Multiomics Analysis of Endocytosis upon HBV Infection and Identification of SCAMP1 as a Novel Host Restriction Factor against HBV Replication
Abstract
1. Introduction
2. Results
2.1. HBV Integration Changes Endocytosis Associated Transcriptome Profile in the HepG2.2.15 Cell Line
2.2. HBV Induced Alterations in Host Endocytosis Associated Proteome
2.3. Changes in Host Endocytosis-Associated Ubiquitylome upon HBV Infection
2.4. Comparison and Correlation among Transcriptome, Proteome, and Ubiquitylome
2.5. Effects of HBV Infection on SCAMP1 mRNA, Protein, and Lysine Ubiquitination
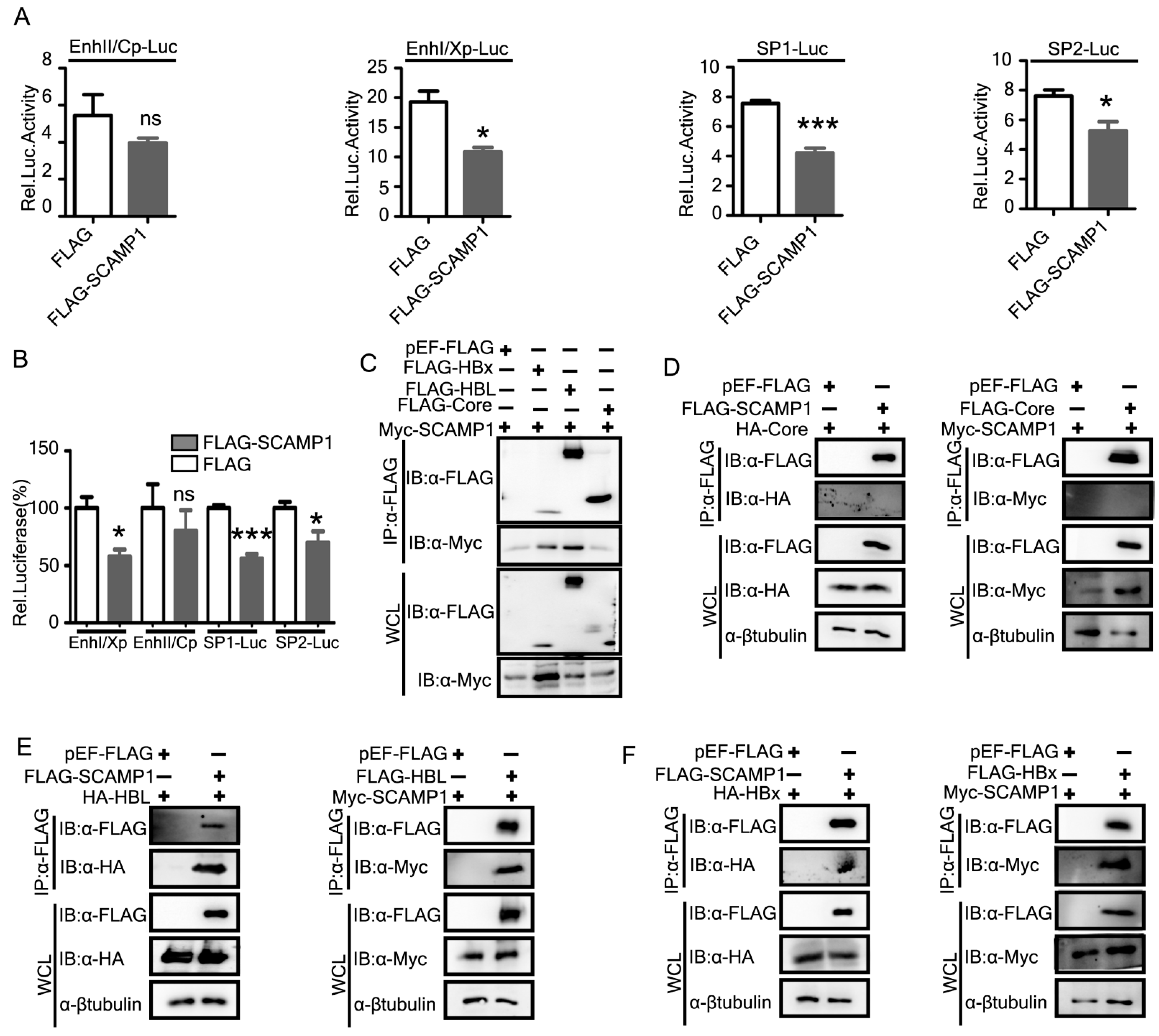
2.6. Identification of SCAMP1 as a Novel Inhibitor of HBV Replication
2.7. Knockdown of Endogenous SCAMP1 Enhances HBV Transcription and Replication
2.8. SCAMP1 Downregulates HBV via Transcriptional Mechanisms
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Transfection
4.2. RNA Extraction, Library Preparation, and Sequencing
4.3. SILAC Labelling, Protein Extraction, and Trypsin Digestion
4.3.1. HPLC Fractionation and Affinity Enrichment
4.3.2. LC-MS/MS Analysis and Database Search
4.4. si-RNAs and Plasmid Construction
4.5. RNA Extraction and qRT-PCR
4.6. In Vivo Ubiquitination Assay
4.7. Immunoprecipitation and Immunoblot Analyses
4.8. Enzyme-Linked Immune-Sorbent Assay
4.9. Northern Blot Analysis
4.10. Reporter Assay
4.11. Protein-Protein Interaction (PPI) Network Analysis Construction
4.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hao, R.; He, J.; Liu, X.; Gao, G.; Liu, D.; Cui, L.; Yu, G.; Yu, W.; Chen, Y.; Guo, D. Inhibition of Hepatitis B Virus Gene Expression and Replication by Hepatocyte Nuclear Factor 6. J. Virol. 2015, 89, 4345–4355. [Google Scholar] [CrossRef]
- Jia, B.; Guo, M.; Li, G.; Yu, D.; Zhang, X.; Lan, K.; Deng, Q. Hepatitis B Virus Core Protein Sensitizes Hepatocytes to Tumor Necrosis Factor-Induced Apoptosis by Suppression of the Phosphorylation of Mitogen-Activated Protein Kinase Kinase 7. J. Virol. 2015, 89, 2041–2051. [Google Scholar] [CrossRef]
- Staring, J.; Raaben, M.; Brummelkamp, T.R. Viral escape from endosomes and host detection at a glance. J. Cell Sci. 2018, 131. [Google Scholar] [CrossRef]
- Boulant, S.; Stanifer, M.; Lozach, P.Y. Dynamics of Virus-Receptor Interactions in Virus Binding, Signaling, and Endocytosis. Viruses 2015, 7, 2794–2815. [Google Scholar] [CrossRef]
