Progress in Methods for Copy Number Variation Profiling
Abstract
:1. Introduction
2. Methods of Cytogenetics
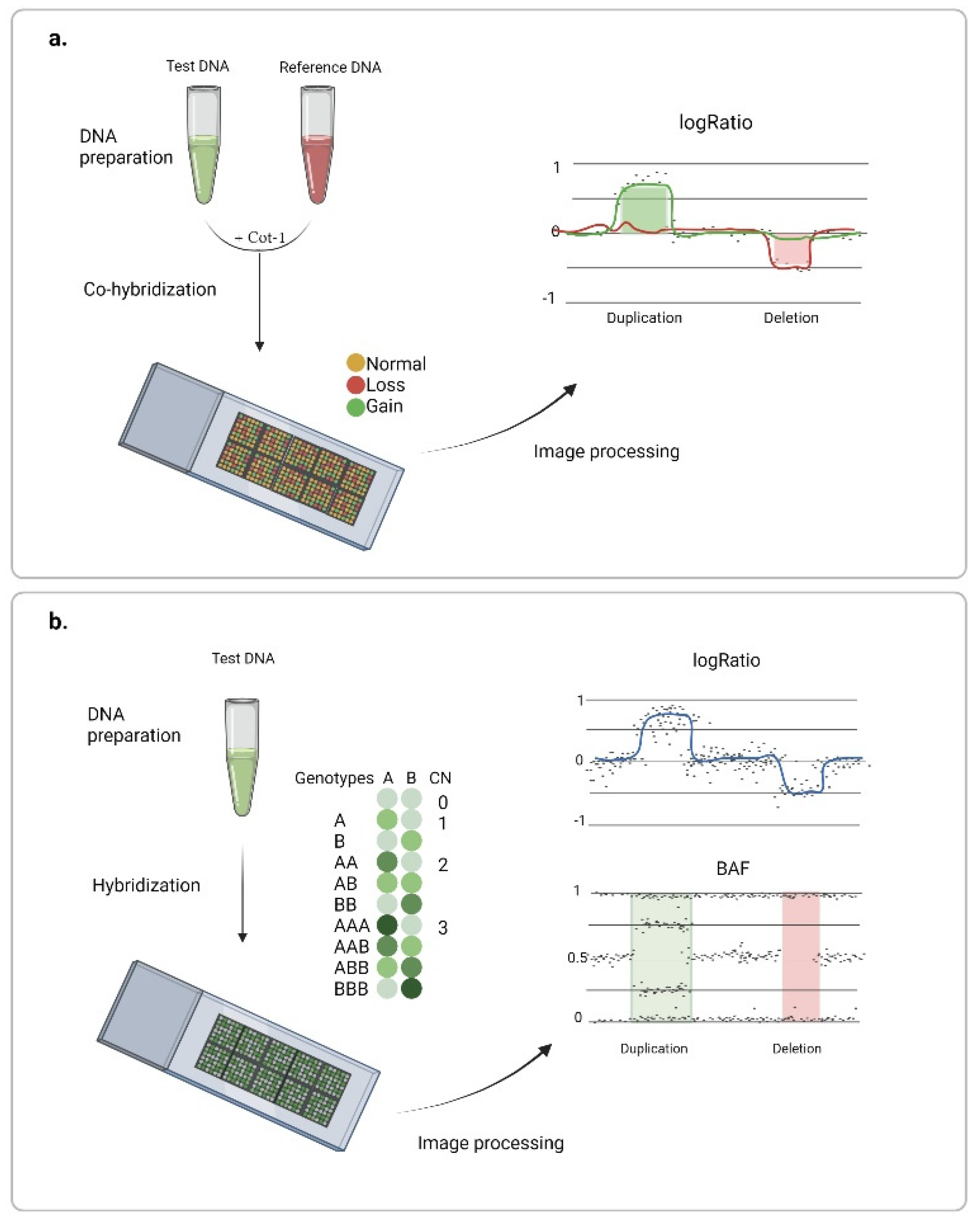
3. Chromosome Microarray Analysis (CMA)
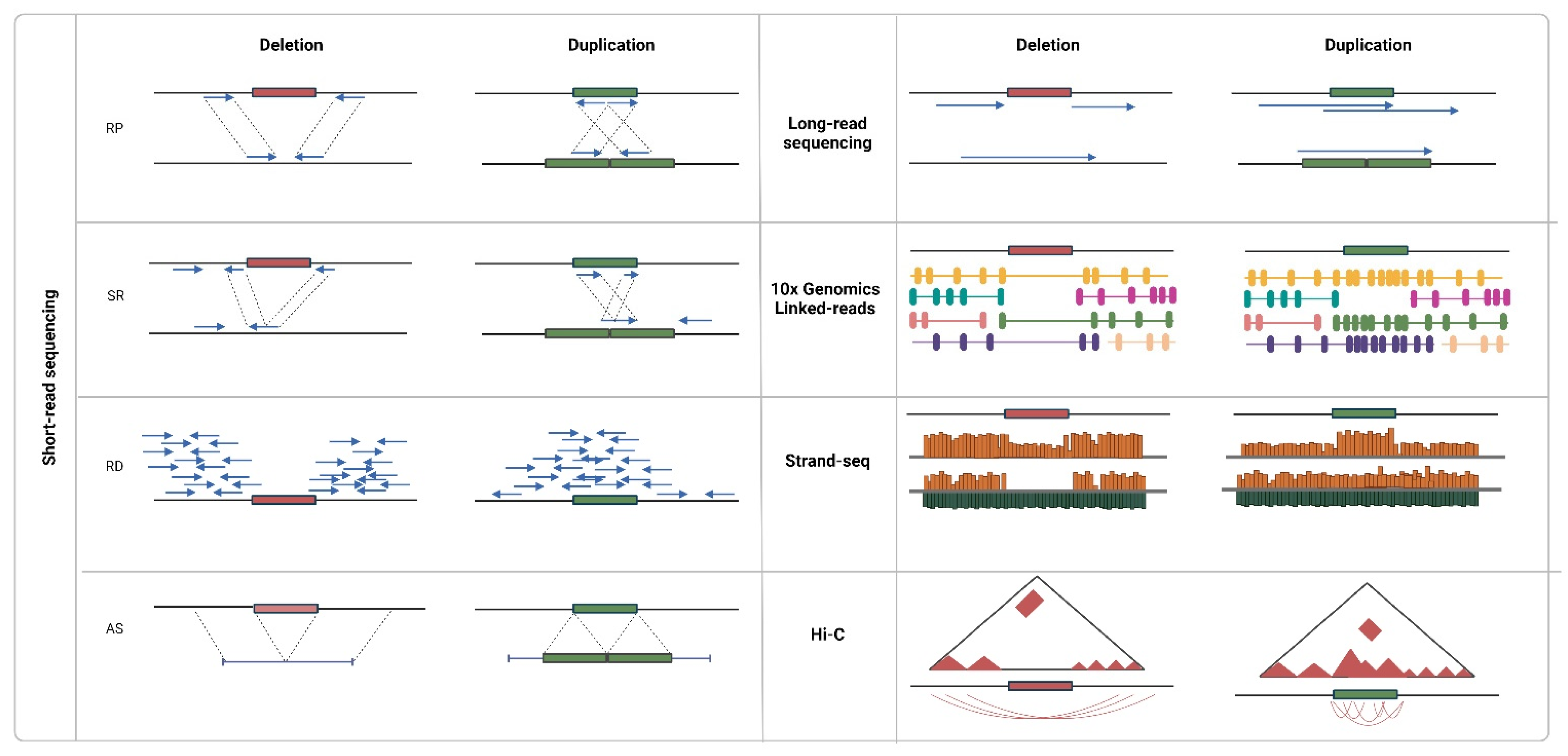
4. Sequencing Data
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Hollox, E.J.; Zuccherato, L.W.; Tucci, S. Genome Structural Variation in Human Evolution. Trends Genet. 2022, 38, 45–58. [Google Scholar] [CrossRef] [PubMed]
- Iafrate, A.J.; Feuk, L.; Rivera, M.N.; Listewnik, M.L.; Donahoe, P.K.; Qi, Y.; Scherer, S.W.; Lee, C. Detection of Large-Scale Variation in the Human Genome. Nat. Genet. 2004, 36, 949–951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Redon, R.; Ishikawa, S.; Fitch, K.R.; Feuk, L.; Perry, G.H.; Andrews, T.D.; Fiegler, H.; Shapero, M.H.; Carson, A.R.; Chen, W.; et al. Global Variation in Copy Number in the Human Genome. Nature 2006, 444, 444–454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zarrei, M.; MacDonald, J.R.; Merico, D.; Scherer, S.W. A Copy Number Variation Map of the Human Genome. Nat. Rev. Genet. 2015, 16, 172–183. [Google Scholar] [CrossRef] [PubMed]
- Shaikh, T.H. Copy Number Variation Disorders. Curr. Genet. Med. Rep. 2017, 5, 183. [Google Scholar] [CrossRef]
- Roca, I.; González-Castro, L.; Fernández, H.; Couce, M.L.; Fernández-Marmiesse, A. Free-Access Copy-Number Variant Detection Tools for Targeted next-Generation Sequencing Data. Mutat. Res./Rev. Mutat. Res. 2019, 779, 114–125. [Google Scholar] [CrossRef]
- Seiser, E.L.; Innocenti, F. Hidden Markov Model-Based CNV Detection Algorithms for Illumina Genotyping Microarrays. Cancer Inform. 2015, 13, CIN-S16345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carter, N.P. Methods and Strategies for Analyzing Copy Number Variation Using DNA Microarrays. Nat. Genet. 2007, 39. [Google Scholar] [CrossRef] [Green Version]
- Mahmoud, M.; Gobet, N.; Cruz-Dávalos, D.I.; Mounier, N.; Dessimoz, C.; Sedlazeck, F.J. Structural Variant Calling: The Long and the Short of It. Genome Biol. 2019, 20, 1–14. [Google Scholar] [CrossRef]
- Salgado, D.; Armean, I.M.; Baudis, M.; Beltran, S.; Capella-Gutierrez, S.; Carvalho-Silva, D.; Del Angel, V.D.; Dopazo, J.; Furlong, L.I.; Gao, B.; et al. The ELIXIR Human Copy Number Variations Community: Building Bioinformatics Infrastructure for Research. F1000Research 2020, 9, 1229. [Google Scholar] [CrossRef]
- Roizen, N.J.; Patterson, D. Down’s Syndrome. Lancet 2003, 361, 1281–1289. [Google Scholar] [CrossRef]
- Lanfranco, F.; Kamischke, A.; Zitzmann, M.; Nieschlag, E. Klinefelter’s Syndrome. Lancet 2004, 364, 273–283. [Google Scholar] [CrossRef]
- Cereda, A.; Carey, J.C. The Trisomy 18 Syndrome. Orphanet J. Rare Dis. 2012, 7, 81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nowell, P.C. The Minute Chromosome (Phl) in Chronic Granulocytic Leukemia. Blut 1962, 8, 65–66. [Google Scholar] [CrossRef]
- Bayani, J.; Squire, J.A. Traditional Banding of Chromosomes for Cytogenetic Analysis. Curr. Protoc. Cell Biol. 2004, 22, 22.3.1–22.3.7. [Google Scholar] [CrossRef]
- Swansbury, J. Introduction to the Analysis of the Human G-Banded Karyotype. Methods Mol. Biol. 2003, 220, 259–269. [Google Scholar] [PubMed]
- Gall, J.G.; Pardue, M.L. Formation and Detection of RNA-DNA Hybrid Molecules in Cytological Preparations. Proc. Natl. Acad. Sci. USA 1969, 63, 378–383. [Google Scholar] [CrossRef] [Green Version]
- Rudkin, G.T.; Stollar, B.D. High Resolution Detection of DNA-RNA Hybrids in Situ by Indirect Immunofluorescence. Nature 1977, 265, 472–473. [Google Scholar] [CrossRef]
- Bauman, J.G.; Wiegant, J.; Borst, P.; van Duijn, P. A New Method for Fluorescence Microscopical Localization of Specific DNA Sequences by in Situ Hybridization of Fluorochromelabelled RNA. Exp. Cell Res. 1980, 128. [Google Scholar] [CrossRef]
- Schrock, E.; du Manoir, S.; Veldman, T.; Schoell, B.; Wienberg, J.; Ferguson-Smith, M.A.; Ning, Y.; Ledbetter, D.H.; Bar-Am, I.; Soenksen, D.; et al. Multicolor Spectral Karyotyping of Human Chromosomes. Science 1996, 273, 494–497. [Google Scholar] [CrossRef] [PubMed]
- Speicher, M.R.; Ballard, S.G.; Ward, D.C. Karyotyping Human Chromosomes by Combinatorial Multi-Fluor FISH. Nat. Genet. 1996, 12, 368–375. [Google Scholar] [CrossRef]
- Kallioniemi, A.; Kallioniemi, O.P.; Sudar, D.; Rutovitz, D.; Gray, J.W.; Waldman, F.; Pinkel, D. Comparative Genomic Hybridization for Molecular Cytogenetic Analysis of Solid Tumors. Science 1992, 258, 818–821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kallioniemi, O.P.; Kallioniemi, A.; Piper, J.; Isola, J.; Waldman, F.M.; Gray, J.W.; Pinkel, D. Optimizing Comparative Genomic Hybridization for Analysis of DNA Sequence Copy Number Changes in Solid Tumors. Genes Chromosomes Cancer 1994, 10, 231–243. [Google Scholar] [CrossRef]
- Gebhart, E. Comparative Genomic Hybridization (CGH): Ten Years of Substantial Progress in Human Solid Tumor Molecular Cytogenetics. Cytogenet. Genome Res. 2004, 104, 352–358. [Google Scholar] [CrossRef] [PubMed]
