MYH10 Governs Adipocyte Function and Adipogenesis through Its Interaction with GLUT4
Abstract
:1. Introduction
2. Results
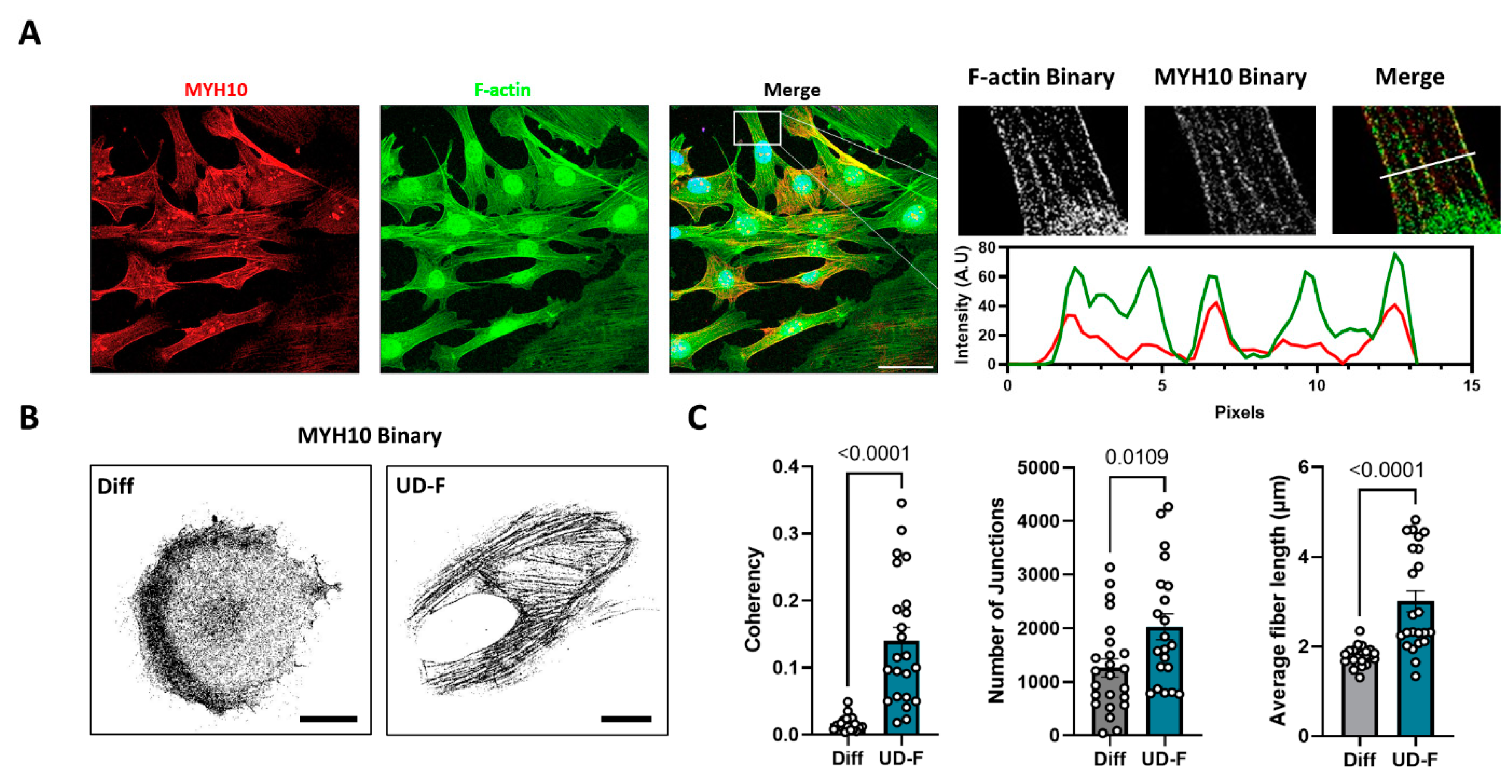
2.1. MYH10 Distribution Is Altered during Adipogenesis in 3T3-L1 Cells
2.2. MYH10 Knockdown Model
2.3. MYH10 KD Affects Cell Motility and Migration
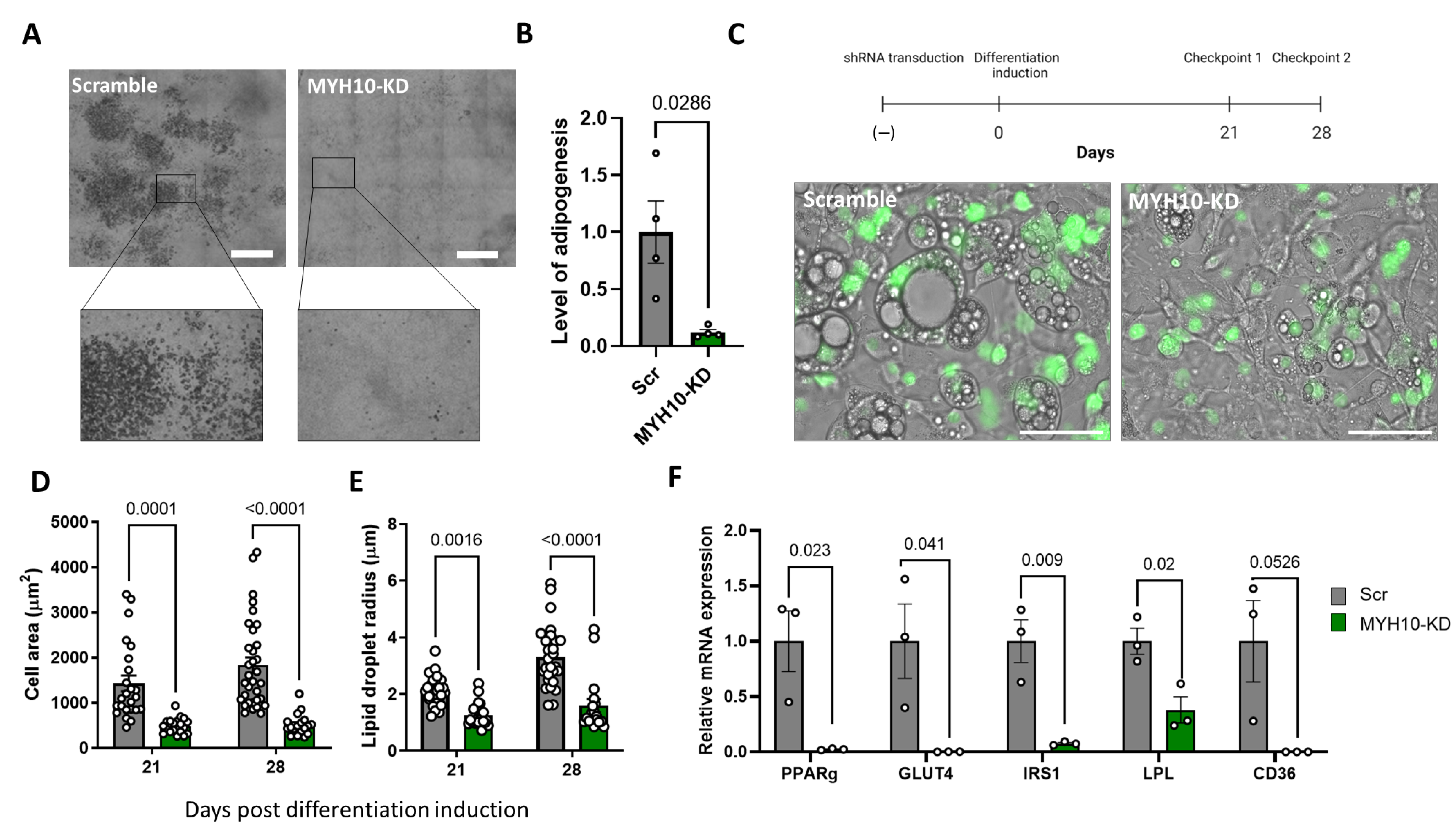
2.4. MYH10 Knockdown Affects Adipogenesis
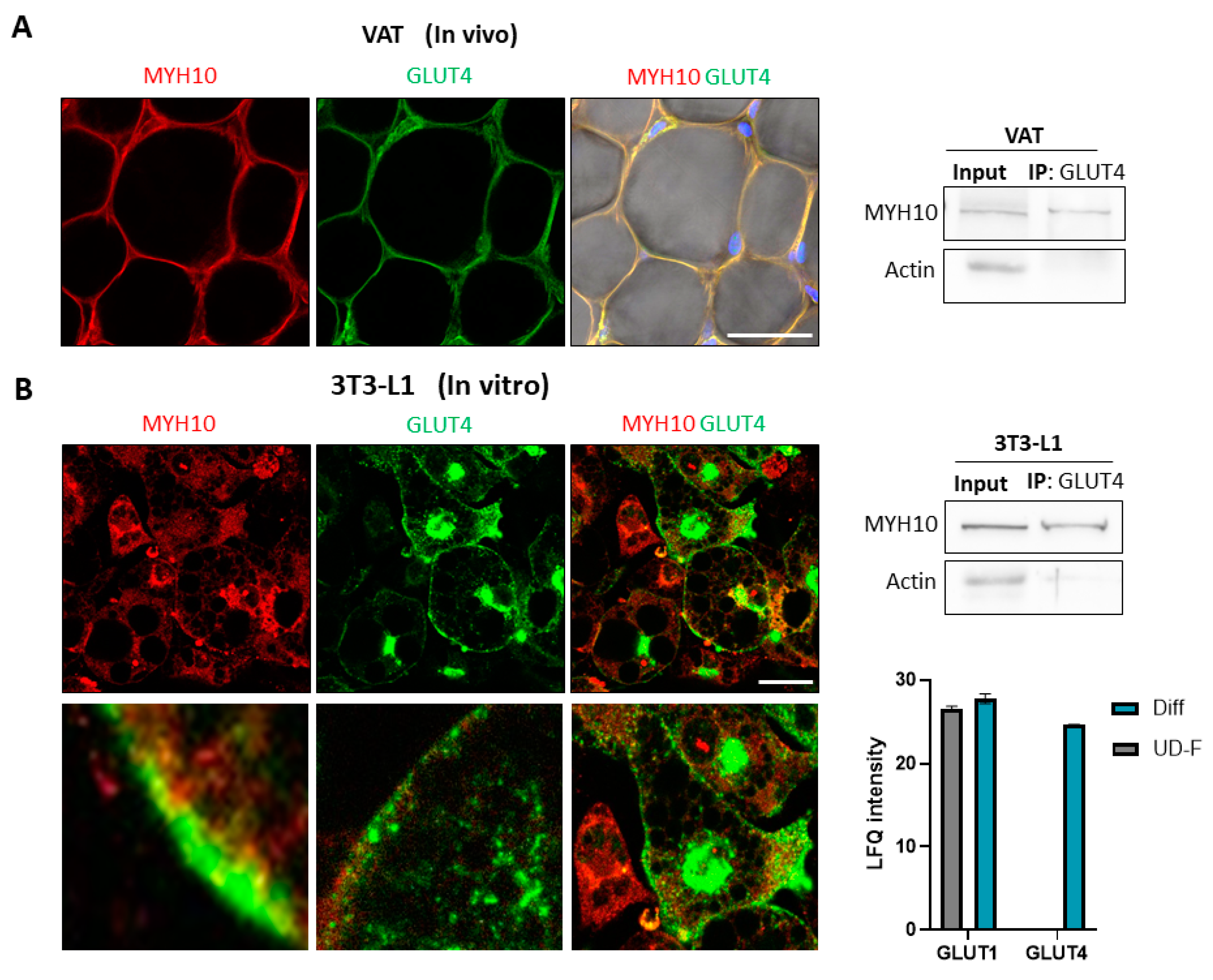
2.5. Co-Expression of Glucose Transporter 4 (GLUT4) and MYH10
2.6. MYH10 Colocalization and Interaction with GLUT4 Is Induced by Insulin
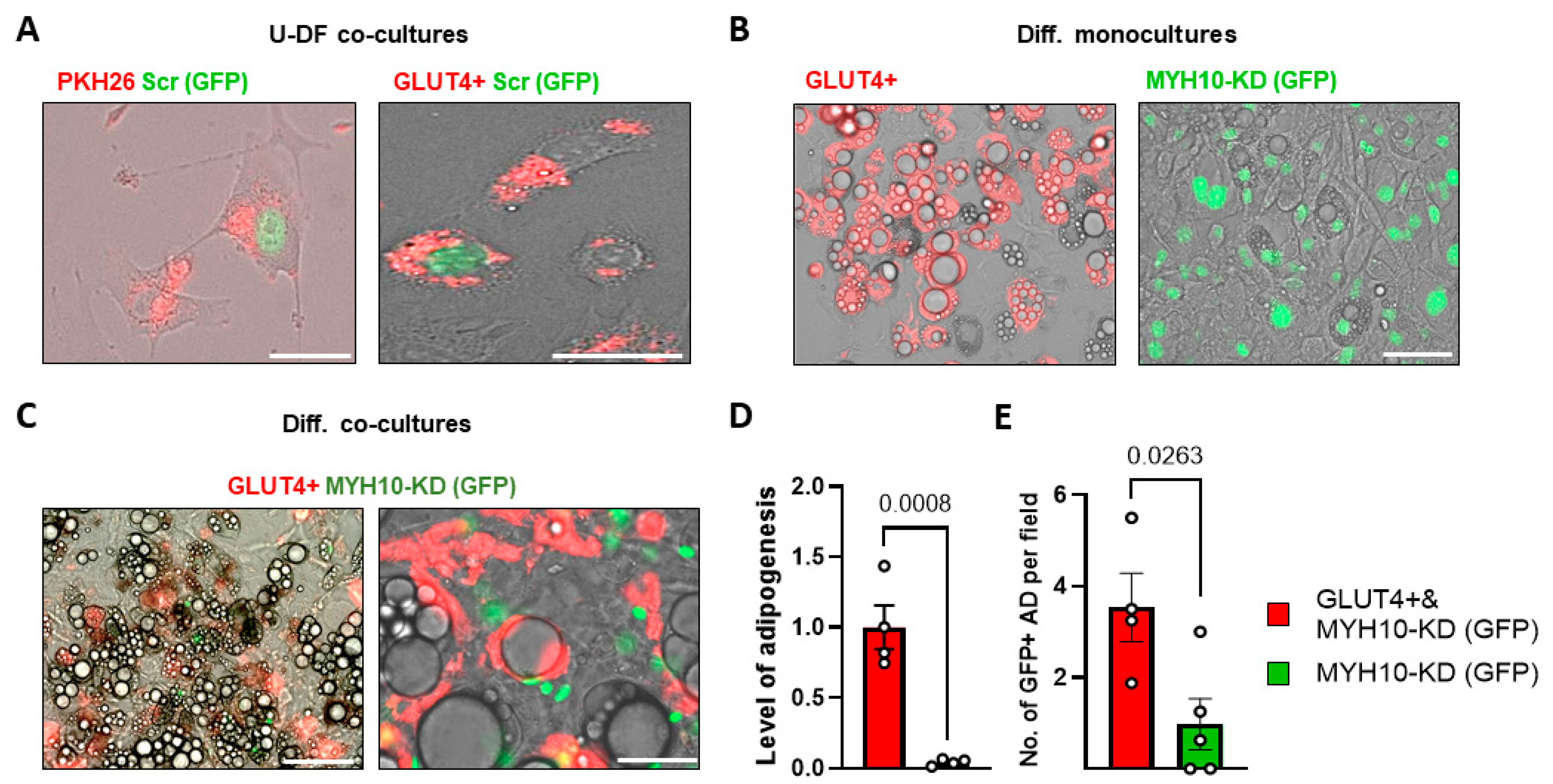
2.7. GLUT4+ Shuttling Can Restore the Adipogenic Capacity of MYH10 KD Cells
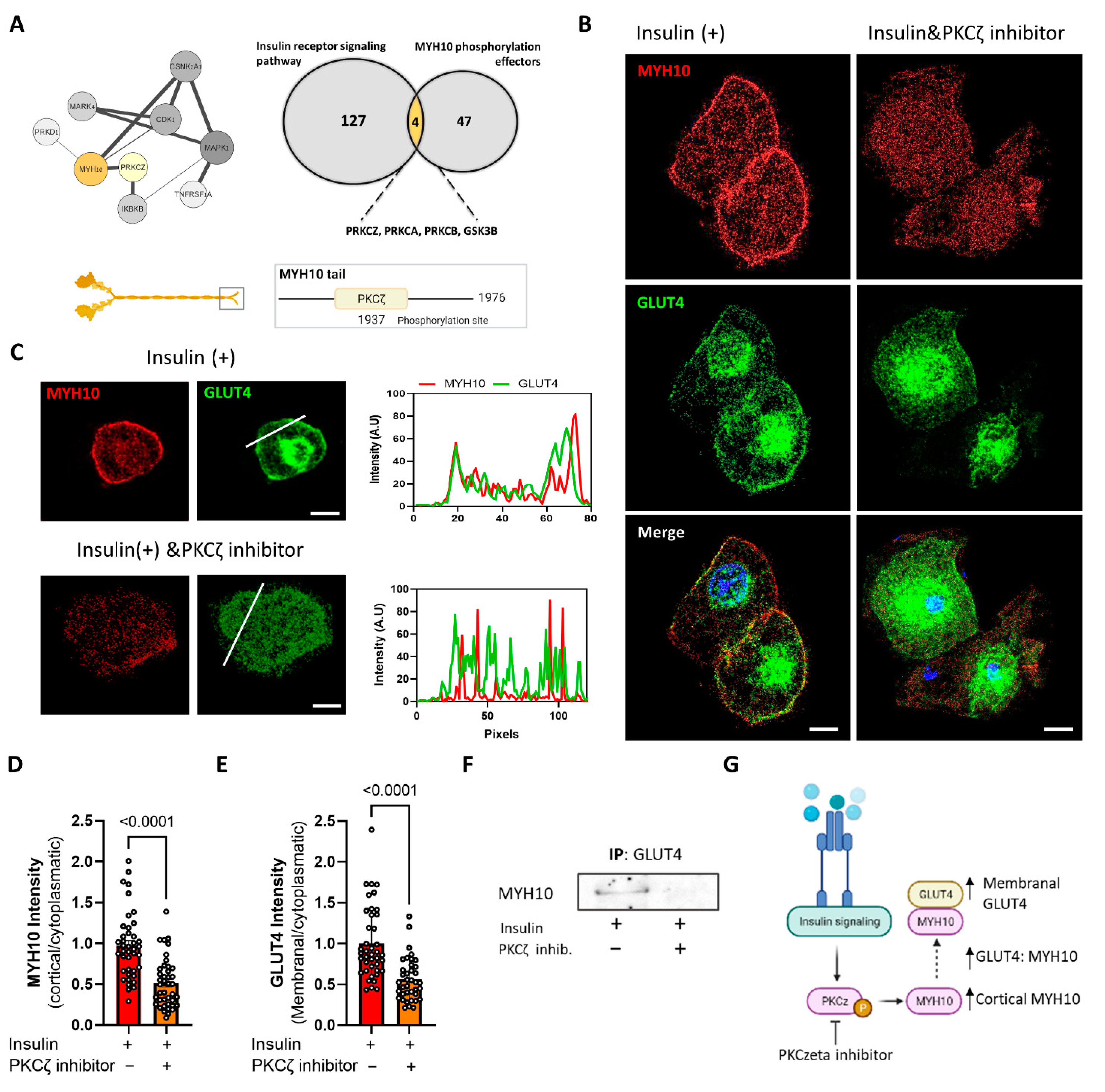
2.8. MYH10 Interaction with GLUT4 Post Insulin Induction Is Mediated by PKCζ
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Cell Lines
4.3. Lentivirus Production and Transduction
4.4. Co-Culturing
4.5. Immunofluorescence Staining
4.6. Whole-Mount Staining
4.7. Image Processing and Analysis Tools
4.8. Live Microscopy
4.9. Adipogenesis Assays
4.10. Immunoblotting
4.11. Mass Spectrometry
4.12. RNA Isolation and qPCR
4.13. Bioinformatics Pathway Analysis
4.14. Statistical Analysis
4.15. Schematic Illustrations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tang, Q.Q.; Lane, M.D. Adipogenesis: From stem cell to adipocyte. Annu. Rev. Biochem. 2012, 81, 715–736. [Google Scholar] [CrossRef] [Green Version]