- Herrscher, C.; Pastor, F.; Burlaud-Gaillard, J.; Dumans, A.; Seigneuret, F.; Moreau, A.; Patient, R.; Eymieux, S.; de Rocquigny, H.; Hourioux, C.; et al. Hepatitis B virus entry into HepG2-NTCP cells requires clathrin-mediated endocytosis. Cell. Microbiol. 2020, 22, e13205. [Google Scholar] [CrossRef]
- Tsukuda, S.; Watashi, K. Hepatitis B virus biology and life cycle. Antivir. Res. 2020, 182, 104925. [Google Scholar] [CrossRef]
- Iwamoto, M.; Saso, W.; Nishioka, K.; Ohashi, H.; Sugiyama, R.; Ryo, A.; Ohki, M.; Yun, J.H.; Park, S.Y.; Ohshima, T.; et al. The machinery for endocytosis of epidermal growth factor receptor coordinates the transport of incoming hepatitis B virus to the endosomal network. J. Biol. Chem. 2020, 295, 800. [Google Scholar] [CrossRef]
- Cossart, P.; Helenius, A. Endocytosis of Viruses and Bacteria. Cold Spring Harb. Perspect. Biol. 2014, 6, a016972. [Google Scholar] [CrossRef]
- Herrscher, C.; Roingeard, P.; Blanchard, E. Hepatitis B Virus Entry into Cells. Cells 2020, 9, 1486. [Google Scholar] [CrossRef]
- Law, A.H.Y.; Chow, C.M.; Jiang, L. Secretory carrier membrane proteins. Protoplasma 2012, 249, 269–283. [Google Scholar] [CrossRef]
- Zheng, J.L.C.; Tham, C.T.; Keatings, K.; Fan, S.; Liou, A.Y.C.; Numata, Y.; Allan, D.; Numata, M. Secretory Carrier Membrane Protein (SCAMP) deficiency influences behavior of adult flies. Front. Cell Dev. Biol. 2014, 2, 64. [Google Scholar] [CrossRef]
- Hubbard, C.; Singleton, D.; Rauch, M.; Jayasinghe, S.; Cafiso, D.; Castle, D. The secretory carrier membrane protein family: Structure and membrane topology. Mol. Biol. Cell 2000, 11, 2933–2947. [Google Scholar] [CrossRef]
- Fernandez-Chacon, R.; Sudhof, T.C. Novel SCAMPs lacking NPF repeats: Ubiquitous and synaptic vesicle-specific forms implicate SCAMPs in multiple membrane-trafficking functions. J. Neurosci. 2000, 20, 7941–7950. [Google Scholar] [CrossRef]
- Yang, S.; Lee, K.T.; Lee, J.Y.; Lee, J.K.; Lee, K.H.; Rhee, J.C. Inhibition of SCAMP1 suppresses cell migration and invasion in human pancreatic and gallbladder cancer cells. Tumor Biol. 2013, 34, 2731–2739. [Google Scholar] [CrossRef]
- Zhang, X.; Sheng, J.; Zhang, Y.; Tian, Y.; Zhu, J.; Luo, N.; Xiao, C.; Li, R. Overexpression of SCAMP3 is an indicator of poor prognosis in hepatocellular carcinoma. Oncotarget 2017, 8, 109247–109257. [Google Scholar] [CrossRef]
- Yue, C.; Xie, S.; Zhong, J.; Zhao, H.; Lin, Z.; Zhang, L.; Xu, B.; Luo, Y. SCAMP2/5 as diagnostic and prognostic markers for acute myeloid leukemia. Sci. Rep. 2021, 11, 17012. [Google Scholar] [CrossRef]