- Solinas-Toldo, S.; Lampel, S.; Stilgenbauer, S.; Nickolenko, J.; Benner, A.; Döhner, H.; Cremer, T.; Lichter, P. Matrix-Based Comparative Genomic Hybridization: Biochips to Screen for Genomic Imbalances. Genes Chromosomes Cancer 1997, 20, 399–407. [Google Scholar] [CrossRef]
- Pinkel, D.; Segraves, R.; Sudar, D.; Clark, S.; Poole, I.; Kowbel, D.; Collins, C.; Kuo, W.-L.; Chen, C.; Zhai, Y.; et al. High Resolution Analysis of DNA Copy Number Variation Using Comparative Genomic Hybridization to Microarrays. Nat. Genet. 1998, 20, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Osoegawa, K.; Mammoser, A.G.; Wu, C.; Frengen, E.; Zeng, C.; Catanese, J.J.; de Jong, P.J. A Bacterial Artificial Chromosome Library for Sequencing the Complete Human Genome. Genome Res. 2001, 11, 483. [Google Scholar] [CrossRef] [Green Version]
- Cowell, J.K.; Nowak, N.J. High-Resolution Analysis of Genetic Events in Cancer Cells Using Bacterial Artificial Chromosome Arrays and Comparative Genome Hybridization. Adv. Cancer Res. 2003, 90, 91–125. [Google Scholar] [CrossRef]
- Malan, V.; Chevallier, S.; Soler, G.; Coubes, C.; Lacombe, D.; Pasquier, L.; Soulier, J.; Morichon-Delvallez, N.; Turleau, C.; Munnich, A.; et al. Array-Based Comparative Genomic Hybridization Identifies a High Frequency of Copy Number Variations in Patients with Syndromic Overgrowth. Eur. J. Hum. Genet. 2009, 18, 227–232. [Google Scholar] [CrossRef]
- Pollack, J.R.; Perou, C.M.; Alizadeh, A.A.; Eisen, M.B.; Pergamenschikov, A.; Williams, C.F.; Jeffrey, S.S.; Botstein, D.; Brown, P.O. Genome-Wide Analysis of DNA Copy-Number Changes Using cDNA Microarrays. Nat. Genet. 1999, 23, 41–46. [Google Scholar] [CrossRef] [Green Version]
- Bashyam; Bair, R.; Kim, Y.H.; Wang, P.; Hernandez-Boussard, T.; Karikari, C.A.; Tibshirani, R.; Maitra, A.; Pollack, J.R. Array-Based Comparative Genomic Hybridization Identifies Localized DNA Amplifications and Homozygous Deletions in Pancreatic Cancer. Neoplasia 2005, 7, 556-IN16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhami, P.; Coffey, A.J.; Abbs, S.; Vermeesch, J.R.; Dumanski, J.P.; Woodward, K.J.; Andrews, R.M.; Langford, C.; Vetrie, D. Exon Array CGH: Detection of Copy-Number Changes at the Resolution of Individual Exons in the Human Genome. Am. J. Hum. Genet. 2005, 76, 750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schena, M.; Shalon, D.; Heller, R.; Chai, A.; Brown, P.O.; Davis, R.W. Parallel Human Genome Analysis: Microarray-Based Expression Monitoring of 1000 Genes. Proc. Natl. Acad. Sci. USA 1996, 93, 10614–10619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeRisi, J.; Penland, L.; Brown, P.O.; Bittner, M.L.; Meltzer, P.S.; Ray, M.; Chen, Y.; Su, Y.A.; Trent, J.M. Use of a cDNA Microarray to Analyse Gene Expression Patterns in Human Cancer. Nat. Genet. 1996, 14, 457–460. [Google Scholar] [CrossRef] [PubMed]
- Lucito, R.; Healy, J.; Alexander, J.; Reiner, A.; Esposito, D.; Chi, M.; Rodgers, L.; Brady, A.; Sebat, J.; Troge, J.; et al. Representational Oligonucleotide Microarray Analysis: A High-Resolution Method to Detect Genome Copy Number Variation. Genome Res. 2003, 13, 2291–2305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrett, M.T.; Scheffer, A.; Ben-Dor, A.; Sampas, N.; Lipson, D.; Kincaid, R.; Tsang, P.; Curry, B.; Baird, K.; Meltzer, P.S.; et al. Comparative Genomic Hybridization Using Oligonucleotide Microarrays and Total Genomic DNA. Proc. Natl. Acad. Sci. USA 2004, 101, 17765–17770. [Google Scholar] [CrossRef] [Green Version]
- Bignell, G.R.; Huang, J.; Greshock, J.; Watt, S.; Butler, A.; West, S.; Grigorova, M.; Jones, K.W.; Wei, W.; Stratton, M.R.; et al. High-Resolution Analysis of DNA Copy Number Using Oligonucleotide Microarrays. Genome Res. 2004, 14, 287–295. [Google Scholar] [CrossRef] [Green Version]
- Peiffer, D.A.; Le, J.M.; Steemers, F.J.; Chang, W.; Jenniges, T.; Garcia, F.; Haden, K.; Li, J.; Shaw, C.A.; Belmont, J.; et al. High-Resolution Genomic Profiling of Chromosomal Aberrations Using Infinium Whole-Genome Genotyping. Genome Res. 2006, 16, 1136–1148. [Google Scholar] [CrossRef] [Green Version]
- Shen, F.; Huang, J.; Fitch, K.R.; Truong, V.B.; Kirby, A.; Chen, W.; Zhang, J.; Liu, G.; McCarroll, S.A.; Jones, K.W.; et al. Improved Detection of Global Copy Number Variation Using High Density, Non-Polymorphic Oligonucleotide Probes. BMC Genet. 2008, 9, 27. [Google Scholar] [CrossRef] [Green Version]
- Miller, D.T.; Adam, M.P.; Aradhya, S.; Biesecker, L.G.; Brothman, A.R.; Carter, N.P.; Church, D.M.; Crolla, J.A.; Eichler, E.E.; Epstein, C.J.; et al. Consensus Statement: Chromosomal Microarray Is a First-Tier Clinical Diagnostic Test for Individuals with Developmental Disabilities or Congenital Anomalies. Am. J. Hum. Genet. 2010, 86, 749. [Google Scholar] [CrossRef] [PubMed]
- Haraksingh, R.R.; Abyzov, A.; Urban, A.E. Comprehensive Performance Comparison of High-Resolution Array Platforms for Genome-Wide Copy Number Variation (CNV) Analysis in Humans. BMC Genom. 2017, 18, 321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinto, D.; Darvishi, K.; Shi, X.; Rajan, D.; Rigler, D.; Fitzgerald, T.; Lionel, A.C.; Thiruvahindrapuram, B.; MacDonald, J.R.; Mills, R.; et al. Comprehensive Assessment of Array-Based Platforms and Calling Algorithms for Detection of Copy Number Variants. Nat. Biotechnol. 2011, 29, 512–520. [Google Scholar] [CrossRef] [Green Version]
