T1121G Point Mutation in the Mitochondrial Gene COX1 Suppresses a Null Mutation in ATP23 Required for the Assembly of Yeast Mitochondrial ATP Synthase
Abstract
:1. Introduction
2. Results
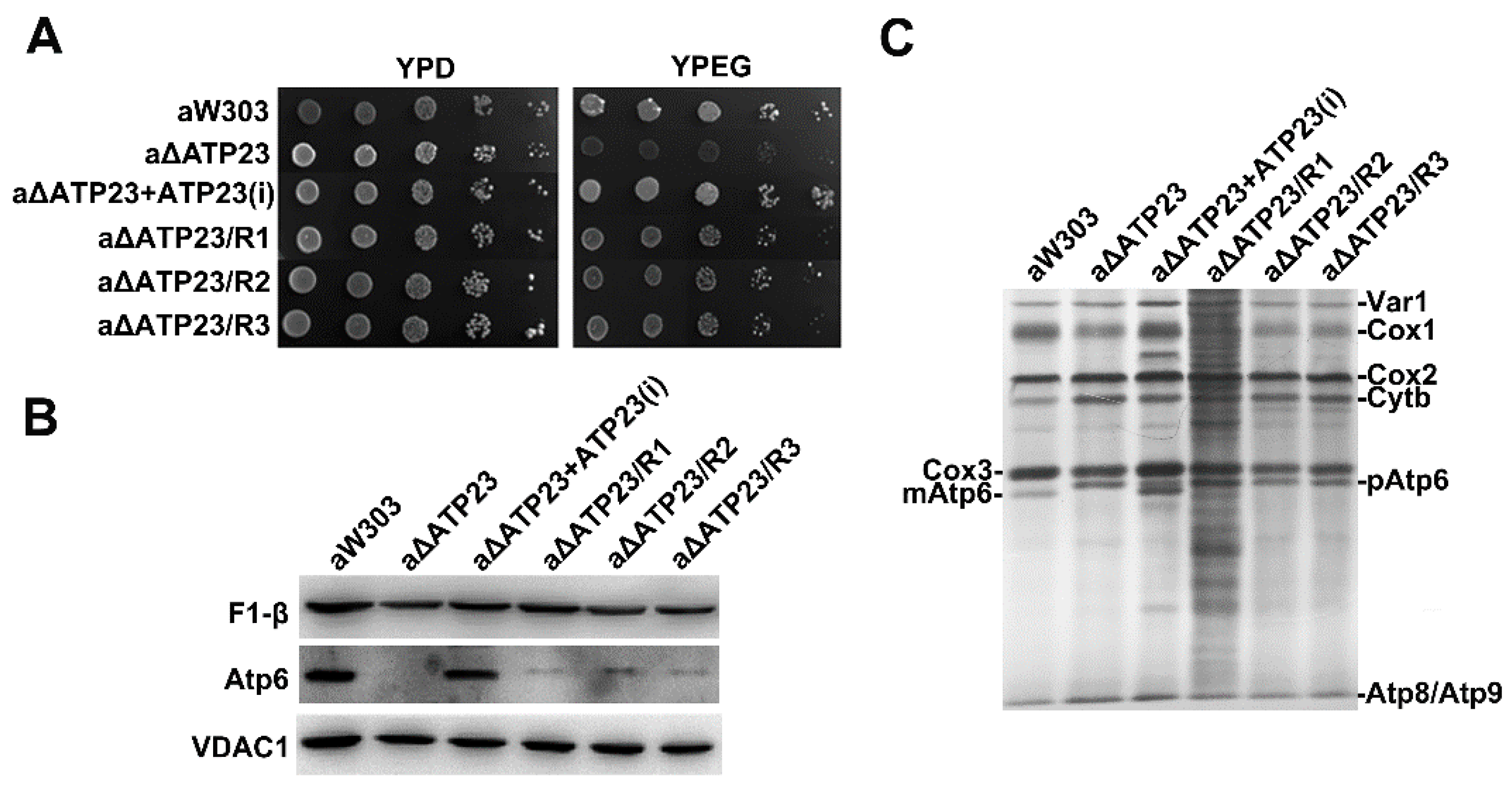
2.1. Isolation and Characterization of Revertants of Atp23 Null Mutant
2.2. The Suppressor of the Revertants Was Localized to T1121G Point Mutation of Mitochondrial Gene COX1 Coding Sequence
2.3. The Suppressor Was Verified by Transformation of Exogenous Hybrid COX1 Plasmid in Atp23 Null Mutant
2.4. Cox1 p.Val374Gly Mutation Did Not Affect the Tertiary Structure of Cox1 Protein
2.5. Stability of Atp6 Increased in the Revertants
3. Discussion
4. Materials and Methods
4.1. Yeast Strains and Growth Media
4.2. Serial Dilution Growth
4.3. Preparation of Yeast Mitochondria and ATPase Activity Assays
4.4. In Vivo and Chase Labeling of Mitochondrial Gene Products
4.5. Mapping of Mitochondrial Suppressors
4.6. Preparation of Mitochondrial DNA for Sequencing
4.7. Construction of Hybrid COX1-HA Plasmid
4.8. Miscellaneous Procedures
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marsy, S.; Frachon, P.; Dujardin, G.; Lombes, A.; Lemaire, C. Respiratory mutations lead to different pleiotropic effects on OXPHOS complexes in yeast and in human cells. FEBS Lett. 2008, 582, 3489–3493. [Google Scholar] [CrossRef] [Green Version]
- Franco, L.V.R.; Su, C.H.; Tzagoloff, A. Modular assembly of yeast mitochondrial ATP synthase and cytochrome oxidase. Biol. Chem. 2020, 401, 835–853. [Google Scholar] [CrossRef]
- Song, J.; Pfanner, N.; Becker, T. Assembling the mitochondrial ATP synthase. Proc. Natl. Acad. Sci. USA 2018, 115, 2850–2852. [Google Scholar] [CrossRef] [Green Version]
- Zeng, X.; Neupert, W.; Tzagoloff, A. The metalloprotease encoded by ATP23 has a dual function in processing and assembly of subunit 6 of mitochondrial ATPase. Mol. Biol. Cell. 2007, 18, 617–626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osman, C.; Wilmes, C.; Tatswta, T.; Langer, T. Prohibitins interact genetically with Atp23, a novel processing peptidase and chaperone for the F1Fo-ATP synthase. Mol. Biol. Cell. 2007, 18, 627–635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Villegas, R.; Camacho-Villasana, Y.; Shingú-Vázquez, M.Á.; Cabrera-Orefice, A.; Uribe-Carvajal, S.; Fox, T.D.; Pérez-Martínez, X. The Cox1 C-terminal domain is a central regulator of cytochrome c oxidase biogenesis in yeast mitochondria. J. Biol. Chem. 2017, 292, 10912–10925. [Google Scholar] [CrossRef] [Green Version]
- Soto, I.C.; Fontanesi, F.; Liu, J.; Barrientos, A. Biogenesis and assembly of eukaryotic cytochrome c oxidase catalytic core. Biochim. Biophys. Acta 2012, 1817, 883–897. [Google Scholar] [CrossRef] [Green Version]
- Barrientos, A.; Zambrano, A.; Tzagoloff, A. Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. EMBO J. 2004, 23, 3472–3482. [Google Scholar] [CrossRef] [Green Version]