- Yuan, S.; Tanzeel, Y.; Tian, X.; Zheng, D.; Wajeeha, N.; Xu, J.; Ke, Y.; Zhang, Z.; Peng, X.; Lu, L.; et al. Global analysis of HBV-mediated host proteome and ubiquitylome change in HepG2.2.15 human hepatoblastoma cell line. Cell Biosci. 2021, 11, 75. [Google Scholar] [CrossRef]
- Naboulsi, W.; Bracht, T.; Megger, D.A.; Reis, H.; Ahrens, M.; Turewicz, M.; Eisenacher, M.; Tautges, S.; Canbay, A.E.; Meyer, H.E.; et al. Quantitative proteome analysis reveals the correlation between endocytosis-associated proteins and hepatocellular carcinoma dedifferentiation. Biochim. Biophys. Acta Proteins Proteom. 2016, 1864, 1579–1585. [Google Scholar] [CrossRef]
- Chen, V.L.; Le, A.K.; Podlaha, O.; Estevez, J.; Li, B.; Vutien, P.; Chang, E.T.; Rosenberg-Hasson, Y.; Pflanz, S.; Jiang, Z.; et al. Soluble intercellular adhesion molecule-1 is associated with hepatocellular carcinoma risk: Multiplex analysis of serum markers. Sci. Rep. 2017, 7, 11169. [Google Scholar] [CrossRef]
- Hu, K.Q.; Yu, C.H.; Vierling, J.M. Up-regulation of intercellular adhesion molecule 1 transcription by hepatitis B virus X protein. Proc. Natl. Acad. Sci. USA 1992, 89, 11441. [Google Scholar] [CrossRef]
- Brito, A.F.; Pinney, J.W. Protein-protein interactions in virus-host systems. Front. Microbiol. 2017, 8, 1557. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Chacón, R.; Achiriloaie, M.; Janz, R.; Albanesi, J.P.; Südhof, T.C. SCAMP1 function in endocytosis. J. Biol. Chem. 2000, 275, 12752–12756. [Google Scholar] [CrossRef] [PubMed]
- Banks, K.E.; Anderson, A.L.; Tang, H.; Hughes, D.E.; Costa, R.H.; McLachlan, A. Hepatocyte Nuclear Factor 3β Inhibits Hepatitis B Virus Replication In Vivo. J. Virol. 2002, 76, 12974–12980. [Google Scholar] [CrossRef][Green Version]
- Mao, R.; Nie, H.; Cai, D.; Zhang, J.; Liu, H.; Yan, R.; Cuconati, A.; Block, T.M.; Guo, J.T.; Guo, H. Inhibition of Hepatitis B Virus Replication by the Host Zinc Finger Antiviral Protein. PLoS Pathog. 2013, 9, e1003494. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.A.; Hu, J. Hepatitis B virus reverse transcriptase: Diverse functions as classical and emerging targets for antiviral intervention. Emerg. Microbes Infect. 2013, 2, e56. [Google Scholar] [CrossRef]
- Quasdorff, M.; Protzer, U. Control of hepatitis B virus at the level of transcription. J. Viral Hepat. 2010, 17, 527–536. [Google Scholar] [CrossRef]