- Veltman, J.A.; Fridlyand, J.; Pejavar, S.; Olshen, A.B.; Korkola, J.E.; DeVries, S.; Carroll, P.; Kuo, W.-L.; Pinkel, D.; Albertson, D.; et al. Array-Based Comparative Genomic Hybridization for Genome-Wide Screening of DNA Copy Number in Bladder Tumors. Cancer Res. 2003, 63, 2872–2880. [Google Scholar] [PubMed]
- Array-Based Comparative Genomic Hybridization for the Genomewide Detection of Submicroscopic Chromosomal Abnormalities. Am. J. Hum. Genet. 2003, 73, 1261–1270. [CrossRef] [PubMed] [Green Version]
- Comparative Genomic Hybridization–Array Analysis Enhances the Detection of Aneuploidies and Submicroscopic Imbalances in Spontaneous Miscarriages. Am. J. Hum. Genet. 2004, 74, 1168–1174. [CrossRef] [PubMed] [Green Version]
- Shaw-Smith, C.; Redon, R.; Rickman, L.; Rio, M.; Willatt, L.; Fiegler, H.; Firth, H.; Sanlaville, D.; Winter, R.; Colleaux, L.; et al. Microarray Based Comparative Genomic Hybridisation (array-CGH) Detects Submicroscopic Chromosomal Deletions and Duplications in Patients with Learning Disability/mental Retardation and Dysmorphic Features. J. Med. Genet. 2004, 41, 241–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwaenen, C.; Nessling, M.; Wessendorf, S.; Salvi, T.; Wrobel, G.; Radlwimmer, B.; Kestler, H.A.; Haslinger, C.; Stilgenbauer, S.; Döhner, H.; et al. Automated Array-Based Genomic Profiling in Chronic Lymphocytic Leukemia: Development of a Clinical Tool and Discovery of Recurrent Genomic Alterations. Proc. Natl. Acad. Sci. USA 2004, 101, 1039–1044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- :Olshen, A.B.; Venkatraman, E.S.; Lucito, R.; Wigler, M. Circular Binary Segmentation for the Analysis of Array-Based DNA Copy Number Data. Biostatistics 2004, 5, 557–572. [Google Scholar] [CrossRef] [PubMed]
- Jong, K.; Marchiori, E.; van der Vaart, A.; Ylstra, B.; Weiss, M.; Meijer, G. Chromosomal Breakpoint Detection in Human Cancer. In Proceedings of the Applications of Evolutionary Computing, Essex, UK, 14–16 April 2003; Springer: Berlin/Heidelberg, Germany, 2003; pp. 54–65. [Google Scholar]
- Hupé, P.; Stransky, N.; Thiery, J.P.; Radvanyi, F.; Barillot, E. Analysis of Array CGH Data: From Signal Ratio to Gain and Loss of DNA Regions. Bioinformatics 2004, 20, 3413–3422. [Google Scholar] [CrossRef] [PubMed]
- Tibshirani, R.; Wang, P. Spatial Smoothing and Hot Spot Detection for CGH Data Using the Fused Lasso. Biostatistics 2008, 9, 18–29. [Google Scholar] [CrossRef]
- Jeng, X.J.; Cai, T.T.; Li, H. Optimal Sparse Segment Identification with Application in Copy Number Variation Analysis. J. Am. Stat. Assoc. 2010, 105, 1156–1166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, Y.S.; Zhang, H. The screening and ranking algorithm to detect dna copy number variations. Ann. Appl. Stat. 2012, 6, 1306–1326. [Google Scholar] [CrossRef] [Green Version]
- de Vries, B.B.A.; Pfundt, R.; Leisink, M.; Koolen, D.A.; Vissers, L.E.L.; Janssen, I.M.; van Reijmersdal, S.; Nillesen, W.M.; Huys, E.H.L.P.; de Leeuw, N.; et al. Diagnostic Genome Profiling in Mental Retardation. Am. J. Hum. Genet. 2005, 77, 606. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Li, C.; Guillermo Paez, J.; Chin, K.; Jänne, P.A.; Chen, T.-H.; Girard, L.; Minna, J.; Christiani, D.; Leo, C.; et al. An Integrated View of Copy Number and Allelic Alterations in the Cancer Genome Using Single Nucleotide Polymorphism Arrays. Cancer Res. 2004, 64, 3060–3071. [Google Scholar] [CrossRef] [Green Version]
- Picard, F.; Robin, S.; Lavielle, M.; Vaisse, C.; Daudin, J.-J. A Statistical Approach for Array CGH Data Analysis. BMC Bioinform. 2005, 6, 27. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Li, M.; Hadley, D.; Liu, R.; Glessner, J.; Grant, S.F.A.; Hakonarson, H.; Bucan, M. PennCNV: An Integrated Hidden Markov Model Designed for High-Resolution Copy Number Variation Detection in Whole-Genome SNP Genotyping Data. Genome Res. 2007, 17, 1665–1674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dellinger, A.E.; Saw, S.M.; Goh, L.K.; Seielstad, M.; Young, T.L.; Li, Y.J. Comparative Analyses of Seven Algorithms for Copy Number Variant Identification from Single Nucleotide Polymorphism Arrays. Nucleic Acids Res. 2010, 38, e105. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Motsinger, R.A. Evaluation of Calling Algorithms for Array-CGH. Front. Genet. 2013, 4, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winchester, L.; Yau, C.; Ragoussis, J. Comparing CNV Detection Methods for SNP Arrays. Brief. Funct. Genom. 2009, 8, 353–366. [Google Scholar] [CrossRef] [Green Version]
- Korn, J.M.; Kuruvilla, F.G.; McCarroll, S.A.; Wysoker, A.; Nemesh, J.; Cawley, S.; Hubbell, E.; Veitch, J.; Collins, P.J.; Darvishi, K.; et al. Integrated Genotype Calling and Association Analysis of SNPs, Common Copy Number Polymorphisms and Rare CNVs. Nat. Genet. 2008, 40, 1253–1260. [Google Scholar] [CrossRef] [Green Version]
- Sun, W.; Wright, F.A.; Tang, Z.; Nordgard, S.H.; Van Loo, P.; Yu, T.; Kristensen, V.N.; Perou, C.M. Integrated Study of Copy Number States and Genotype Calls Using High-Density SNP Arrays. Nucleic Acids Res. 2009, 37, 5365–5377. [Google Scholar] [CrossRef] [PubMed]
