The Ubiquitin E3 Ligase PRU2 Modulates Phosphate Uptake in Arabidopsis
Abstract
:1. Introduction
2. Results
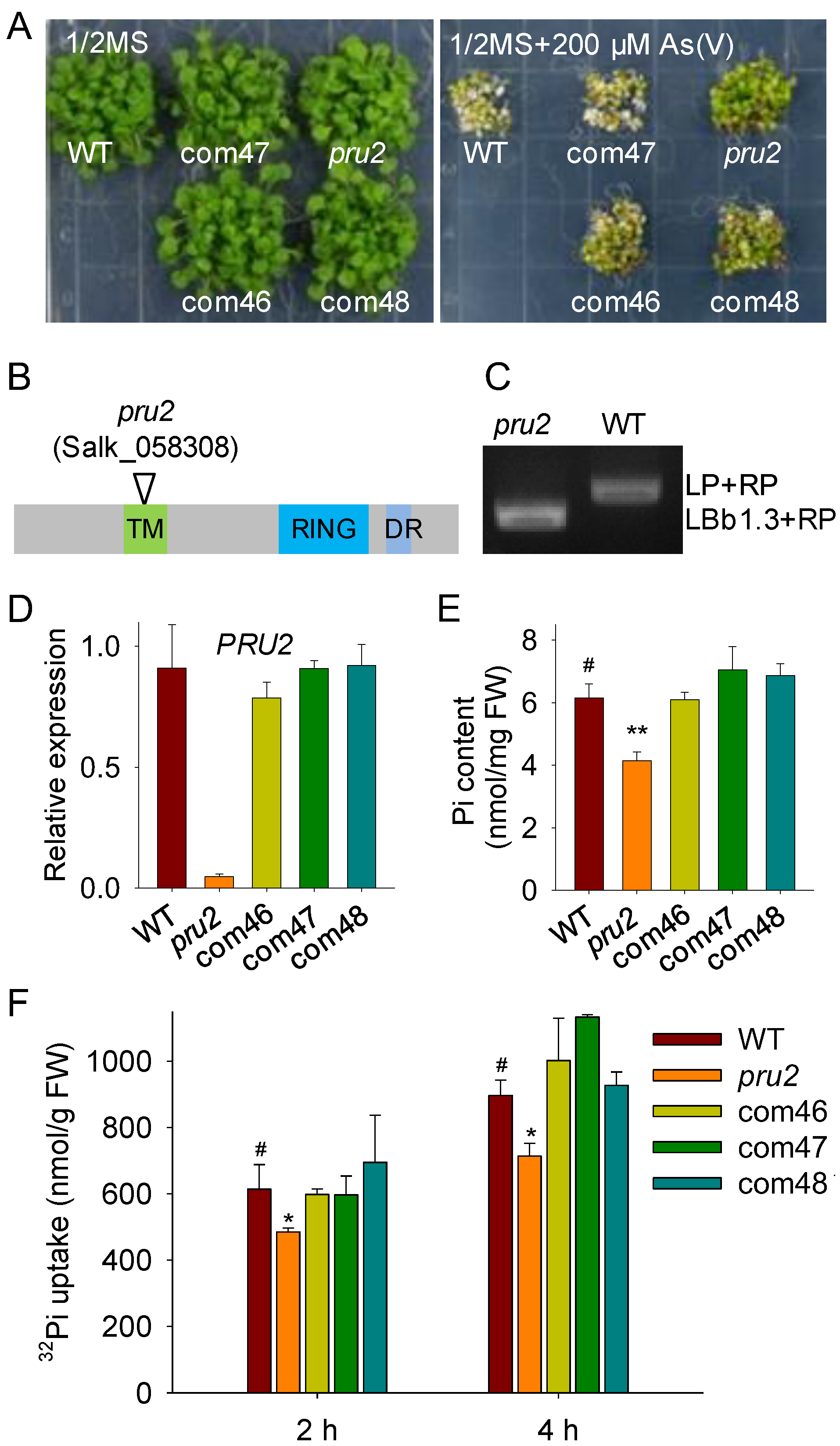
2.1. The pru2 Mutant Was Defective in Pi Uptake
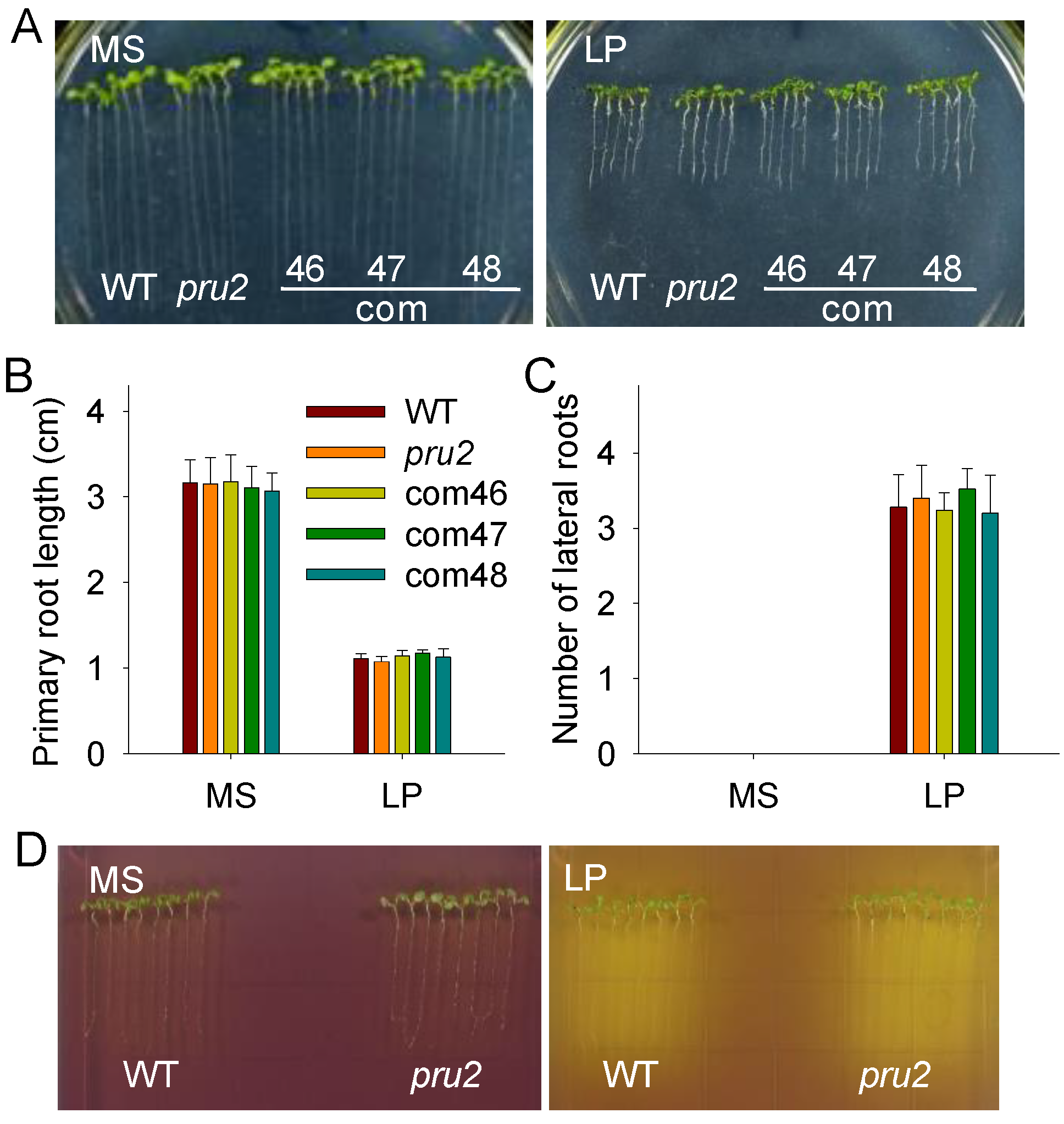
2.2. RSA and Organic Acids Exudation in the pru2 Mutant under LP Stress
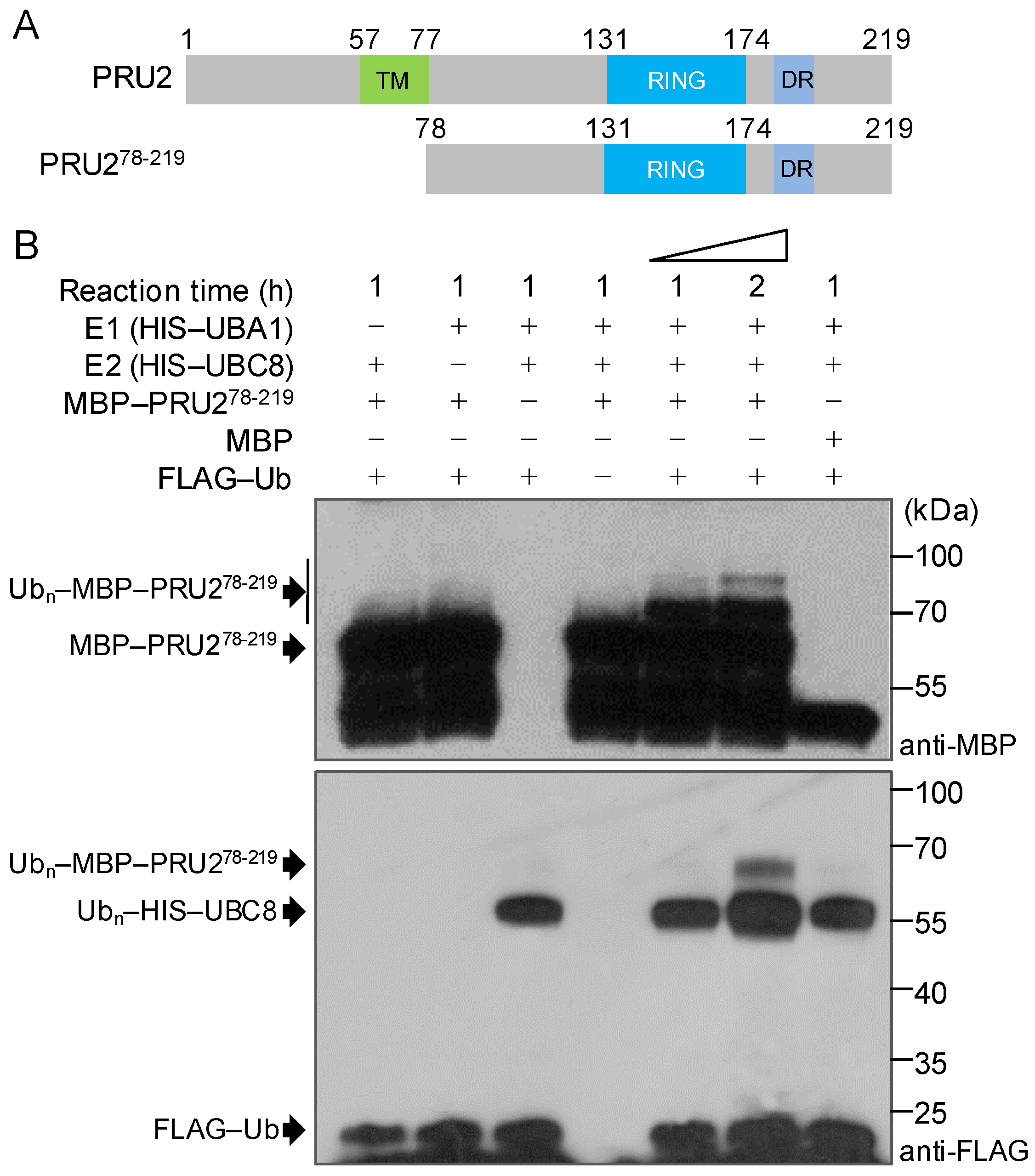
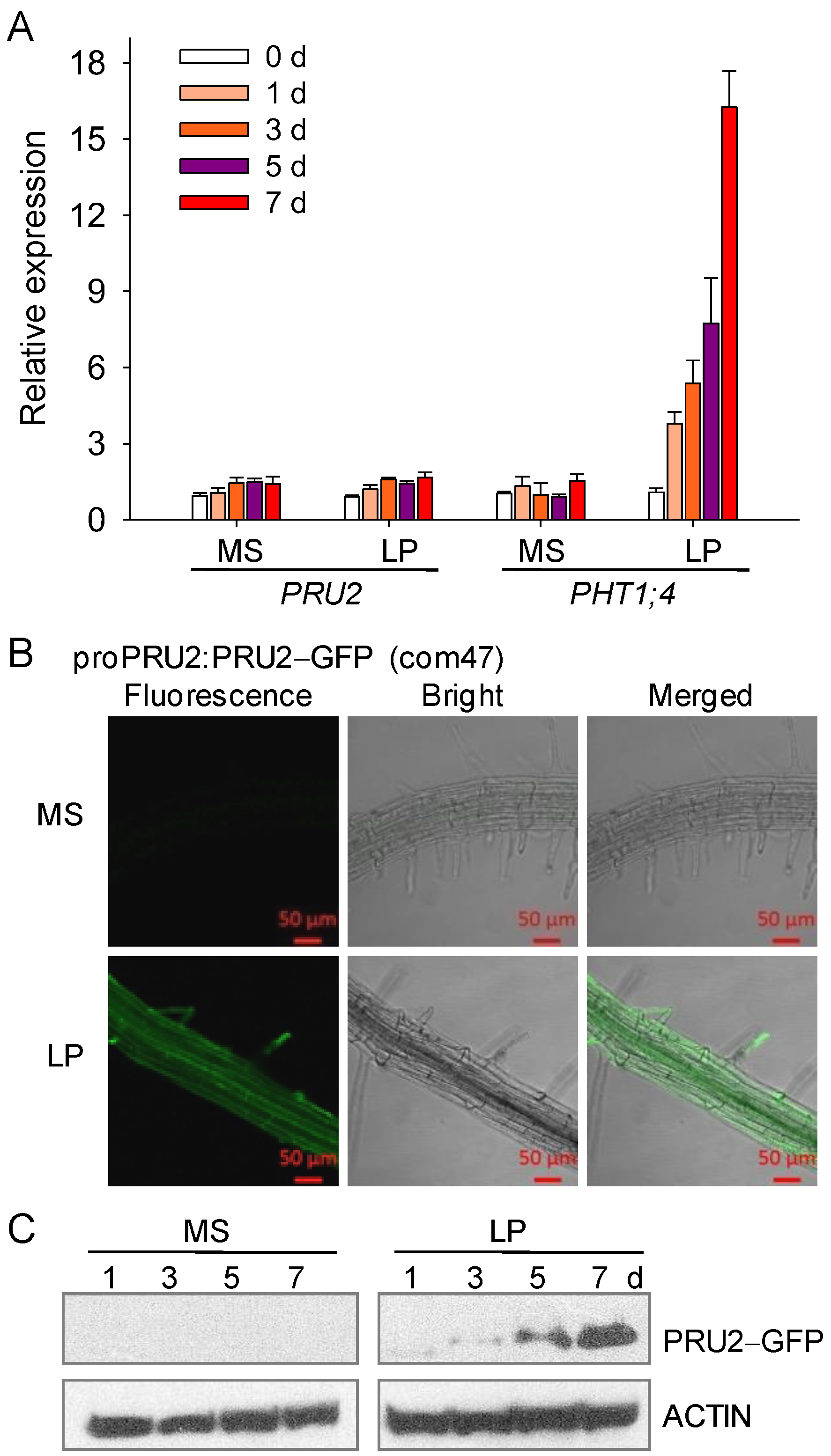
2.3. PRU2 Was a LP-Response Ubiquitin E3 Ligase
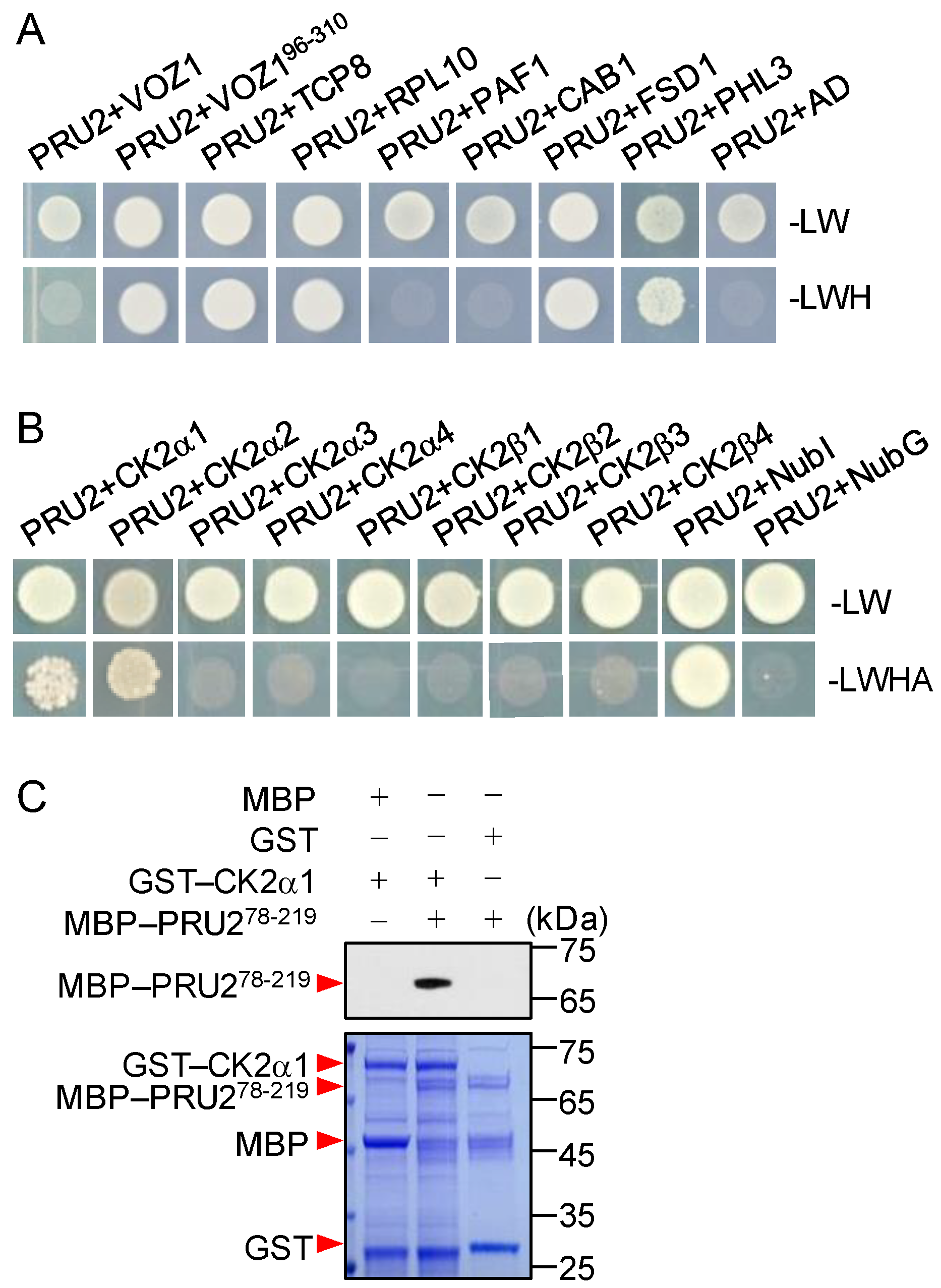
2.4. Identification Targets of PRU2
2.5. RPL10 and CK2α1 Were Targets of PRU2 under LP Stress
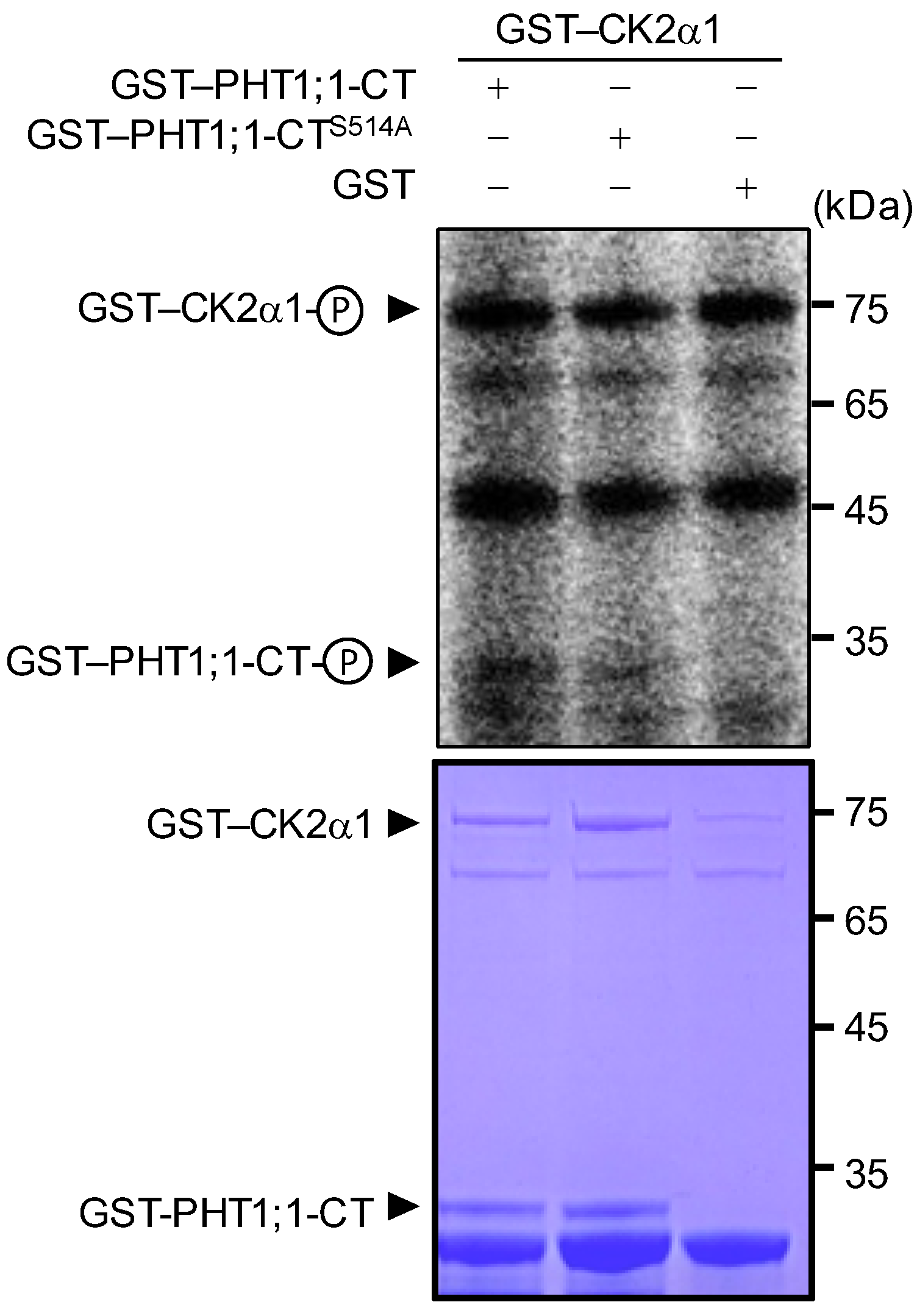
2.6. Arabidopsis CK2α1 Phosphorylated PHT1;1 at Ser-514
3. Discussion
3.1. PRU2 Was a Vital Regulator in Arabidopsis Response to Pi Starvation
3.2. PRU2 Was Involved in a Cross-Talk between the Stress and the Nutrient Deficiency
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Pi Content Measurement and 32Pi Uptake Assay
4.3. RT-qPCR Assay
4.4. In Vitro Self-Ubiquitination Assay
4.5. Yeast Two-Hybrid Assay
4.6. Protein Expression and Cell-Free Degradation Assay
4.7. In Vitro Pull-Down Assay
4.8. Phosphorylation Assay
4.9. Accession Numbers
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Raghothama, K.G. Phosphate acquisition. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 665–693. [Google Scholar] [CrossRef] [PubMed]
- Nelson, N.O.; Janke, R.R. Phosphorus sources and management in organic production systems. Horttechnology 2007, 17, 442–454. [Google Scholar] [CrossRef] [Green Version]
- Cordell, D.; Drangert, J.O.; White, S. The story of phosphorus: Global food security and food for thought. Glob. Environ. Change 2009, 19, 292–305. [Google Scholar] [CrossRef]