- Yang, H.; Zhou, Y.; Mo, J.; Xiang, Q.; Qin, M.; Liu, W.; Shang, J. SOX9 represses hepatitis B virus replication through binding to HBV EnhII / Cp and inhibiting the promoter activity. Antivir. Res. 2020, 177, 104761. [Google Scholar] [CrossRef] [PubMed]
- Piracha, Z.Z.; Kwon, H.; Saeed, U.; Kim, J.; Jung, J.; Chwae, Y.-J.; Park, S.; Shin, H.-J.; Kim, K. Sirtuin 2 Isoform 1 Enhances Hepatitis B Virus RNA Transcription and DNA Synthesis through the AKT/GSK-3β/β-Catenin Signaling Pathway. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Tang, H.; McLachlan, A. Transcriptional regulation of hepatitis B virus by nuclear hormone receptors is a critical determinant of viral tropism. Proc. Natl. Acad. Sci. USA 2001, 98, 1841–1846. [Google Scholar] [CrossRef]
- Li, J.; Ou, J. Differential Regulation of Hepatitis B Virus Gene Expression by the Sp1 Transcription Factor. J. Virol. 2001, 75, 8400–8406. [Google Scholar] [CrossRef]
- Tan, G.; Xu, F.; Song, H.; Yuan, Y.; Xiao, Q.; Ma, F.; Qin, F.X.F.; Cheng, G. Identification of TRIM14 as a type I IFN-stimulated gene controlling hepatitis B virus replication by targeting HBx. Front. Immunol. 2018, 9, 1872. [Google Scholar] [CrossRef] [PubMed]
- Shin, G.C.; Ahn, S.H.; Choi, H.S.; Lim, K.H.; Choi, D.Y.; Kim, K.P.; Kim, K.H. Hepatocystin/80K-H inhibits replication of hepatitis B virus through interaction with HBx protein in hepatoma cell. Biochim. Biophys. Acta Mol. Basis Dis. 2013, 1832, 1569–1581. [Google Scholar] [CrossRef]
- Lamontagne, J.; Mell, J.C.; Bouchard, M.J. Transcriptome-Wide Analysis of Hepatitis B Virus-Mediated Changes to Normal Hepatocyte Gene Expression. PLoS Pathog. 2016, 12, e1005438. [Google Scholar] [CrossRef] [PubMed]
- Xie, N.; Chen, X.; Zhang, T.; Liu, B.; Huang, C. Using proteomics to identify the HBx interactome in hepatitis B virus: How can this inform the clinic? Expert Rev. Proteom. 2014, 11, 59–74. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Fan, F.; Wei, W.; Liu, Y.; Xu, Z.; Zhai, L.; Qi, Y.; Ye, B.; Zhang, Y.; Basu, S.; et al. Multi-omics analyses reveal metabolic alterations regulated by hepatitis B virus core protein in hepatocellular carcinoma cells. Sci. Rep. 2017, 7, 41089. [Google Scholar]
- Lamontagne, R.J.; Bagga, S.; Bouchard, M.J. Hepatitis B virus molecular biology and pathogenesis. Hepatoma Res. 2016, 2, 163. [Google Scholar] [CrossRef]
- McBrearty, N.; Arzumanyan, A.; Bichenkov, E.; Merali, S.; Merali, C.; Feitelson, M. Short chain fatty acids delay the development of hepatocellular carcinoma in HBx transgenic mice. Neoplasia 2021, 23, 529. [Google Scholar] [CrossRef]