- Darvishi, K. Application of Nexus Copy Number Software for CNV Detection and Analysis. Curr. Protoc. Hum. Genet. 2010, 65, 4–14. [Google Scholar] [CrossRef]
- Colella, S.; Yau, C.; Taylor, J.M.; Mirza, G.; Butler, H.; Clouston, P.; Bassett, A.S.; Seller, A.; Holmes, C.C.; Ragoussis, J. QuantiSNP: An Objective Bayes Hidden-Markov Model to Detect and Accurately Map Copy Number Variation Using SNP Genotyping Data. Nucleic Acids Res. 2007, 35, 2013–2025. [Google Scholar] [CrossRef] [Green Version]
- Zhao, M.; Wang, Q.; Wang, Q.; Jia, P.; Zhao, Z. Computational Tools for Copy Number Variation (CNV) Detection Using next-Generation Sequencing Data: Features and Perspectives. BMC Bioinform. 2013, 14, S1. [Google Scholar] [CrossRef]
- Korbel, J.O.; Urban, A.E.; Affourtit, J.P.; Godwin, B.; Grubert, F.; Simons, J.F.; Kim, P.M.; Palejev, D.; Carriero, N.J.; Du, L.; et al. Paired-End Mapping Reveals Extensive Structural Variation in the Human Genome. Science 2007, 318, 420–426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, K.; Wallis, J.W.; McLellan, M.D.; Larson, D.E.; Kalicki, J.M.; Pohl, C.S.; McGrath, S.D.; Wendl, M.C.; Zhang, Q.; Locke, D.P.; et al. BreakDancer: An Algorithm for High-Resolution Mapping of Genomic Structural Variation. Nat. Methods 2009, 6, 677–681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korbel, J.O.; Abyzov, A.; Mu, X.J.; Carriero, N.; Cayting, P.; Zhang, Z.; Snyder, M.; Gerstein, M.B. PEMer: A Computational Framework with Simulation-Based Error Models for Inferring Genomic Structural Variants from Massive Paired-End Sequencing Data. Genome Biol. 2009, 10, R23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.; Hormozdiari, F.; Alkan, C.; Brudno, M. MoDIL: Detecting Small Indels from Clone-End Sequencing with Mixtures of Distributions. Nat. Methods 2009, 6, 473–474. [Google Scholar] [CrossRef] [PubMed]
- Hayes, M.; Pyon, Y.S.; Li, J. A Model-Based Clustering Method for Genomic Structural Variant Prediction and Genotyping Using Paired-End Sequencing Data. PLoS ONE 2012, 7, e52881. [Google Scholar] [CrossRef] [Green Version]
- Marschall, T.; Costa, I.G.; Canzar, S.; Bauer, M.; Klau, G.W.; Schliep, A.; Schönhuth, A. CLEVER: Clique-Enumerating Variant Finder. Bioinformatics 2012, 28, 2875–2882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trappe, K.; Emde, A.-K.; Ehrlich, H.-C.; Reinert, K. Gustaf: Detecting and Correctly Classifying SVs in the NGS Twilight Zone. Bioinformatics 2014, 30, 3484–3490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, K.; Schulz, M.H.; Long, Q.; Apweiler, R.; Ning, Z. Pindel: A Pattern Growth Approach to Detect Break Points of Large Deletions and Medium Sized Insertions from Paired-End Short Reads. Bioinformatics 2009, 25, 2865–2871. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; Xuan, Z.; Makarov, V.; Ye, K.; Sebat, J. Sensitive and Accurate Detection of Copy Number Variants Using Read Depth of Coverage. Genome Res. 2009, 19, 1586–1592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, C.; Tammi, M.T. CNV-Seq, a New Method to Detect Copy Number Variation Using High-Throughput Sequencing. BMC Bioinform. 2009, 10, 80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gusnanto, A.; Taylor, C.C.; Nafisah, I.; Wood, H.M.; Rabbitts, P.; Berri, S. Estimating Optimal Window Size for Analysis of Low-Coverage next-Generation Sequence Data. Bioinformatics 2014, 30, 1823–1829. [Google Scholar] [CrossRef] [Green Version]
- Benjamini, Y.; Speed, T.P. Summarizing and Correcting the GC Content Bias in High-Throughput Sequencing. Nucleic Acids Res. 2012, 40, e72. [Google Scholar] [CrossRef] [Green Version]
- Talevich, E.; Hunter Shain, A.; Botton, T.; Bastian, B.C. CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing. PLoS Comput. Biol. 2016, 12, e1004873. [Google Scholar] [CrossRef]
- Abyzov, A.; Urban, A.E.; Snyder, M.; Gerstein, M. CNVnator: An Approach to Discover, Genotype, and Characterize Typical and Atypical CNVs from Family and Population Genome Sequencing. Genome Res. 2011, 21, 974–984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dharanipragada, P.; Vogeti, S.; Parekh, N. iCopyDAV: Integrated Platform for Copy Number variations—Detection, Annotation and Visualization. PLoS ONE 2018, 13, e0195334. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Wang, W.; Sun, W.; Crowley, J.J.; Szatkiewicz, J.P. Allele-Specific Copy-Number Discovery from Whole-Genome and Whole-Exome Sequencing. Nucleic Acids Res. 2015, 43, e90. [Google Scholar] [CrossRef] [Green Version]
- Xi, R.; Lee, S.; Xia, Y.; Kim, T.-M.; Park, P.J. Copy Number Analysis of Whole-Genome Data Using BIC-seq2 and Its Application to Detection of Cancer Susceptibility Variants. Nucleic Acids Res. 2016, 44, 6274–6286. [Google Scholar] [CrossRef]
- Boeva, V.; Zinovyev, A.; Bleakley, K.; Vert, J.P.; Janoueix-Lerosey, I.; Delattre, O.; Barillot, E. Control-Free Calling of Copy Number Alterations in Deep-Sequencing Data Using GC-Content Normalization. Bioinformatics 2011, 27, 268–269. [Google Scholar] [CrossRef] [Green Version]