- Vitousek, P.M.; Naylor, R.; Crews, T.; David, M.B.; Drinkwater, L.E.; Holland, E.; Johnes, P.J.; Katzenberger, J.; Martinelli, L.A.; Matson, P.A.; et al. Agriculture. Nutrient imbalances in agricultural development. Science 2009, 324, 1519–1520. [Google Scholar] [CrossRef] [PubMed]
- Crombez, H.; Motte, H.; Beeckman, T. Tackling plant phosphate starvation by the roots. Dev. Cell 2019, 48, 599–615. [Google Scholar] [CrossRef] [Green Version]
- López-Arredondo, D.L.; Leyva-González, M.A.; González-Morales, S.I.; López-Bucio, J.; Herrera-Estrella, L. Phosphate nutrition: Improving low-phosphate tolerance in crops. Annu. Rev. Plant Biol. 2014, 65, 95–123. [Google Scholar] [CrossRef]
- Hurley, B.A.; Tran, H.T.; Marty, N.J.; Park, J.; Snedden, W.A.; Mullen, R.T.; Plaxton, W.C. The dual-targeted purple acid phosphatase isozyme AtPAP26 is essential for efficient acclimation of Arabidopsis to nutritional phosphate deprivation. Plant Physiol. 2010, 153, 1112–1122. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.S.; Li, Z.; Qian, W.Q.; Guo, W.L.; Gao, X.; Huang, L.L.; Wang, H.; Zhu, H.F.; Wu, J.W.; Wang, D.W.; et al. The Arabidopsis purple acid phosphatase AtPAP10 is predominantly associated with the root surface and plays an important role in plant tolerance to phosphate limitation. Plant Physiol. 2011, 157, 1283–1299. [Google Scholar] [CrossRef] [Green Version]
- Mudge, S.R.; Rae, A.L.; Diatloff, E.; Smith, F.W. Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J. 2002, 31, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Chang, X.J.; Ye, Y.; Xie, W.B.; Wu, P.; Lian, X.M. Comprehensive sequence and whole-life-cycle expression profile analysis of the phosphate transporter gene family in rice. Mol. Plant 2011, 4, 1105–1122. [Google Scholar] [CrossRef] [Green Version]