- Su, C.H.; McStay, G.P.; Tzagoloff, A. The Cox3p assembly module of yeast cytochrome oxidase. Mol. Biol. Cell. 2014, 25, 965–976. [Google Scholar] [CrossRef]
- Williams, S.L.; Valnot, I.; Rustin, P.; Taanman, J.W. Cytochrome c oxidase subassemblies in fibroblast cultures from patients carrying mutations in COX10, SCO1, or SURF1. J. Biol. Chem. 2004, 279, 7462–7469. [Google Scholar] [CrossRef] [Green Version]
- Simon, M.; Faye, G. Organization and processing of the mitochondrial oxi3/oli2 multigenic transcript in yeast. Mol. Gen. Genet. 1984, 196, 266–274. [Google Scholar] [CrossRef] [PubMed]
- Soto, I.C.; Fontanesi, F.; Valledor, M.; Horn, D.; Singh, R.; Barrientos, A. Synthesis of cytochrome c oxidase subunit 1 is translationally downregulated in the absence of functional F1Fo-ATP synthase. Biochim. Biophys. Acta 2009, 1793, 1776–1786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marzuki, S.; Watkins, L.C.; Choo, W.M. Mitochondrial H+-ATPase in mutants of Saccharomyces cerevisiae with defective subunit 8 of the enzyme complex. Biochim. Biophys. Acta 1989, 975, 222–230. [Google Scholar] [CrossRef]
- Paul, M.F.; Velours, J.; Arselin de Chateaubodeau, G.; Aigle, M.; Guerin, B. The role of subunit 4, a nuclear-encoded protein of the Fo sector of yeast mitochondrial ATP synthase, in the assembly of the whole complex. Eur. J. Biochem. 1989, 185, 163–171. [Google Scholar] [CrossRef]
- Spannagel, C.; Vaillier, J.; Arselin, G.; Graves, P.V.; Velours, J. The subunit f of mitochondrial yeast ATP synthase--characterization of the protein and disruption of the structural gene ATP17. Eur. J. Biochem. 1997, 247, 1111–1117. [Google Scholar] [CrossRef]
- Bietenhader, M.; Martos, A.; Tetaud, E.; Aiyar, R.S.; Sellem, C.H.; Kucharczyk, R.; Clauder-Münster, S.; Giraud, M.F.; Godard, F.; Salin, B.; et al. Experimental relocation of the mitochondrial ATP9 gene to the nucleus reveals forces underlying mitochondrial genome evolution. PLoS Genet. 2012, 8, e1002876. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Rak, M.; Tetaud, E.; Godard, F.; Sardin, E.; Bouhier, M.; Gombeau, K.; Caetano-Anollés, D.; Salin, B.; Chen, H.; et al. Deregulating mitochondrial metabolite and ion transport has beneficial effects in yeast and human cellular models for NARP syndrome. Hum. Mol. Genet. 2019, 28, 3792–3804. [Google Scholar] [CrossRef]
- Boyle, G.M.; Roucou, X.; Nagley, P.; Devenish, R.J.; Prescott, M. Identification of subunit g of yeast mitochondrial F1Fo-ATP synthase, a protein required for maximal activity of cytochrome c oxidase. Eur. J. Biochem. 1999, 262, 315–323. [Google Scholar] [CrossRef] [Green Version]
- Rak, M.; Tetaud, E.; Duvezin Caubet, S.; Ezkurdia, E.; Bietenhader, M.; Rytka, J.; di Rago, J.P. A yeast model of the neurogenic ataxia retinitis pigmentosa (NARP) T8993G mutation in the mitochondrial ATP synthase-6 gene. J. Biol. Chem. 2007, 282, 34039–34047. [Google Scholar] [CrossRef] [Green Version]