- Miller, C.A.; Hampton, O.; Coarfa, C.; Milosavljevic, A. ReadDepth: A Parallel R Package for Detecting Copy Number Alterations from Short Sequencing Reads. PLoS ONE 2011, 6, e16327. [Google Scholar] [CrossRef] [Green Version]
- Gordeeva, V.; Sharova, E.; Babalyan, K.; Sultanov, R.; Govorun, V.M.; Arapidi, G. Benchmarking Germline CNV Calling Tools from Exome Sequencing Data. Sci. Rep. 2021, 11, 14416. [Google Scholar] [CrossRef]
- Fromer, M.; Moran, J.L.; Chambert, K.; Banks, E.; Bergen, S.E.; Ruderfer, D.M.; Handsaker, R.E.; McCarroll, S.A.; O’Donovan, M.C.; Owen, M.J.; et al. Discovery and Statistical Genotyping of Copy-Number Variation from Whole-Exome Sequencing Depth. Am. J. Hum. Genet. 2012, 91, 597–607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.; Oldridge, D.A.; Diskin, S.J.; Zhang, N.R. CODEX: A Normalization and Copy Number Variation Detection Method for Whole Exome Sequencing. Nucleic Acids Res. 2015, 43, e39. [Google Scholar] [CrossRef] [Green Version]
- Plagnol, V.; Curtis, J.; Epstein, M.; Mok, K.Y.; Stebbings, E.; Grigoriadou, S.; Wood, N.W.; Hambleton, S.; Burns, S.O.; Thrasher, A.J.; et al. A Robust Model for Read Count Data in Exome Sequencing Experiments and Implications for Copy Number Variant Calling. Bioinformatics 2012, 28, 2747–2754. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Myšičková, A.; Sun, R.; Kalscheuer, V.; Vingron, M.; Haas, S.A. Modeling Read Counts for CNV Detection in Exome Sequencing Data. Stat. Appl. Genet. Mol. Biol. 2011, 10. [Google Scholar] [CrossRef] [Green Version]
- D’Aurizio, R.; Pippucci, T.; Tattini, L.; Giusti, B.; Pellegrini, M.; Magi, A. Enhanced Copy Number Variants Detection from Whole-Exome Sequencing Data Using EXCAVATOR2. Nucleic Acids Res. 2016, 44, e154. [Google Scholar] [CrossRef] [PubMed]
- Kuśmirek, W.; Szmurło, A.; Wiewiórka, M.; Nowak, R.; Gambin, T. Comparison of kNN and K-Means Optimization Methods of Reference Set Selection for Improved CNV Callers Performance. BMC Bioinform. 2019, 20, 266. [Google Scholar] [CrossRef] [Green Version]
- Klambauer, G.; Schwarzbauer, K.; Mayr, A.; Clevert, D.-A.; Mitterecker, A.; Bodenhofer, U.; Hochreiter, S. cn.MOPS: Mixture of Poissons for Discovering Copy Number Variations in next-Generation Sequencing Data with a Low False Discovery Rate. Nucleic Acids Res. 2012, 40, e69. [Google Scholar] [CrossRef] [PubMed]
- Krumm, N.; Sudmant, P.H.; Ko, A.; O’Roak, B.J.; Malig, M.; Coe, B.P.; Quinlan, A.R.; Nickerson, D.A.; Eichler, E.E. Copy Number Variation Detection and Genotyping from Exome Sequence Data. Genome Res. 2012, 22, 1525–1532. [Google Scholar] [CrossRef] [Green Version]
- Johansson, L.F.; van Dijk, F.; de Boer, E.N.; van Dijk-Bos, K.K.; Jongbloed, J.D.; van der Hout, A.H.; Westers, H.; Sinke, R.J.; Swertz, M.A.; Sijmons, R.H.; et al. CoNVaDING: Single Exon Variation Detection in Targeted NGS Data. Hum. Mutat. 2016, 37, 457–464. [Google Scholar] [CrossRef] [PubMed]
- Fowler, A.; Mahamdallie, S.; Ruark, E.; Seal, S.; Ramsay, E.; Clarke, M.; Uddin, I.; Wylie, H.; Strydom, A.; Lunter, G.; et al. Accurate Clinical Detection of Exon Copy Number Variants in a Targeted NGS Panel Using DECoN. Wellcome Open Res. 2016, 1, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pevzner, P.A.; Tang, H.; Waterman, M.S. An Eulerian Path Approach to DNA Fragment Assembly. Proc. Natl. Acad. Sci. USA 2001, 98, 9748–9753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nijkamp, J.F.; van den Broek, M.A.; Geertman, J.-M.A.; Reinders, M.J.T.; Daran, J.-M.G.; de Ridder, D. De Novo Detection of Copy Number Variation by Co-Assembly. Bioinformatics 2012, 28, 3195–3202. [Google Scholar] [CrossRef] [Green Version]
- Iqbal, Z.; Caccamo, M.; Turner, I.; Flicek, P.; McVean, G. De Novo Assembly and Genotyping of Variants Using Colored de Bruijn Graphs. Nat. Genet. 2012, 44, 226–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cameron, D.L.; Schröder, J.; Penington, J.S.; Do, H.; Molania, R.; Dobrovic, A.; Speed, T.P.; Papenfuss, A.T. GRIDSS: Sensitive and Specific Genomic Rearrangement Detection Using Positional de Bruijn Graph Assembly. Genome Res. 2017, 27, 2050–2060. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Mullighan, C.G.; Easton, J.; Roberts, S.; Heatley, S.L.; Ma, J.; Rusch, M.C.; Chen, K.; Harris, C.C.; Ding, L.; et al. CREST Maps Somatic Structural Variation in Cancer Genomes with Base-Pair Resolution. Nat. Methods 2011, 8, 652–654. [Google Scholar] [CrossRef] [PubMed]
- Using ERDS to Infer Copy-Number Variants in High-Coverage Genomes. Am. J. Hum. Genet. 2012, 91, 408–421. [CrossRef] [PubMed] [Green Version]
- Layer, R.M.; Chiang, C.; Quinlan, A.R.; Hall, I.M. LUMPY: A Probabilistic Framework for Structural Variant Discovery. Genome Biol. 2014, 15, e1004572. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Schulz-Trieglaff, O.; Shaw, R.; Barnes, B.; Schlesinger, F.; Källberg, M.; Cox, A.J.; Kruglyak, S.; Saunders, C.T. Manta: Rapid Detection of Structural Variants and Indels for Germline and Cancer Sequencing Applications. Bioinformatics 2015, 32, 1220–1222. [Google Scholar] [CrossRef] [PubMed]