- Rosen, E.D.; MacDougald, O.A. Adipocyte differentiation from the inside out. Nat. Rev. Mol. Cell Biol. 2006, 72, 885–896. [Google Scholar] [CrossRef] [PubMed]
- Goldrick, R.B. Morphological changes in the adipocyte during fat deposition and mobilization. Am. J. Physiol. 1967, 212, 777–782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Novikoff, A.B.; Novikoff, P.M.; Rosen, O.M.; Rubin, C.S. Organelle relationships in cultured 3T3-L1 preadipocytes. J. Cell Biol. 1980, 87, 180–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chalut, K.J.; Paluch, E.K. The Actin Cortex: A Bridge between Cell Shape and Function. Dev. Cell. 2016, 38, 571–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mor-Yossef Moldovan, L.; Lustig, M.; Naftaly, A.; Mardamshina, M.; Geiger, T.; Gefen, A.; Benayahu, D. Cell shape alteration during adipogenesis is associated with coordinated matrix cues. J. Cell Physiol. 2019, 234, 3850–3863. [Google Scholar] [CrossRef]
- Kunitomi, H.; Oki, Y.; Onishi, N.; Kano, K.; Banno, K.; Aoki, D.; Saya, H.; Nobusue, H. The insulin-PI3K-Rac1 axis contributes to terminal adipocyte differentiation through regulation of actin cytoskeleton dynamics. Genes Cells 2020, 25, 165–174. [Google Scholar] [CrossRef]
- Spiegelman, B.M.; Farmer, S.R. Decreases in tubulin and actin gene expression prior to morphological differentiation of 3T3 adipocytes. Cell 1982, 29, 53–60. [Google Scholar] [CrossRef]