- Rak, M.; Tetaud, E.; Godard, F.; Sagot, I.; Salin, B.; Duvezin-Caubet, S.; Slonimski, P.P.; Rytka, J.; di Rago, J.P. Yeast cells lacking the mitochondrial gene encoding the ATP synthase subunit 6 exhibit a selective loss of complex IV and unusual mitochondrial morphology. J. Biol. Chem. 2007, 282, 10853–10864. [Google Scholar] [CrossRef] [Green Version]
- Su, C.H.; McStay, G.P.; Tzagoloff, A. Assembly of the rotor component of yeast mitochondrial ATP synthase is enhanced when Atp9p is supplied by Atp9p-Cox6p complexes. J. Biol. Chem. 2014, 289, 31605–31616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franco, L.V.R.; Su, C.H.; Burnett, J.; Teixeira, L.S.; Tzagoloff, A. Atco, a yeast mitochondrial complex of Atp9 and Cox6, is an assembly intermediate of the ATP synthase. PLoS ONE 2020, 15, e0233177. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Ding, Y.; Shang, X.; Zhao, T.; Lu, S.; Tian, J.; Weng, J.; Zeng, X. Atp23 and Atp10p coordinate to regulate the assembly of yeast mitochondrial ATP synthase. FASEB J. 2021, 35, 21538. [Google Scholar]
- Tzagoloff, A.; Barrientos, A.; Neupert, W.; Herrmann, J.M. Atp10p assists assembly of Atp6p into the Fo unit of the yeast mitochondrial ATPase. J. Biol. Chem. 2004, 279, 19775–19780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paul, M.F.; Barrientos, A.; Tzagoloff, A. A single amino acid change in subunit 6 of the yeast mitochondrial ATPase suppresses a null mutation in ATP10. J. Biol. Chem. 2000, 275, 29238–29243. [Google Scholar] [CrossRef] [Green Version]
- Barrientos, A.; Korr, D.; Tzagoloff, A. Shy1p is necessary for full expression of mitochondrial COX1 in the yeast model of Leigh’s syndrome. EMBO J. 2002, 21, 43–52. [Google Scholar] [CrossRef]
- Malkamäki, A.; Meunier, B.; Reidelbach, M.; Rich, P.R.; Sharma, V. The H channel is not a proton transfer path in yeast cytochrome c oxidase. Biochim. Biophys. Acta Bioenerg. 2019, 1860, 717–723. [Google Scholar] [CrossRef]
- Khalimonchuk, O.; Bestwick, M.; Meunier, B.; Watts, T.C.; Winge, D.R. Formation of the redox cofactor centers during Cox1 maturation in yeast cytochrome oxidase. Mol. Cell. Biol. 2010, 30, 1004–1017. [Google Scholar] [CrossRef] [Green Version]
- Meunier, B.; Rich, P.R. Second-site reversion analysis is not a reliable method to determine distances in membrane proteins: An assessment using mutations in yeast cytochrome c oxidase subunits I and II. J. Mol. Biol. 1998, 283, 727–730. [Google Scholar] [CrossRef]
- Bruno, C.; Martinuzzi, A.; Tang, Y.; Andreu, A.L.; Pallotti, F.; Bonilla, E.; Shanske, S.; Fu, J.; Sue, C.M.; Angelini, C.; et al. A stop-codon mutation in the human mtDNA cytochrome c oxidase I gene disrupts the functional structure of complex IV. Am. J. Hum. Genet. 1999, 65, 611–620. [Google Scholar] [CrossRef] [Green Version]
- Afkhami, E.; Heidari, M.M.; Khatami, M.; Ghadamyari, F.; Dianatpour, S. Detection of novel mitochondrial mutations in cytochrome c oxidase subunit 1 (COX1) in patients with familial adenomatous polyposis (FAP). Clin. Transl. Oncol. 2020, 22, 908–918. [Google Scholar] [CrossRef] [PubMed]