- Handsaker, R.E.; Korn, J.M.; Nemesh, J.; McCarroll, S.A. Discovery and Genotyping of Genome Structural Polymorphism by Sequencing on a Population Scale. Nat. Genet. 2011, 43, 269–276. [Google Scholar] [CrossRef]
- Rausch, T.; Zichner, T.; Schlattl, A.; Stütz, A.M.; Benes, V.; Korbel, J.O. DELLY: Structural Variant Discovery by Integrated Paired-End and Split-Read Analysis. Bioinformatics 2012, 28, i333–i339. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Clark, R.A.; Sokolova, S.; Leibowitz, M.L.; Zhang, Y.; Hurles, M.E.; Mell, J.C.; Hall, I.M. Genome-Wide Mapping and Assembly of Structural Variant Breakpoints in the Mouse Genome. Genome Res. 2010, 20, 623–635. [Google Scholar] [CrossRef] [Green Version]
- Mohiyuddin, M.; Mu, J.C.; Li, J.; Bani Asadi, N.; Gerstein, M.B.; Abyzov, A.; Wong, W.H.; Lam, H.Y.K. MetaSV: An Accurate and Integrative Structural-Variant Caller for next Generation Sequencing. Bioinformatics 2015, 31, 2741–2744. [Google Scholar] [CrossRef] [Green Version]
- Michaelson, J.J.; Sebat, J. forestSV: Structural Variant Discovery through Statistical Learning. Nat. Methods 2012, 9, 819–821. [Google Scholar] [CrossRef]
- Parikh, H.; Mohiyuddin, M.; Lam, H.Y.K.; Iyer, H.; Chen, D.; Pratt, M.; Bartha, G.; Spies, N.; Losert, W.; Zook, J.M.; et al. Svclassify: A Method to Establish Benchmark Structural Variant Calls. BMC Genom. 2016, 17, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, L.; Wu, Y.; Gao, J. DeepSV: Accurate Calling of Genomic Deletions from High-Throughput Sequencing Data Using Deep Convolutional Neural Network. BMC Bioinform. 2019, 20, 665. [Google Scholar] [CrossRef] [Green Version]
- Mills, R.E.; Walter, K.; Stewart, C.; Handsaker, R.E.; Chen, K.; Alkan, C.; Abyzov, A.; Yoon, S.C.; Ye, K.; Cheetham, R.K.; et al. Mapping Copy Number Variation by Population-Scale Genome Sequencing. Nature 2011, 470, 59–65. [Google Scholar] [CrossRef]
- Sudmant, P.H.; Rausch, T.; Gardner, E.J.; Handsaker, R.E.; Abyzov, A.; Huddleston, J.; Zhang, Y.; Ye, K.; Jun, G.; Fritz, M.H.-Y.; et al. An Integrated Map of Structural Variation in 2504 Human Genomes. Nature 2015, 526, 75–81. [Google Scholar] [CrossRef] [Green Version]
- Collins, R.L.; Brand, H.; Karczewski, K.J.; Zhao, X.; Alföldi, J.; Francioli, L.C.; Khera, A.V.; Lowther, C.; Gauthier, L.D.; Wang, H.; et al. A Structural Variation Reference for Medical and Population Genetics. Nature 2020, 581, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Cretu Stancu, M.; van Roosmalen, M.J.; Renkens, I.; Nieboer, M.M.; Middelkamp, S.; de Ligt, J.; Pregno, G.; Giachino, D.; Mandrile, G.; Espejo Valle-Inclan, J.; et al. Mapping and Phasing of Structural Variation in Patient Genomes Using Nanopore Sequencing. Nat. Commun. 2017, 8, 1326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- English, A.C.; Salerno, W.J.; Reid, J.G. PBHoney: Identifying Genomic Variants via Long-Read Discordance and Interrupted Mapping. BMC Bioinform. 2014, 15, 180. [Google Scholar] [CrossRef] [Green Version]
- Jiang, T.; Liu, S.; Cao, S.; Liu, Y.; Cui, Z.; Wang, Y.; Guo, H. Long-Read Sequencing Settings for Efficient Structural Variation Detection Based on Comprehensive Evaluation. BMC Bioinform. 2021, 22, 552. [Google Scholar] [CrossRef] [PubMed]
- Spies, N.; Weng, Z.; Bishara, A.; McDaniel, J.; Catoe, D.; Zook, J.M.; Salit, M.; West, R.B.; Batzoglou, S.; Sidow, A. Genome-Wide Reconstruction of Complex Structural Variants Using Read Clouds. Nat. Methods 2017, 14, 915–920. [Google Scholar] [CrossRef] [Green Version]
- Elyanow, R.; Wu, H.T.; Raphael, B.J. Identifying Structural Variants Using Linked-Read Sequencing Data. Bioinformatics 2018, 34, 353–360. [Google Scholar] [CrossRef] [Green Version]
- Hills, M.; O’Neill, K.; Falconer, E.; Brinkman, R.; Lansdorp, P.M. BAIT: Organizing Genomes and Mapping Rearrangements in Single Cells. Genome Med. 2013, 5, 82. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Lee, S.; Chu, C.; Jain, D.; Kerpedjiev, P.; Nelson, G.M.; Walsh, J.M.; Alver, B.H.; Park, P.J. HiNT: A Computational Method for Detecting Copy Number Variations and Translocations from Hi-C Data. Genome Biol. 2020, 21, 73. [Google Scholar] [CrossRef] [Green Version]
- Schwartz, D.C.; Li, X.; Hernandez, L.I.; Ramnarain, S.P.; Huff, E.J.; Wang, Y.K. Ordered Restriction Maps of Saccharomyces Cerevisiae Chromosomes Constructed by Optical Mapping. Science 1993, 262, 110–114. [Google Scholar] [CrossRef]
- Latreille, P.; Norton, S.; Goldman, B.S.; Henkhaus, J.; Miller, N.; Barbazuk, B.; Bode, H.B.; Darby, C.; Du, Z.; Forst, S.; et al. Optical Mapping as a Routine Tool for Bacterial Genome Sequence Finishing. BMC Genom. 2007, 8, 321. [Google Scholar] [CrossRef] [Green Version]
- Pendleton, M.; Sebra, R.; Pang, A.W.C.; Ummat, A.; Franzen, O.; Rausch, T.; Stütz, A.M.; Stedman, W.; Anantharaman, T.; Hastie, A.; et al. Assembly and Diploid Architecture of an Individual Human Genome via Single-Molecule Technologies. Nat. Methods 2015, 12, 780–786. [Google Scholar] [CrossRef] [PubMed]
- Optical Genome Mapping Enables Constitutional Chromosomal Aberration Detection. Am. J. Hum. Genet. 2021, 108, 1409–1422. [CrossRef]