- Becker, A.C.; Gannagé, M.; Giese, S.; Hu, Z.; Abou-Eid, S.; Roubaty, C.; Paul, P.; Bühler, L.; Gretzmeier, C.; Dumit, V.I.; et al. Influenza a virus induces autophagosomal targeting of ribosomal proteins. Mol. Cell. Proteom. 2018, 17, 1909–1921. [Google Scholar] [CrossRef]
- Stieler, J.T.; Prange, R. Involvement of ESCRT-II in Hepatitis B Virus Morphogenesis. PLoS ONE 2014, 9, e91279. [Google Scholar] [CrossRef]
- Probst, O.C.; Puxbaum, V.; Svoboda, B.; Leksa, V.; Stockinger, H.; Mikula, M.; Mikulits, W.; Mach, L. The mannose 6-phosphate/insulin-like growth factor II receptor restricts the tumourigenicity and invasiveness of squamous cell carcinoma cells. Int. J. Cancer 2009, 124, 2559–2567. [Google Scholar] [CrossRef]
- Li, J.; Sahagian, G.G. Demonstration of tumor suppression by mannose 6-phosphate/insulin-like growth factor 2 receptor. Oncogene 2004, 23, 9359–9368. [Google Scholar] [CrossRef] [PubMed]
- Hébert, E. Mannose-6-phosphate/insulin-like growth factor II receptor expression and tumor development. Biosci. Rep. 2006, 26, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Enguita-Germán, M.; Fortes, P. Targeting the insulin-like growth factor pathway in hepatocellular carcinoma. World J. Hepatol. 2014, 6, 716. [Google Scholar] [CrossRef] [PubMed]
- Besharat, S.; Katoonizadeh, A.; Moossavi, S.; Darvishi, Z.; Roshandel, G.; Poustchi, H.; Mohamadkhani, A. The possible impact of sortilin in reducing HBsAg expression in chronic hepatitis B. J. Med. Virol. 2016, 88, 647–652. [Google Scholar] [CrossRef]
- Rodriguez-Gil, J.L.; Bianconi, S.E.; Farhat, N.; Kleiner, D.E.; Nelson, M.; Porter, F.D. Hepatocellular carcinoma as a complication of Niemann-Pick disease type C1. Am. J. Med. Genet. Part A 2021, 185, 3111–3117. [Google Scholar] [CrossRef]
- Mo, C.-F.; Li, J.; Yang, S.-X.; Guo, H.-J.; Liu, Y.; Luo, X.-Y.; Wang, Y.-T.; Li, M.-H.; Li, J.Y.; Zou, Q. IQGAP1 promotes anoikis resistance and metastasis through Rac1-dependent ROS accumulation and activation of Src/FAK signalling in hepatocellular carcinoma. Br. J. Cancer 2020, 123, 1154–1163. [Google Scholar] [CrossRef]
- Xu, G.; Xia, Z.; Deng, F.; Liu, L.; Wang, Q.; Yu, Y.; Wang, F.; Zhu, C.; Liu, W.; Cheng, Z.; et al. Inducible LGALS3BP/90K activates antiviral innate immune responses by targeting TRAF6 and TRAF3 complex. PLOS Pathog. 2019, 15, e1008002. [Google Scholar] [CrossRef]