- Spiegelman, B.M.; Ginty, C.A. Fibronectin modulation of cell shape and lipogenic gene expression in 3t3-adipocytes. Cell 1983, 35, 657–666. [Google Scholar] [CrossRef]
- Tong, P.; Khayat, Z.A.; Huang, C.; Patel, N.; Ueyama, A.; Klip, A. Insulin-induced cortical actin remodeling promotes GLUT4 insertion at muscle cell membrane ruffles. J. Clin. Investig. 2001, 108, 371–381. [Google Scholar] [CrossRef]
- Tsakiridis, T.; Tong, P.; Matthews, B.; Tsiani, E.; Bilan, P.J.; Klip, A.; Downey, G.P. Role of the actin cytoskeleton in insulin action. Microsc. Res. Tech. 1999, 47, 79–92. [Google Scholar] [CrossRef]
- Kim, J.I.; Park, J.; Ji, Y.; Jo, K.; Han, S.M.; Sohn, J.H.; Shin, K.C.; Han, J.S.; Jeon, Y.G.; Nahmgoong, H.; et al. During Adipocyte Remodeling, Lipid Droplet Configurations Regulate Insulin Sensitivity through F-Actin and G-Actin Reorganization. Mol. Cell Biol. 2019, 39, e00210-19. [Google Scholar] [CrossRef] [Green Version]
- Brozinick, J.T., Jr.; Berkemeier, B.A.; Elmendorf, J.S. “Acting” on GLUT4: Membrane & Cytoskeletal Components of Insulin Action. Curr. Diabetes Rev. 2007, 3, 111. [Google Scholar] [CrossRef]
- Kanzaki, M.; Pessin, J.E. Insulin-stimulated GLUT4 translocation in adipocytes is dependent upon cortical actin remodeling. J. Biol. Chem. 2001, 276, 42436–42444. [Google Scholar] [CrossRef] [Green Version]
