in porcine parthenotes using a CRISPR-Cas9 system. Biochem. Biophys. Res. Commun. 2016, 476, 225–229. [Google Scholar] [CrossRef] [PubMed]
- Tanihara, F.; Takemoto, T.; Kitagawa, E.; Rao, S.; Do, L.T.K.; Onishi, A.; Yamashita, Y.; Kosugi, C.; Suzuki, H.; Sembon, S.; et al. Somatic cell reprogramming-free generation of genetically modified pigs. Sci. Adv. 2016, 2, e1600803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliveros, J.C.; Franch, M.; Tabas-Madrid, D.; San-León, D.; Montoliu, L.; Cubas, P.; Pazos, F. Breaking-Cas-interactive design of guide RNAs for CRISPR-Cas experiments for ENSEMBL genomes. Nucleic Acids Res. 2016, 44, W267–W271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cánovas, S.; Ivanova, E.; Romar, R.; García-Martínez, S.; Soriano-Úbeda, C.; García-Vázquez, F.A.; Saadeh, H.; Andrews, S.; Kelsey, G.; Coy, P. DNA methylation and gene expression changes derived from assisted reproductive technologies can be decreased by reproductive fluids. elife 2017, 6, e23670. [Google Scholar] [CrossRef]
- Petters, R.M.; Wells, K.D. Culture of pig embryos. J. Reprod. Fertil. Suppl. 1993, 48, 61–73. [Google Scholar] [CrossRef]
- Qin, W.; Dion, S.L.; Kutny, P.M.; Zhang, Y.; Cheng, A.W.; Jillette, N.L.; Malhotra, A.; Geurts, A.M.; Chen, Y.-G.; Wang, H. Efficient CRISPR/Cas9-Mediated Genome Editing in Mice by Zygote Electroporation of Nuclease. Genetics 2015, 200, 423–430. [Google Scholar] [CrossRef] [Green Version]
- Rath, D.; Long, C.R.; Dobrinsky, J.R.; Welch, G.R.; Schreier, L.L.; Johnson, L.A. In vitro production of sexed embryos for gender preselection: High-speed sorting of X-chromosome-bearing sperm to produce pigs after embryo transfer. J. Anim. Sci. 1999, 77, 3346–3352. [Google Scholar] [CrossRef] [Green Version]
- Navarro-Serna, S.; París-Oller, E.; Simonik, O.; Romar, R.; Gadea, J. Replacement of Albumin by Preovulatory Oviductal Fluid in Swim-Up Sperm Preparation Method Modifies Boar Sperm Parameters and Improves In Vitro Penetration of Oocytes. Animals 2021, 11, 1202. [Google Scholar] [CrossRef]
- Limsirichaikul, S.; Niimi, A.; Fawcett, H.; Lehmann, A.; Yamashita, S.; Ogi, T. A rapid non-radioactive technique for measurement of repair synthesis in primary human fibroblasts by incorporation of ethynyl deoxyuridine (EdU). Nucleic Acids Res. 2009, 37, e31. [Google Scholar] [CrossRef]
- Ramlee, M.K.; Yan, T.; Cheung, A.M.S.; Chuah, C.T.H.; Li, S. High-throughput genotyping of CRISPR/Cas9-mediated mutants using fluorescent PCR-capillary gel electrophoresis. Sci. Rep. 2015, 5, 15587. [Google Scholar] [CrossRef]






| Aphidicolin | [Cas9/sgRNA] | Aphidicolin × [Cas9/sgRNA] | |
|---|---|---|---|
| Cleavage rate 1 | <0.01 | 0.23 | 0.41 |
| Blastocyst rate 2 | <0.01 | 0.75 | 0.93 |
| Mutation rate 3 | 0.09 | <0.01 | 0.52 |
| Mosaicism/total 4 | 0.06 | 0.15 | 0.39 |
| Number of alleles 5 | 0.13 | 0.89 | 098 |
| Mosaicism/mutant 6 | 0.18 | 0.68 | 0.78 |
| Biallelic KO/total 7 | 0.81 | 0.53 | 0.91 |
| E1 | EAp1 | E2 | EAp2 | p Value | |
|---|---|---|---|---|---|
| [Cas9/sgRNA] (ng/µL) | 12.5/6.75 | 12.5/6.75 | 25/12.5 | 25/12.5 | |