- Uluca, Ü.; Şen, V.; Ece, A.; Tan, İ.; Karabel, D.; Aktar, F.; Karabel, M.; Balık, H.; Güneş, A. Serum galectin-3 levels in children with chronic hepatitis B infection and inactive hepatitis B carriers. Med. Sci. Monit. 2015, 21, 1376–1380. [Google Scholar] [PubMed]
- Kindrat, I.; Tryndyak, V.; de Conti, A.; Shpyleva, S.; Mudalige, T.K.; Kobets, T.; Erstenyuk, A.M.; Beland, F.A.; Pogribny, I.P.; Kindrat, I.; et al. MicroRNA-152-mediated dysregulation of hepatic transferrin receptor 1 in liver carcinogenesis. Oncotarget 2015, 7, 1276–1287. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Peña, R.; El Mounadi, K.; Garcia-Ruiz, H. Changes in Subcellular Localization of Host Proteins Induced by Plant Viruses. Viruses 2021, 13, 677. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Zhu, H.; Dong, L.; Shi, W.; Chen, R.; Song, Z.; Huang, C.; Li, J.; Dong, X.; Zhou, Y.; et al. Integrated Proteogenomic Characterization of HBV-Related Hepatocellular Carcinoma. Cell 2019, 179, 561.e22–577.e22. [Google Scholar] [CrossRef]
- Chen, S.W.; Himeno, M.; Koui, Y.; Sugiyama, M.; Nishitsuji, H.; Mizokami, M.; Shimotohno, K.; Miyajima, A.; Kido, T. Modulation of hepatitis B virus infection by epidermal growth factor secreted from liver sinusoidal endothelial cells. Sci. Rep. 2020, 10, 14349. [Google Scholar] [CrossRef] [PubMed]
- Bakker, J.; Spits, M.; Neefjes, J.; Berlin, I. The EGFR odyssey—From activation to destruction in space and time. J. Cell Sci. 2017, 130, 4087–4096. [Google Scholar] [CrossRef] [PubMed]
- Arzumanyan, A.; Reis, H.M.G.P.V.; Feitelson, M.A. Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nat. Rev. Cancer 2013, 13, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Wu, C.; Wang, X.; Liu, S.; Kemper, T.; Li, F.; Squire, A.; Zhu, Y.; Zhang, J.; Chen, X.; et al. Synaptosomal-associated protein 29 is required for the autophagic degradation of hepatitis B virus. FASEB J. 2019, 33, 6023–6034. [Google Scholar] [CrossRef] [PubMed]
- Chou, S.F.; Tsai, M.L.; Huang, J.Y.; Chang, Y.S.; Shih, C. The Dual Role of an ESCRT-0 Component HGS in HBV Transcription and Naked Capsid Secretion. PLoS Pathog. 2015, 11, e1005123. [Google Scholar] [CrossRef]
- Beck, J.; Nassal, M. Hepatitis B virus replication. World J. Gastroenterol. 2007, 13, 48–64. [Google Scholar] [CrossRef] [PubMed]
- Clark, D.N.; Hu, J. Unveiling the roles of HBV polymerase for new antiviral strategies. Future Virol. 2015, 10, 283–295. [Google Scholar] [CrossRef]