- Guilherme, A.; Emoto, M.; Buxton, J.M.; Bose, S.; Sabini, R.; Theurkauf, W.E.; Leszyk, J.; Czech, M.P. Perinuclear localization and insulin responsiveness of GLUT4 requires cytoskeletal integrity in 3T3-L1 adipocytes. J. Biol. Chem. 2000, 275, 38151–38159. [Google Scholar] [CrossRef] [Green Version]
- Klip, A.; McGraw, T.E.; James, D.E. Thirty sweet years of GLUT4. J. Biol. Chem. 2019, 294, 11369–11381. [Google Scholar] [CrossRef] [Green Version]
- Antonescu, C.N.; Foti, M.; Sauvonnet, N.; Klip, A. Ready, set, internalize: Mechanisms and regulation of GLUT4 endocytosis. Biosci. Rep. 2009, 29, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Abel, E.D.; Peroni, O.; Kim, J.K.; Kim, Y.B.; Boss, O.; Hadro, E.; Minnemann, T.; Shulman, G.I.; Kahn, B.B. Adipose-selective targeting of the GLUT4 gene impairs insulin action in muscle and liver. Nature 2001, 409, 729–733. [Google Scholar] [CrossRef]
- Garvey, W.T.; Maianu, L.; Huecksteadt, T.P.; Birnbaum, M.J.; Molina, J.M.; Ciaraldi, T.P. Pretranslational suppression of a glucose transporter protein causes insulin resistance in adipocytes from patients with non-insulin-dependent diabetes mellitus and obesity. J. Clin. Investig. 1991, 87, 1072–1081. [Google Scholar] [CrossRef] [Green Version]