- Gu, M.; Chen, A.Q.; Sun, S.B.; Xu, G.H. Complex regulation of plant phosphate transporters and the gap between molecular mechanisms and practical application: What is missing? Mol. Plant 2016, 9, 396–416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, F.; Deng, M.J.; Xu, J.M.; Zhu, X.L.; Mao, C.Z. Molecular mechanisms of phosphate transport and signaling in higher plants. Semin. Cell Dev. Biol. 2018, 74, 114–122. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Y.F.; Wu, W.H. Potassium and phosphorus transport and signaling in plants. J. Integr. Plant Biol. 2021, 63, 34–52. [Google Scholar] [CrossRef] [PubMed]
- González, E.; Solano, R.; Rubio, V.; Leyva, A.; Paz-Ares, J. PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 is a plant-specific SEC12-related protein that enables the endoplasmic reticulum exit of a high-affinity phosphate transporter in Arabidopsis. Plant Cell 2005, 17, 3500–3512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bayle, V.; Arrighi, J.F.; Creff, A.; Nespoulous, C.; Vialaret, J.; Rossignol, M.; González, E.; Paz-Ares, J.; Nussaume, L. Arabidopsis thaliana high-affinity phosphate transporters exhibit multiple levels of posttranslational regulation. Plant Cell 2011, 23, 1523–1535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.Y.; Liu, Y.; Ni, J.; Wang, Y.F.; Bai, Y.H.; Shi, J.; Gan, J.; Wu, Z.C.; Wu, P. OsPHF1 regulates the plasma membrane localization of low- and high-affinity inorganic phosphate transporters and determines inorganic phosphate uptake and translocation in rice. Plant Physiol. 