- Li, L.; Leung, A.K.-Y.; Kwok, T.-P.; Lai, Y.Y.Y.; Pang, I.K.; Chung, G.T.-Y.; Mak, A.C.Y.; Poon, A.; Chu, C.; Li, M.; et al. OMSV Enables Accurate and Comprehensive Identification of Large Structural Variations from Nanochannel-Based Single-Molecule Optical Maps. Genome Biol. 2017, 18, 230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Cheng, H.; Hong, X.; Di Narzo, A.F.; Franzen, O.; Peng, S.; Ruusalepp, A.; Kovacic, J.C.; Bjorkegren, J.L.M.; Wang, X.; et al. EnsembleCNV: An Ensemble Machine Learning Algorithm to Identify and Genotype Copy Number Variation Using SNP Array Data. Nucleic Acids Res. 2019, 47, e39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pounraja, V.K.; Jayakar, G.; Jensen, M.; Kelkar, N.; Girirajan, S. A Machine-Learning Approach for Accurate Detection of Copy-Number Variants from Exome Sequencing. Genome Res. 2019, 29, 1134–1143. [Google Scholar] [CrossRef] [Green Version]
- Akbarinejad, S.; Hadadian Nejad Yousefi, M.; Goudarzi, M. SVNN: An Efficient PacBio-Specific Pipeline for Structural Variations Calling Using Neural Networks. BMC Bioinform. 2021, 22, 335. [Google Scholar] [CrossRef]
- Kosugi, S.; Momozawa, Y.; Liu, X.; Terao, C.; Kubo, M.; Kamatani, Y. Comprehensive Evaluation of Structural Variation Detection Algorithms for Whole Genome Sequencing. Genome Biol. 2019, 20, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Zook, J.M.; Hansen, N.F.; Olson, N.D.; Chapman, L.; Mullikin, J.C.; Xiao, C.; Sherry, S.; Koren, S.; Phillippy, A.M.; Boutros, P.C.; et al. A Robust Benchmark for Detection of Germline Large Deletions and Insertions. Nat. Biotechnol. 2020, 38, 1347–1355. [Google Scholar] [CrossRef]

| Array Platform | Specification * | Resolution ** | Description |
|---|---|---|---|
| Agilent SurePrint G3 Human CGH | 1 × 1 M | 2.1 kb | enhanced coverage on known genes, promoters, miRNAs, PAR, and telomeric regions |
| 2 × 400 K | 5.3 kb | ||
| 4 × 180 K | 13 kb | ||
| 8 × 60 K | 41 kb | ||
| Agilent Human Genome CGH | 2 × 105 | 35 kb | |
| 4 × 44 K | 43 kb | ||
| Agilent SurePrint G3 Human Genome CGH + SNP | 2 × 400 K | 7.2 Kb | |
| 4 × 180 K | 25.3 kb | ||
| Agilent SurePrint G3 Unrestricted CGH ISCA v2 | 4 × 180 K | 25 kb | enhanced coverage on ISCA (International Standards for Cytogenomic Arrays) regions |
| 8 × 60 K | 60 kb | ||
| 4 × 44 K | 75 kb | ||
| Agilent SurePrint G3 ISCA v2 CGH + SNP | 4 × 180 K | 25.3 kb | |
| Agilent SurePrint G3 Human High-Resolution Discovery | 1 × 1 M | 2.6 kb | association studies |
| Agilent SurePrint G3 Human CNV | 2 × 400 K | 1 kb | |
| Agilent Human CNV Association | 2 × 105 K | 232 b | |
| Agilent SurePrint G3 CGH Postnatal Research | 4 × 180 K | 2.4 kb | regions identified by Baylor College of Medicine experts |
| 8 × 60 K | 3.7 kb | ||
| Agilent GenetiSure Postnatal Research CGH + SNP | 2 × 400 K | 9.8 kb | disease-associated regions (The Clinical Genome/ISCA database) |
| Agilent GenetiSure Pre-Screen | 4 × 180 K | 31 kb | CNV identification from embryo biopsies and single-cell samples; increased density on chromosomes 13, 18, 20, 21, 22, and X |
| 8 × 60 K | 50 kb | ||
| Agilent GenetiSure Cyto CGH | 4 × 180 K | 3.5 kb | disease-associated regions linked to developmental delay, intellectual disability, neuropsychiatric disorders, congenital anomalies, or dysmorphic features |
| 8 × 60 K | 7.1 kb | ||
| Agilent GenetiSure Cyto CGH + SNP | 4 × 180 K | 7.3 kb | |
| Agilent GenetiSure Cancer Research CGH + SNP | 2 × 400 K | 9.8 kb | cancer regions of the genome COSMIC (Catalogue of Somatic Mutation in Cancer) CGC (Cancer Genetics Consortium) databases |
| Illumina HumanCytoSNP | 12 × 300 K | 6.2 kb | enhanced coverage of ~250 disease regions, including subtelomeric regions, pericentromeric regions, and sex chromosomes |
| Illumina Infinium CytoSNP-850 K | 8 × 850 K | 1.8 kb | comprehensive coverage of cytogenetically relevant genes for congenital disorders and cancer research ICCG (International Collaboration for Clinical Genomics) and CCMC (Cancer Cytogenomics Microarray Consortium) |
| Illumina Infinium Core | 24 × 300 K | 5.8 kb | genome-wide tag SNPs found across diverse world populations |
| Illumina Infinium Exome | 24 × 300 K | 0.21 kb | comprehensive coverage of putative functional exonic variants (including markers representing a range of common conditions, such as type 2 diabetes, cancer, and metabolic, and psychiatric disorders) |
| Illumina Infinium CoreExome | 24 × 600 K | 1.82 kb | all of the markers from the Infinium Core-24 BeadChip and the Infinium Exome-24 BeadChip |
| Illumina Infinium Global Diversity Array | 8 × 2 M | 0.63 kb | common and low frequency variants in global populations, curated clinical research variants |
| Illumina Infinium Global Screening Array | 24 × 700 K | 2.3 kb | multiethnic genome-wide content, curated clinical research variants |
| Illumina Infinium Omni2.5 | 8 × 2.4 M | 0.65 kb | common and rare SNP content from the 1000 Genomes Project (MAF > 2.5%) |
| Illumina Infinium Omni2.5Exome | 8 × 2.7 M | 0.56 kb | combined Infinum Omni2.5 and Infinium Exome-24 markers |
| Illumina Infinium Omni5 | 4 × 4.3 M | 0.36 kb | comprehensive coverage of the genome including common, intermediate, and rare SNPs |