- Liao, W.; Nguyen, M.T.A.; Imamura, T.; Singer, O.; Verma, I.M.; Olefsky, J.M. Lentiviral Short Hairpin Ribonucleic Acid-Mediated Knockdown of GLUT4 in 3T3-L1 Adipocytes. Endocrinology 2006, 147, 2245–2252. [Google Scholar] [CrossRef]
- Hauner, H.; Röhrig, K.; Spelleken, M.; Liu, L.; Eckel, J. Development of insulin-responsive glucose uptake and GLUT4 expression in differentiating human adipocyte precursor cells. Int. J. Obes. 1998, 22, 448–453. [Google Scholar] [CrossRef] [Green Version]
- Fletcher, L.M.; Welsh, G.I.; Oatey, P.B.; Tavaré, J.M. Role for the microtubule cytoskeleton in GLUT4 vesicle trafficking and in the regulation of insulin-stimulated glucose uptake. Biochem, J. 2000, 352, 267, Erratum in Biochem. J. 2001, 353, 735. [Google Scholar] [CrossRef]
- Boguslavsky, S.; Chiu, T.; Foley, K.P.; Osorio-Fuentealba, C.; Antonescu, C.N.; Bayer, K.U.; Bilan, P.J.; Klip, A. Myo1c binding to submembrane actin mediates insulin-induced tethering of GLUT4 vesicles. Mol. Biol. Cell 2012, 23, 4065. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, Y.; Zhang, J.; Deng, Y.; Jiang, L.; Song, E.; Wu, X.S.; Hammer, J.A.; Xu, T.; Lippincott-Schwartz, J. Rab10 and myosin-Va mediate insulin-stimulated GLUT4 storage vesicle translocation in adipocytes. J. Cell Biol. 2012, 198, 545–560. [Google Scholar] [CrossRef] [Green Version]