2011, 157, 269–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.Y.; Wang, Y.F.; Wang, F.; Yang, J.; Gao, M.X.; Li, C.Y.; Liu, Y.Y.; Liu, Y.; Yamaji, N.; Ma, J.F.; et al. The rice CK2 kinase regulates trafficking of phosphate transporters in response to phosphate levels. Plant Cell 2015, 27, 711–723. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Yang, J.; Wang, Y.; Wang, F.; Mao, W.; He, Q.; Xu, J.; Wu, Z.; Mao, C.Z. PROTEIN PHOSPHATASE95 regulates phosphate homeostasis by affecting phosphate transporter trafficking in rice. Plant Cell 2020, 32, 740–757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, H.; Shin, H.S.; Dewbre, G.R.; Harrison, M.J. Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J. 2004, 39, 629–642. [Google Scholar] [CrossRef]
- Catarecha, P.; Segura, M.D.; Franco-Zorrilla, J.M.; Garcia-Ponce, B.; Lanza, M.; Solano, R.; Paz-Ares, J.; Leyva, A. A mutant of the Arabidopsis phosphate transporter PHT1;1 displays enhanced arsenic accumulation. Plant Cell 2007, 19, 1123–1133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castrillo, G.; Sanchez-Bermejo, E.; de Lorenzo, L.; Crevillen, P.; Fraile-Escanciano, A.; Tc, M.; Mouriz, A.; Catarecha, P.; Sobrino-Plata, J.; Olsson, S.; et al. WRKY6 transcription factor restricts arsenate uptake and transposon activation in Arabidopsis. Plant Cell 2013, 25, 2944–2957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DiTusa, S.F.; Fontenot, E.B.; Wallace, R.W.; Silvers, M.A.; Steele, T.N.; Elnagar, A.H.; Dearman, K.M.; Smith, A.P. A member of the Phosphate transporter 1 (Pht1) family from the arsenic-hyperaccumulating fern Pteris vittata is a high-affinity arsenate transporter. New Phytol. 2016, 209, 762–772. [Google Scholar] [CrossRef]