- Rahman, S.; Taanman, J.W.; Cooper, J.M.; Nelson, I.; Hargreaves, I.; Meunier, B.; Hanna, M.G.; García, J.J.; Capaldi, R.A.; Lake, B.D.; et al. A missense mutation of cytochrome oxidase subunit II causes defective assembly and myopathy. Am. J. Hum. Genet. 1999, 65, 1030–1039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keightley, J.A.; Hoffbuhr, K.C.; Burton, M.D.; Salas, V.M.; Johnston, W.S.; Penn, A.M.; Buist, N.R.; Kennaway, N.G. A microdeletion in cytochrome c oxidase (COX) subunit III associated with COX deficiency and recurrent myoglobinuria. Nat. Genet. 1996, 12, 410–416. [Google Scholar] [CrossRef] [PubMed]
- Hanna, M.G.; Nelson, I.P.; Rahman, S.; Lane, R.J.; Land, J.; Heales, S.; Cooper, M.J.; Schapira, A.H.; Morgan-Hughes, J.A.; Wood, N.W. Cytochrome c oxidase deficiency associated with the first stop-codon point mutation in human mtDNA. Am. J. Hum. Genet. 1998, 63, 29–36. [Google Scholar] [CrossRef] [Green Version]
- Cwerman-Thibault, H.; Augustin, S.; Lechauve, C.; Ayache, J.; Ellouze, S.; Sahel, J.A.; Corral-Debrinski, M. Nuclear expression of mitochondrial ND4 leads to the protein assembling in complex I and prevents optic atrophy and visual loss. Mol. Ther. Methods. Clin. Dev. 2015, 2, 15003. [Google Scholar] [CrossRef]
- Tzagoloff, A.; Foury, F.; Akai, A. Assembly of the mitochondrial membrane system. XVIII. Genetic loci on mitochondrial DNA involved in cytochrome b biosynthesis. Mol. Gen. Genet. 1976, 149, 33–42. [Google Scholar] [CrossRef]
- Foury, F.; Tzagoloff, A. Localization on mitochondria1 DNA of mutations leading to a loss of rutamycin-sensitive adenosine triphosphatase. Eur. J. Biochem. 1976, 68, 113–119. [Google Scholar] [CrossRef]
- McStay, G.P.; Su, C.H.; Tzagoloff, A. Modular assembly of yeast cytochrome oxidase. Mol. Biol. Cell. 2013, 24, 440–452. [Google Scholar] [CrossRef]
- Faye, G.; Kujawa, C.; Fukuhara, H. Physical and genetic organization of petite and grande yeast mitochondrial DNA. IV. In vivo transcription products of mitochondrial DNA and localization of 23S ribosomal RNA in petite mutants of Saccharomyces cerevisiae. J. Mol. Biol. 1974, 88, 185–203. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- King, E.J. The colorimetric determination of phosphorus. Biochem. J. 1932, 26, 292–297. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Slonimski, P.P.; Tzagoloff, A. Localization in yeast mitochondrial DNA of mutations expressed in a deficiency of cytochrome oxidase and/or coenzyme QH2-Cytochrome c reductase. Eur. J. Biochem. 1976, 61, 27–41. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Schiestl, R.H.; Gietz, R.D. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 1989, 16, 339–346. [Google Scholar] [CrossRef]





| ATPase Activity (μmol/mg/min) | ||||
|---|---|---|---|---|
| Strain | % ρ+ | −Oligomycin | +Oligomycin | Inhibition (%) |