- Le Chung, T.K.; Hosaka, T.; Harada, N.; Jambaldorj, B.; Fukunaga, K.; Nishiwaki, Y.; Teshigawara, K.; Sakai, T.; Nakaya, Y.; Funaki, M. Myosin IIA participates in docking of Glut4 storage vesicles with the plasma membrane in 3T3-L1 adipocytes. Biochem. Biophys. Res. Commun. 2010, 391, 995–999. [Google Scholar] [CrossRef]
- Steimle, P.A.; Fulcher, F.K.; Patel, Y.M. A novel role for myosin II in insulin-stimulated glucose uptake in 3T3-L1 adipocytes. Biochem. Biophys. Res. Commun. 2005, 331, 1560–1565. [Google Scholar] [CrossRef]
- Choi, Y.O.; Ryu, H.J.; Kim, H.R.; Song, Y.S.; Kim, C.; Lee, W.; Choe, H.; Leem, C.H.; Jang, Y.J. Implication of phosphorylation of the myosin II regulatory light chain in insulin-stimulated GLUT4 translocation in 3T3-F442A adipocytes. Exp. Mol. Med. 2006, 38, 180–189. [Google Scholar] [CrossRef]
- Yoshizaki, T.; Imamura, T.; Babendure, J.L.; Lu, J.-C.; Sonoda, N.; Olefsky, J.M. Myosin 5a Is an Insulin-Stimulated Akt2 (Protein Kinase Bβ) Substrate Modulating GLUT4 Vesicle Translocation. Mol. Cell Biol. 2007, 274, 5172. [Google Scholar] [CrossRef] [Green Version]
- Fulcher, F.K.; Smith, B.T.; Russ, M.; Patel, Y.M. Dual role for myosin II in GLUT4-mediated glucose uptake in 3T3-L1 adipocytes. Exp. Cell Res. 2008, 3147, 3264–3274. [Google Scholar] [CrossRef] [Green Version]
- Heissler, S.M.; Manstein, D.J. Nonmuscle myosin-2: Mix and match. Cell. Mol. Life Sci. 2012, 70, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Wang, A.; Ma, X.; Conti, M.A.; Adelstein, R.S. Distinct and redundant roles of the non-muscle myosin II isoforms and functional domains. Biochem. Soc. Trans. 2011, 39, 1131. [Google Scholar] [CrossRef] [Green Version]