- Ye, Q.; Wang, H.; Su, T.; Wu, W.H.; Chen, Y.F. The ubiquitin E3 ligase PRU1 regulates WRKY6 degradation to modulate phosphate homeostasis in response to low-Pi stress in Arabidopsis. Plant Cell 2018, 30, 1062–1076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-Bucio, J.; Cruz-Ramírez, A.; Herrera-Estrella, L. The role of nutrient availability in regulating root architecture. Curr. Opin. Plant Biol. 2003, 6, 280–287. [Google Scholar] [CrossRef]
- Devaiah, B.N.; Karthikeyan, A.S.; Raghothama, K.G. WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol. 2007, 143, 1789–1801. [Google Scholar] [CrossRef] [Green Version]
- Merlot, S.; Leonhardt, N.; Fenzi, F.; Valon, C.; Costa, M.; Piette, L.; Vavasseur, A.; Genty, B.; Boivin, K.; Müller, A.; et al. Constitutive activation of a plasma membrane H+-ATPase prevents abscisic acid-mediated stomatal closure. EMBO J. 2007, 26, 3216–3226. [Google Scholar] [CrossRef] [Green Version]
- Neumann, G.; Rmheld, V. Root excretion of carboxylic acids and protons in phosphorus-deficient plants. Plant Soil 1999, 211, 121–130. [Google Scholar] [CrossRef]
- Ryan, P.; Delhaize, E.; Jones, D. Function and mechanism of organic anion exudation from plant roots. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 527–560. [Google Scholar] [CrossRef]
- Kim, S.J.; Kim, W.T. Suppression of Arabidopsis RING E3 ubiquitin ligase AtATL78 increases tolerance to cold stress and decreases tolerance to drought stress. FEBS Lett. 2013, 587, 2584–2590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Portolés, S.; Más, P. The functional interplay between protein kinase CK2 and CCA1 transcriptional activity is essential for clock temperature compensation in Arabidopsis. PLoS Genet. 