| 0.5 μM aphidicolin | - | + | - | + | |
| Mutation rate 1 | 48.78 a (20/41) | 40.63 a (13/32) | 76.47 b (39/51) | 58.06 ab (18/31) | 0.01 |
| Mosaicism/total 2 | 12.20 ab (5/41) | 6.25 a (2/32) | 25.49 b (13/51) | 9.68 ab (3/31) | 0.06 |
| Number of alleles 3 | 2.32 | 2.15 | 2.33 | 2.17 | 0.46 |
| Mosaicism/mutant 4 | 26.32 (5/19) | 15.38 (2/13) | 33.33 (13/39) | 16.67 (3/18) | 0.45 |
| Biallelic KO/total 5 | 5.26 (1/19) | 7.69 (1/13) | 10.26 (4/39) | 11.11 (2/18) | 0.91 |
| Aphidicolin | Method | Aphidicolin × Method | |
|---|---|---|---|
| Cleavage rate 1 | 0.39 | <0.01 | 0.83 |
| Blastocyst rate 2 | 0.03 | <0.01 | 0.71 |
| Mutation rate 3 | 0.37 | 0.53 | 0.41 |
| Mosaicism/total 4 | 0.03 | 0.82 | 0.91 |
| Number of alleles 5 | 0.40 | 0.63 | 0.56 |
| Mosaicism/mutant 6 | 0.05 | 0.85 | 0.68 |
| Biallelic KO/total 7 | 0.11 | 0.31 | 0.69 |
| E | EAp | M | MAp | p Value | |
|---|---|---|---|---|---|
| Method | Electroporation | Microinjection | |||
| 0.5 μM aphidicolin | - | + | - | + | |
| Mutation rate 1 | 67.44 ± 7.23 (29/43) | 52.50 ± 8.00 (21/40) | 54.84 ± 9.09 (17/31) | 54.17 ± 10.3 (13/24) | 0.508 |
| Mosaicism/total 2 | 34.88 ± 7.35 (15/43) | 17.50 ± 6.08 (7/40) | 32.26 ± 8.53 (10/31) | 16.67 ± 7.77 (4/24) | 0.177 |
| Number of alleles 3 | 2.47 ± 0.11 | 2.28 ± 0.10 | 2.45 ± 0.12 | 2.42 ± 0.21 | 0.317 |
| Mosaicism/mutant 4 | 51.72 ± 9.44 (15/29) | 33.33 ± 10.54 (7/21) | 58.82 ± 12.30 (10/17) | 30.77 ± 0.31 (4/13) | 0.260 |
| Biallelic KO/total 5 | 11.63 ± 4.94 (5/43) | 5.00 ± 3.49 (2/40) | 19.35 ± 7.21 (6/31) | 8.33 ± 5.76 (2/24) | 0.274 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Navarro-Serna, S.; Piñeiro-Silva, C.; Luongo, C.; Parrington, J.; Romar, R.; Gadea, J. Effect of Aphidicolin, a Reversible Inhibitor of Eukaryotic Nuclear DNA Replication, on the Production of Genetically Modified Porcine Embryos by CRISPR/Cas9. Int. J. Mol. Sci. 2022, 23, 2135. https://doi.org/10.3390/ijms23042135
Navarro-Serna S, Piñeiro-Silva C, Luongo C, Parrington J, Romar R, Gadea J. Effect of Aphidicolin, a Reversible Inhibitor of Eukaryotic Nuclear DNA Replication, on the Production of Genetically Modified Porcine Embryos by CRISPR/Cas9. International Journal of Molecular Sciences. 2022; 23(4):2135. https://doi.org/10.3390/ijms23042135
Chicago/Turabian StyleNavarro-Serna, Sergio, Celia Piñeiro-Silva, Chiara Luongo, John Parrington, Raquel Romar, and Joaquín Gadea. 2022. "Effect of Aphidicolin, a Reversible Inhibitor of Eukaryotic Nuclear DNA Replication, on the Production of Genetically Modified Porcine Embryos by CRISPR/Cas9" International Journal of Molecular Sciences 23, no. 4: 2135. https://doi.org/10.3390/ijms23042135
APA StyleNavarro-Serna, S., Piñeiro-Silva, C., Luongo, C., Parrington, J., Romar, R., & Gadea, J. (2022). Effect of Aphidicolin, a Reversible Inhibitor of Eukaryotic Nuclear DNA Replication, on the Production of Genetically Modified Porcine Embryos by CRISPR/Cas9. International Journal of Molecular Sciences, 23(4), 2135. https://doi.org/10.3390/ijms23042135