- Song, F.; Wei, M.; Wang, J.; Liu, Y.; Guo, M.; Li, X.; Luo, J.; Zhou, J.; Wang, M.; Guo, D.; et al. Hepatitis B virus-regulated growth of liver cancer cells occurs through the microRNA-340-5p-activating transcription factor 7-heat shock protein A member 1B axis. Cancer Sci. 2019, 110, 1633–1643. [Google Scholar] [CrossRef]
- Wang, J.; Chen, J.; Liu, Y.; Zeng, X.; Wei, M.; Wu, S.; Xiong, Q.; Song, F.; Yuan, X.; Xiao, Y.; et al. Hepatitis B Virus Induces Autophagy to Promote its Replication by the Axis of miR-192-3p-XIAP Through NF kappa B Signaling. Hepatology 2019, 69, 974–992. [Google Scholar] [CrossRef]
- Laney, J.D.; Hochstrasser, M. Analysis of protein ubiquitination. Curr. Protoc. Protein Sci. 2011, 1. [Google Scholar] [CrossRef] [PubMed]

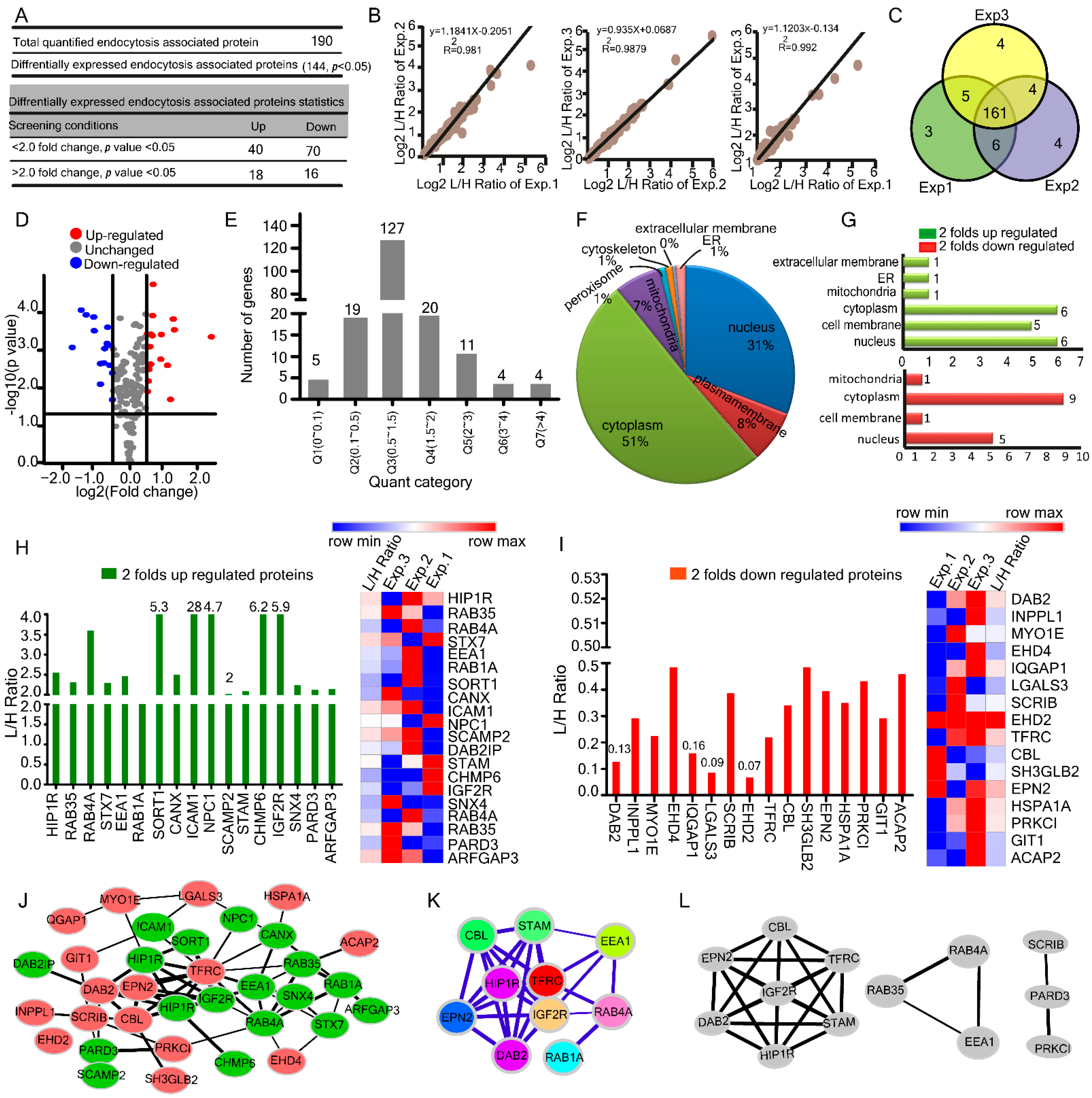
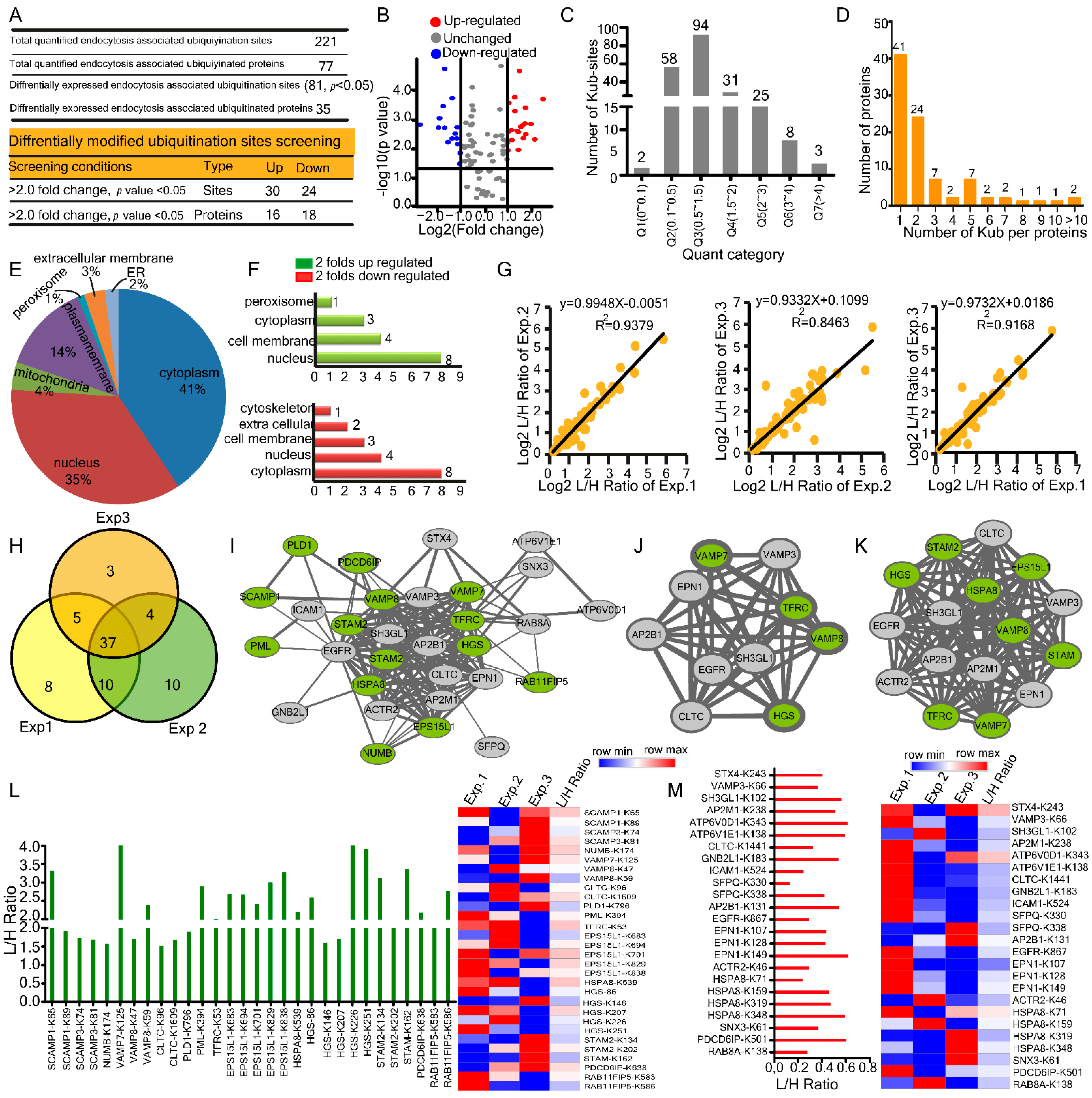
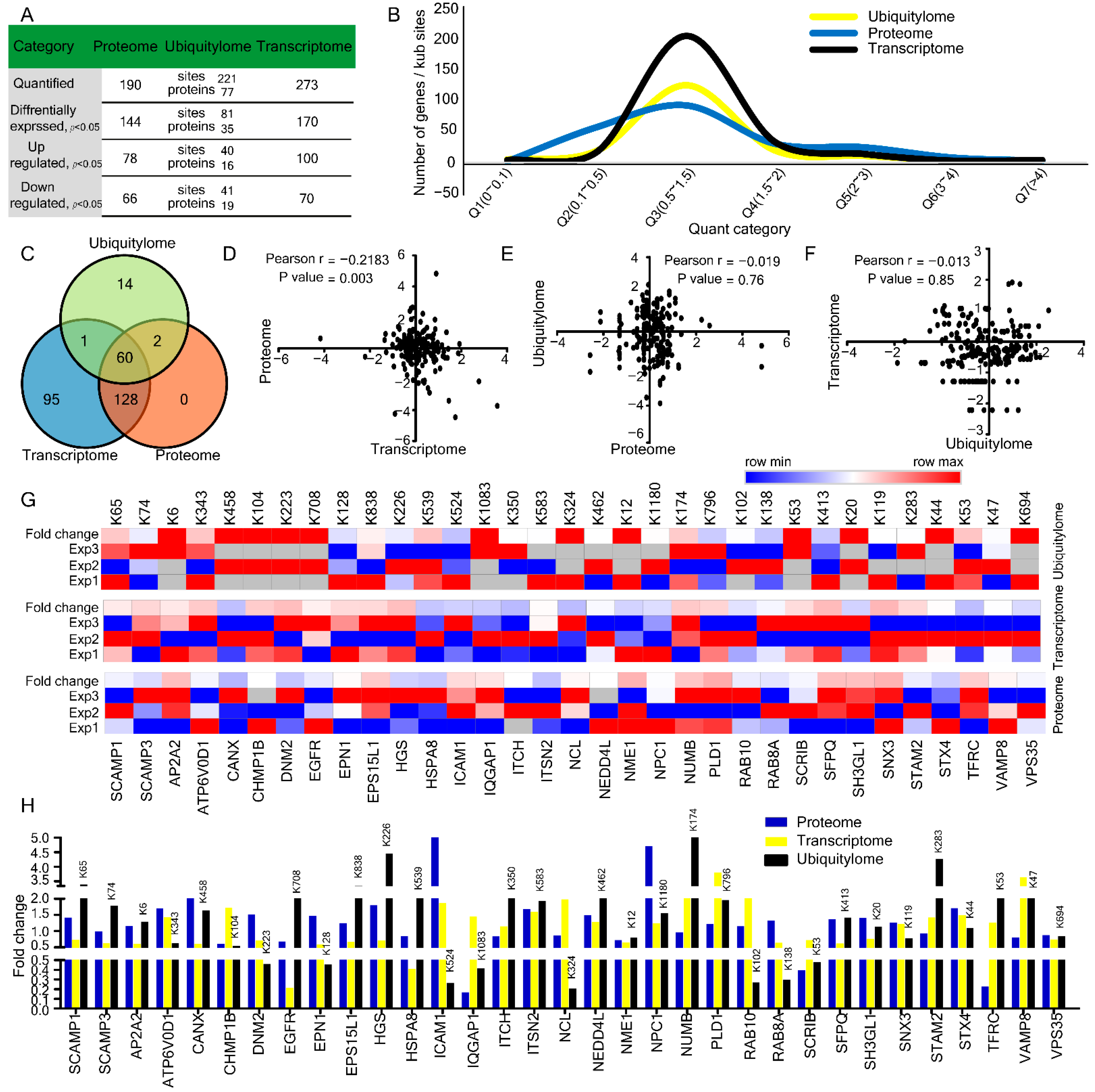
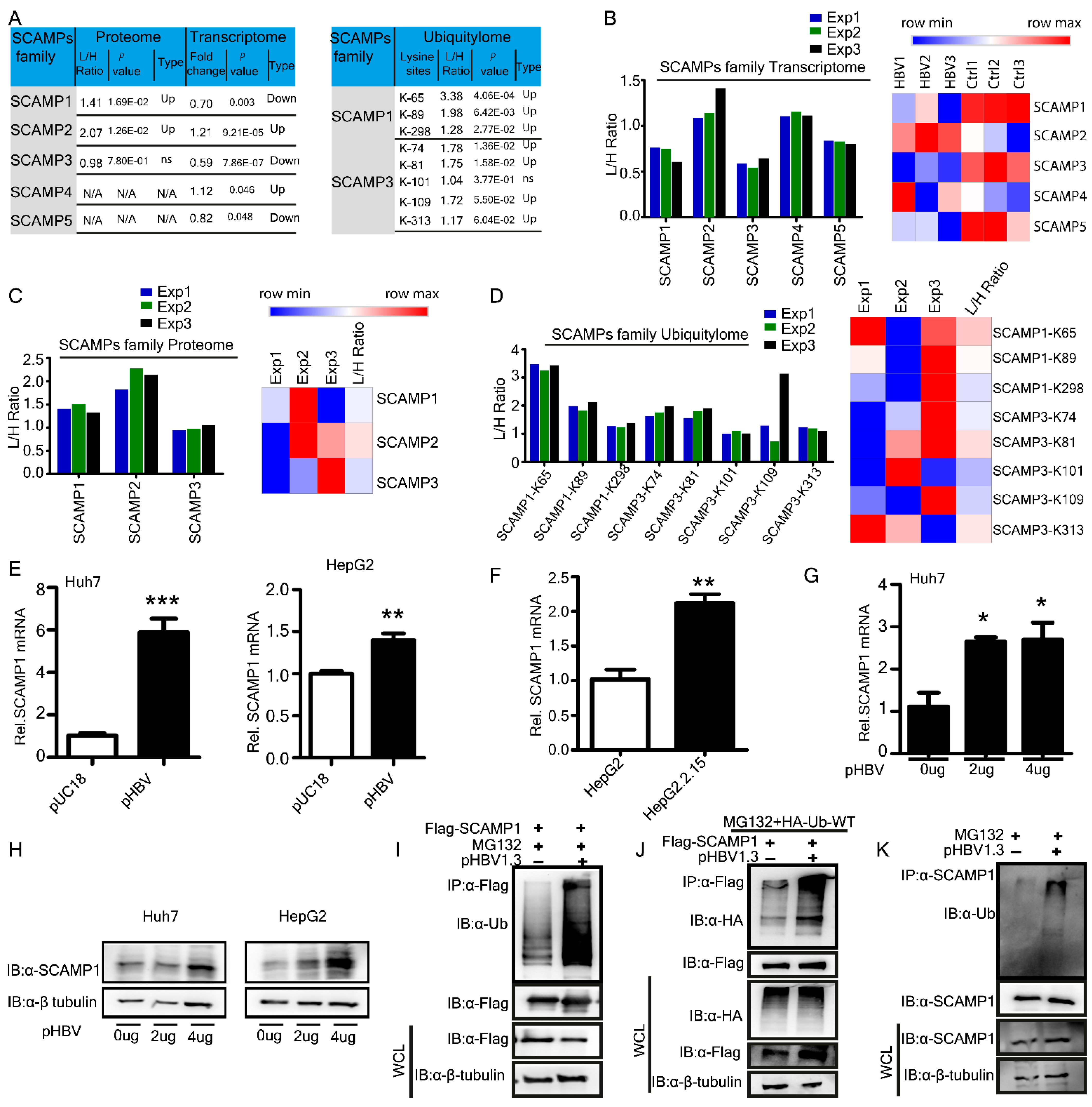


| Parameters | Proteome−Transcriptome | Ubiquitylome−Proteome | Transcriptome−Ubiquitylome |
|---|---|---|---|
| Pearson r | −0.2183 | −0.01983 | −0.01331 |
| p value | 0.003 | 0.7634 | 0.8505 |
| Spearman r | −0.1456 | −0.03623 | −0.00135 |
| p value | 0.0492 | 0.5822 | 0.9846 |
| R squared | 0.04765 | 0.00039 | 0.00017 |
| Number of XY pairs | 183 | 233 | 203 |
| Common Genes | Transcriptome | Proteome | Ubiquitylome | |||||
|---|---|---|---|---|---|---|---|---|
| Protein Accession | Gene Name | FCs | Regulation | L/H Ratio | Regulation | Lysine Position | L/H Ratio | Regulation |
| O15126 | SCAMP1 | 0.74 | Down | 1.41 | Up | 65 | 3.38 | Up |
| O14828 | SCAMP3 | 0.63 | Down | 0.98 | Down | 74 | 1.78 | Up |
| O94973 | AP2A2 | 0.6 | Down | 1.15 | Up | 6 | 1.29 | Up |
| P61421 | ATP6V0D1 | 1.42 | Up | 1.69 | Up | 343 | 0.63 | Down |
| P27824 | CANX | 0.6 | Down | 2.56 | Up | 458 | 1.64 | Up |
| Q7LBR1 | CHMP1B | 1.72 | Up | 0.61 | Down | 104 | 0.54 | Down |
| P50570 | DNM2 | 0.7 | Down | 1.51 | Up | 223 | 0.46 | Down |
| P00533 | EGFR | 0.21 | Down | 0.68 | Down | 708 | 2.34 | Up |
| Q9Y6I3 | EPN1 | 0.58 | Down | 1.47 | Up | 128 | 0.45 | Down |
| Q9UBC2 | EPS15L1 | 0.67 | Down | 1.24 | Down | 838 | 3.34 | Up |
| O14964 | HGS | 0.7 | Down | 1.79 | Up | 226 | 4.45 | Up |
| P11142 | HSPA8 | 0.41 | Down | 0.84 | Down | 539 | 2.26 | Up |
| P05362 | ICAM1 | 1.86 | Up | 28.4 | Up | 524 | 0.26 | Down |
| P46940 | IQGAP1 | 1.45 | Up | 0.16 | Down | 1083 | 0.41 | Down |
| Q96J02 | ITCH | 1.14 | Up | 0.83 | Down | 350 | 3.23 | Up |
| Q9NZM3 | ITSN2 | 1.59 | Up | 1.68 | Up | 583 | 1.93 | Up |
| P19338 | NCL | 1.98 | Up | 0.86 | Down | 324 | 0.2 | Down |