2010, 6, e1001201. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Zheng, Z.; Song, L.; Liu, D. Functional characterization of Arabidopsis PHL4 in plant response to phosphate starvation. Front Plant Sci. 2018, 9, 1432. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Ren, H.; Gu, M.; Zhao, J.; Sun, S.; Zhang, X.; Chen, J.; Wu, P.; Xu, G. The phosphate transporter gene OsPht1;8 is involved in phosphate homeostasis in rice. Plant Physiol. 2011, 156, 1164–1175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, F.; Cui, P.J.; Tian, Y.; Huang, Y.; Wang, H.F.; Liu, F.; Chen, Y.F. Maize ZmPT7 regulates Pi uptake and redistribution which is modulated by phosphorylation. Plant Biotechnol. J. 2020, 18, 2406–2419. [Google Scholar] [CrossRef]
- Devaiah, B.N.; Madhuvanthi, R.; Karthikeyan, A.S.; Raghothama, K.G. Phosphate starvation responses and gibberellic acid biosynthesis are regulated by the MYB62 transcription factor in Arabidopsis. Mol. Plant 2009, 2, 43–58. [Google Scholar] [CrossRef] [Green Version]
- Bustos, R.; Castrillo, G.; Linhares, F.; Puga, M.I.; Rubio, V.; Pérez-Pérez, J.; Solano, R.; Leyva, A.; Paz-Ares, J. A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet. 2010, 6, e1001102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Xu, Q.; Kong, Y.H.; Chen, Y.; Duan, J.Y.; Wu, W.H.; Chen, Y.F. Arabidopsis WRKY45 transcription factor activates PHOSPHATE TRANSPORTER1;1 expression in response to phosphate starvation. Plant Physiol. 2014, 164, 2020–2029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Wang, H.F.; Chen, Y.; Sun, M.M.; Wang, Y.; Chen, Y.F. The transcription factor NIGT1.2 modulates both phosphate uptake and nitrate influx during phosphate starvation in Arabidopsis and maize. Plant Cell 2020, 32, 3519–3534. [Google Scholar] [CrossRef]