- Vicente-Manzanares, M.; Ma, X.; Adelstein, R.S.; Horwitz, A.R. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 2009, 101, 778–790. [Google Scholar] [CrossRef] [Green Version]
- Burridge, K.; Guilluy, C. Focal adhesions, stress fibers and mechanical tension. Exp. Cell Res. 2016, 343, 14. [Google Scholar] [CrossRef] [Green Version]
- Kuragano, M.; Uyeda, T.Q.P.; Kamijo, K.; Murakami, Y.; Takahashi, M. Different contributions of nonmuscle myosin IIA and IIB to the organization of stress fiber subtypes in fibroblasts. Mol. Biol. Cell. 2018, 29, 911–922. [Google Scholar] [CrossRef]
- Tharp, K.M.; Kang, M.S.; Timblin, G.A.; Saijo, K.; Kumar, S.; Stahl Correspondence, A. Actomyosin-Mediated Tension Orchestrates Uncoupled Respiration in Adipose Tissues. Cell Metab. 2018, 27, 602–615. [Google Scholar] [CrossRef] [Green Version]
- Al-Sayegh, M.; Ali, H.; Jamal, M.H.; ElGindi, M.; Chanyong, T.; Al-Awadi, K.; Abu-Farha, M. Mouse Embryonic Fibroblast Adipogenic Potential: A Comprehensive Transcriptome Analysis. Adipocyte 2021, 10, 1–20. [Google Scholar] [CrossRef]
- Woody, S.; Stall, R.; Ramos, J.; Patel, Y.M. Regulation of Myosin Light Chain Kinase during Insulin-Stimulated Glucose Uptake in 3T3-L1 Adipocytes. PLoS ONE 2013, 80, e77248. [Google Scholar] [CrossRef] [Green Version]