| Illumina Infinium Omni5 Exome | 4 × 4.6 M | 0.33 kb | comprehensive genome-wide backbone combined with putative functional exonic variants |
| Illumina Infinium OmniExpress | 24 × 700 K | 2.23 kb | high coverage of common variants for genome-wide association studies |
| Illumina Infinium OmniExpressExome | 8 × 1 M | 1.36 kb | tag SNPs and functional exonic content |
| Illumina Infinium OncoArray | 24 × 500 K | 5.4 kb | genetic variants associated with five common cancers |
| Illumina Infinium PsychArray | 24 × 700 K | 1.74 kb | genetic variants associated with common psychiatric disorders |
| Affymetrix Genome-Wide Human SNP Array 6.0 | 1 × 1.8 M | 0.68 kb | comprehensive coverage of the genome |
| Affymetrix CytoScan XON Suite | 24 × 6.85 M | 0.5 kb | enhanced coverage in 7000 clinically relevant gene, exon-level copy number changes |
| Affymetrix CytoScan HD | 24 × 2.7 M | 1.3 kb | enhanced coverage on cytogenetic relevant region |
| Tool | Description | aCGH | SNP-Array | Reference | |
|---|---|---|---|---|---|
| Affymetrix | Illumina | ||||
| ADM-2 | search for intervals in which a Z-score based on the average weighted log ratio exceeds a user-specified threshold | ✓ | technical documentation (Agilent) | ||
| Birdsuite | integration of common CNP genotypes and CNVs discovered using HMM | ✓ | [61] | ||
| ChAS | HMM on the log2 ratios processed through a Bayes wavelet shrinkage estimator | ✓ | technical documentation (Affymetrix) | ||
| cnvPartition | recursive partitioning approach based on preliminary copy number estimates | ✓ | technical documentation (Illumina) | ||
| DNAcopy | circular binary segmentation | ✓ | [48] | ||
| GenoCN | estimation of HMM, parameters from data, germline, and somatic modes | ✓ | ✓ | [62] | |
| iPattern | normalization of the total intensities across individuals, Gaussian mixture model fitting | ✓ | ✓ | [42] | |
| Nexus | the probe’s log-ratio rank segmentation | ✓ | ✓ | ✓ | [63] |
| PennCN | HMM, also counted for the population frequency of the B allele | ✓ | ✓ | [57] | |
| QuantiSNP | objective Bayes-HMM, fixed rate of heterozygosity for each SNP | ✓ | ✓ | [64] | |
| Tool | Description | Data | Mode | Reference | ||
|---|---|---|---|---|---|---|
| WES | Targeted | Germline | Somatic | |||
| cn.MOPS | mixture Poisson model and Bayes approach | ✓ | ✓ | ✓ | ✓ | [92] |
| CNVkit | in- and off-target regions, rolling median bias correction, CBS | ✓ | ✓ | ✓ | [78] | |
| CODEX | log-linear decomposition-based normalization, Poisson likelihood-based segmentation | ✓ | ✓ | ✓ | ✓ | [87] |
| CoNIFER | singular value decomposition-based normalization, ± 1.5 SVD-ZRPKM threshold | ✓ | ✓ | [93] | ||
| CoNVaDING | ratio scores and Z-scores of the sample of interest compared to the selected control | ✓ | ✓ | [94] | ||
| DECoN | ExomeDepth modification (the distance between exons is taken into account) | ✓ | ✓ | [95] | ||
| ExomeDepth | beta-binomial distribution, optimized reference set, HMM | ✓ | ✓ | ✓ | [88] | |
| XHMM | principal component analysis normalization, HMM | ✓ | ✓ | [86] | ||
| Approach | Tool | Description | Reference |
|---|---|---|---|
| RP | BreakDancer | search for regions that include more anomalous read pairs than expected | [67] |
| SR | Pindel | pattern growth approach for breakpoint identification | [73] |
| RD | CNVnator | mean-shift technique, multiple-bandwidth partitioning, and GC correction | [79] |
| AS | Cortex | bubble-calling in the colored de Bruijn graph | [98] |
| RP + RD | GenomeSTRiP | connected components algorithm for read pair clustering, Gaussian mixture model for read depth genotyping | [104] |
| RP + SR | DELLY | graph-based paired-end clustering, breakpoints refinement using split-read alignment | [105] |
| RP + AS | Hydra | assembly of discordant mate pairs and aligned to the reference genome with MEGABLAST | [106] |
| RP + SR + AS | Manta | breakend graph construction, independent for each edge variation hypothesis refinement and scoring with diploid model | [103] |
| RP + SR + RD | Lumpy | probabilistic representation of an SV breakpoint | [102] |
| Ensemble | MetaSV | merging calls from tools (BreakDancer, CNVnator, BreakSeq, Pindel), breakpoint refinement by aligning the assembled CNV regions | [107] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gordeeva, V.; Sharova, E.; Arapidi, G. Progress in Methods for Copy Number Variation Profiling. Int. J. Mol. Sci. 2022, 23, 2143. https://doi.org/10.3390/ijms23042143
Gordeeva V, Sharova E, Arapidi G. Progress in Methods for Copy Number Variation Profiling. International Journal of Molecular Sciences. 2022; 23(4):2143. https://doi.org/10.3390/ijms23042143
Chicago/Turabian StyleGordeeva, Veronika, Elena Sharova, and Georgij Arapidi. 2022. "Progress in Methods for Copy Number Variation Profiling" International Journal of Molecular Sciences 23, no. 4: 2143. https://doi.org/10.3390/ijms23042143
APA StyleGordeeva, V., Sharova, E., & Arapidi, G. (2022). Progress in Methods for Copy Number Variation Profiling. International Journal of Molecular Sciences, 23(4), 2143. https://doi.org/10.3390/ijms23042143