| aW303 | >99 | 4.30 | 0.57 | 86.7 |
| aΔATP23 | 22 | 1.54 | 1.26 | 18.2 |
| aΔATP23 + ATP23(i) | >99 | 4.19 | 0.75 | 82.1 |
| aΔATP23/R1 | 81 | 3.03 | 1.59 | 47.5 |
| aΔATP23/R2 | 74 | 2.80 | 1.74 | 37.9 |
| aΔATP23/R3 | 71 | 1.79 | 0.92 | 46.9 |
| ATPase Activity (μmol/mg/min) | ||||
|---|---|---|---|---|
| Strain | % ρ+ | −Oligomycin | +Oligomycin | Inhibition (%) |
| aW303 | >99 | 4.16 | 0.78 | 81.3 |
| aΔATP23 | 25 | 1.58 | 1.35 | 16.5 |
| aΔATP23 + ATP23(i) | >99 | 4.01 | 0.74 | 81.5 |
| aΔATP23 + COX1-HA (T1121T) (e) | 27 | 1.77 | 1.43 | 19.2 |
| aΔATP23 + COX1-HA (T1121G) (e) | 87 | 2.82 | 1.57 | 44.3 |
| Strain | Genotype | Source |
|---|---|---|
| aW303 | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 | Dr. Rodney Rothstein a |
| aW303ΔATP23 | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 atp23::HIS3 | [4] |
| W303ΔATP23 | MATα ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 atp23::HIS3 | [4] |
| aW303ΔATP23 + ATP23(i) | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 atp23::HIS3 ATP23-HA | [4] |
| aW303ΔATP23/R1, -R2, -R3 | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 atp23::HIS3 (ρ+supR) | This study |
| aW303ΔATP23R1/ρ−supR | MATa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 atp23::HIS3 (ρ−supR) | This study |
| M7-40/A1 | MATα ade1 cob | [36] |
| M28-82 | MATα met6 Atp6 | [37] |
| MR6 | ΜAΤa ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 arg8::HIS3 | [38] |
| MRSI0ΔCOX1 | MATα ade1 his-1,15 leu2-3,112 trp1-1 ura3-1 arg8::HIS3 intronless cox1::ARG8m | [38] |
| MRSΔATP8 | MATα ade2-1 leu2-3,112 trp1-1 ura3-1 arg8::URA3 atp8::ARG8m | This study |
| MRSΔATP9 | MATα leu2Δ ura3-52 ade2-101 arg8::HIS3 atp9::ARG8m | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, G.; Zhao, T.; Lu, S.; Weng, J.; Zeng, X. T1121G Point Mutation in the Mitochondrial Gene COX1 Suppresses a Null Mutation in ATP23 Required for the Assembly of Yeast Mitochondrial ATP Synthase. Int. J. Mol. Sci. 2022, 23, 2327. https://doi.org/10.3390/ijms23042327
Yang G, Zhao T, Lu S, Weng J, Zeng X. T1121G Point Mutation in the Mitochondrial Gene COX1 Suppresses a Null Mutation in ATP23 Required for the Assembly of Yeast Mitochondrial ATP Synthase. International Journal of Molecular Sciences. 2022; 23(4):2327. https://doi.org/10.3390/ijms23042327
Chicago/Turabian StyleYang, Guangying, Tong Zhao, Shan Lu, Jun Weng, and Xiaomei Zeng. 2022. "T1121G Point Mutation in the Mitochondrial Gene COX1 Suppresses a Null Mutation in ATP23 Required for the Assembly of Yeast Mitochondrial ATP Synthase" International Journal of Molecular Sciences 23, no. 4: 2327. https://doi.org/10.3390/ijms23042327
APA StyleYang, G., Zhao, T., Lu, S., Weng, J., & Zeng, X. (2022). T1121G Point Mutation in the Mitochondrial Gene COX1 Suppresses a Null Mutation in ATP23 Required for the Assembly of Yeast Mitochondrial ATP Synthase. International Journal of Molecular Sciences, 23(4), 2327. https://doi.org/10.3390/ijms23042327