- Suh, J.Y.; Kim, S.J.; Oh, T.R.; Cho, S.K.; Yang, S.W.; Kim, W.T. Arabidopsis Tóxicos en Levadura78 (AtATL78) mediates ABA-dependent ROS signaling in response to drought stress. Biochem. Biophys. Res. Commun. 2016, 469, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Morales, E.; Aguilar-Hernández, V.; Aguilar-Henonin, L.; Guzmán, P. Molecular basis for neofunctionalization of duplicated E3 ubiquitin ligases underlying adaptation to drought tolerance in Arabidopsis thaliana. Plant J. 2020, 104, 474–492. [Google Scholar] [CrossRef]
- Monteclaro, F.S.; Vogt, P.K. A Jun-binding protein related to a putative tumor suppressor. Proc. Natl. Acad. Sci. USA 1993, 90, 6726–6730. [Google Scholar] [CrossRef] [Green Version]
- Oh, H.S.; Kwon, H.; Sun, S.K.; Yang, C.H. QM, a putative tumor suppressor, regulates proto-oncogene c-yes. J. Biol. Chem. 2002, 277, 36489–36498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zorzatto, C.; Machado, J.P.; Lopes, K.V.; Nascimento, K.J.; Pereira, W.A.; Brustolini, O.J.; Reis, P.A.; Calil, I.P.; Deguchi, M.; Sachetto-Martins, G.; et al. NIK1-mediated translation suppression functions as a plant antiviral immunity mechanism. Nature 2015, 520, 679–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, T.; Xu, Q.; Zhang, F.C.; Chen, Y.; Li, L.Q.; Wu, W.H.; Chen, Y.F. WRKY42 modulates phosphate homeostasis through regulating phosphate translocation and acquisition in Arabidopsis. Plant Physiol. 2015, 167, 1579–1591. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Feng, C.Z.; Ye, Q.; Wu, W.H.; Chen, Y.F. Arabidopsis WRKY6 transcription factor acts as a positive regulator of abscisic acid signaling during seed germination and early seedling development. PLoS Genet. 2016, 12, e1005833. [Google Scholar] [CrossRef] [Green Version]
- Ryu, M.Y.; Cho, S.K.; Kim, W.T. The Arabidopsis C3H2C3-type RING E3 ubiquitin ligase AtAIRP1 is a positive regulator of an abscisic acid dependent response to drought stress. Plant Physiol. 2010, 154, 1983–1997. [Google Scholar] [CrossRef] [Green Version]


| Gene | Full Name of the Gene | Gene Number |
|---|---|---|
| VOZ1 | VASCULAR PLANT ONE ZINC FINGER PROTEIN 1 | AT1G28520 |
| FSD1 | FE SUPEROXIDE DISMUTASE 1 | AT4G25100 |
| CAB1 | CHLOROPHYLL A/B BINDING PROTEIN 1 | AT1G29930 |
| TCP8 | TCP DOMAIN PROTEIN 8 | AT1G58100 |
| RPL10 | RIBOSOMAL PROTEIN L10 | AT1G14320 |
| PAF1 | PROTEASOME ALPHA SUBUNIT F1 | AT5G42790 |
| PHL3 | PHR1-LIKE 3 | AT4G13640 |
| CK2α1 | CASEIN KINASE II, ALPHA CHAIN 1 | AT5G67380 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, M.-M.; Tian, Y.; Chun, M.; Chen, Y.-F. The Ubiquitin E3 Ligase PRU2 Modulates Phosphate Uptake in Arabidopsis. Int. J. Mol. Sci. 2022, 23, 2273. https://doi.org/10.3390/ijms23042273
Sun M-M, Tian Y, Chun M, Chen Y-F. The Ubiquitin E3 Ligase PRU2 Modulates Phosphate Uptake in Arabidopsis. International Journal of Molecular Sciences. 2022; 23(4):2273. https://doi.org/10.3390/ijms23042273
Chicago/Turabian StyleSun, Mi-Mi, Yan Tian, Mei Chun, and Yi-Fang Chen. 2022. "The Ubiquitin E3 Ligase PRU2 Modulates Phosphate Uptake in Arabidopsis" International Journal of Molecular Sciences 23, no. 4: 2273. https://doi.org/10.3390/ijms23042273