| Q96PU5 | NEDD4L | 1.27 | Up | 1.49 | Up | 462 | 2.33 | Up |
| P15531 | NME1 | 0.65 | Down | 0.72 | Down | 12 | 0.79 | Down |
| O15118 | NPC1 | 1.24 | Up | 4.71 | Up | 1180 | 1.54 | Up |
| P49757 | NUMB | 2.01 | Up | 0.95 | Down | 174 | 5.7 | Up |
| Q13393 | PLD1 | 3.8 | Up | 1.21 | Up | 796 | 1.96 | Up |
| P61026 | RAB10 | 2.23 | Up | 1.14 | Up | 102 | 0.27 | Down |
| P61006 | RAB8A | 0.64 | Down | 1.31 | Up | 138 | 0.29 | Down |
| Q14160 | SCRIB | 0.72 | Down | 0.39 | Down | 53 | 0.47 | Down |
| P23246 | SFPQ | 0.62 | Down | 1.36 | Up | 413 | 1.41 | Up |
| Q99961 | SH3GL1 | 0.76 | Down | 1.4 | Up | 20 | 1.13 | Up |
| O60493 | SNX3 | 1.21 | Up | 1.25 | Up | 119 | 0.77 | Down |
| O75886 | STAM2 | 1.41 | Up | 0.91 | Down | 202 | 2 | Up |
| Q12846 | STX4 | 1.49 | Up | 1.7 | Up | 243 | 0.42 | Down |
| P02786 | TFRC | 1.26 | Up | 0.22 | Down | 53 | 2.06 | Up |
| Q9BV40 | VAMP8 | 3.63 | Up | 0.8 | Down | 47 | 2.46 | Up |
| Q96QK1 | VPS3 | 0.74 | Down | 0.87 | Down | 694 | 0.84 | Down |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yousaf, T.; Sun, Y.; Naz, W.; Liu, Y.; Xu, J.; Yuan, S.; Wu, K.; Wang, M.; Wang, J.; Guo, M.; et al. Multiomics Analysis of Endocytosis upon HBV Infection and Identification of SCAMP1 as a Novel Host Restriction Factor against HBV Replication. Int. J. Mol. Sci. 2022, 23, 2211. https://doi.org/10.3390/ijms23042211
Yousaf T, Sun Y, Naz W, Liu Y, Xu J, Yuan S, Wu K, Wang M, Wang J, Guo M, et al. Multiomics Analysis of Endocytosis upon HBV Infection and Identification of SCAMP1 as a Novel Host Restriction Factor against HBV Replication. International Journal of Molecular Sciences. 2022; 23(4):2211. https://doi.org/10.3390/ijms23042211
Chicago/Turabian StyleYousaf, Tanzeel, Yuting Sun, Wajeeha Naz, Yang Liu, Jiaqi Xu, Sen Yuan, Kangwei Wu, Min Wang, Jun Wang, Mingxiong Guo, and et al. 2022. "Multiomics Analysis of Endocytosis upon HBV Infection and Identification of SCAMP1 as a Novel Host Restriction Factor against HBV Replication" International Journal of Molecular Sciences 23, no. 4: 2211. https://doi.org/10.3390/ijms23042211
APA StyleYousaf, T., Sun, Y., Naz, W., Liu, Y., Xu, J., Yuan, S., Wu, K., Wang, M., Wang, J., Guo, M., & Sun, G. (2022). Multiomics Analysis of Endocytosis upon HBV Infection and Identification of SCAMP1 as a Novel Host Restriction Factor against HBV Replication. International Journal of Molecular Sciences, 23(4), 2211. https://doi.org/10.3390/ijms23042211