- Bedi, D.; Dennis, J.C.; Morrison, E.E.; Braden, T.D.; Judd, R.L. Regulation of intracellular trafficking and secretion of adiponectin by myosin II. Biochem. Biophys. Res. Commun. 2017, 490, 202–208. [Google Scholar] [CrossRef]
- Standaert, M.L.; Galloway, L.; Karnam, P.; Bandyopadhyay, G.; Moscat, J.; Farese, R.V. Protein kinase C-ζ as a downstream effector of phosphatidylinositol 3- kinase during insulin stimulation in rat adipocytes. Potential role in glucose transport. J. Biol. Chem. 1997, 272, 30075–30082. [Google Scholar] [CrossRef] [Green Version]
- Bandyopadhyay, G.; Sajan, M.P.; Kanoh, Y.; Standaert, M.L.; Quon, M.J.; Lea-Currie, R.; Sen, A.; Farese, R.V. PKC-zeta mediates insulin effects on glucose transport in cultured preadipocyte-derived human adipocytes. J. Clin. Endocrinol. Metab. 2002, 87, 716–723. [Google Scholar] [CrossRef] [Green Version]
- Discher, D.E.; Janmey, P.; Wang, Y.L. Tissue cells feel and respond to the stiffness of their substrate. Science 2005, 310, 1139–1143. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Hu, H.; Qiu, W.; Shi, K.; Kassem, M. Actin depolymerization enhances adipogenic differentiation in human stromal stem cells. Stem Cell Res. 2018, 29, 76–83. [Google Scholar] [CrossRef]
- Zang, J.-H.; Cavet, G.; Sabry, J.H.; Wagner, P.; Moores, S.L.; Spudich, J.A. On the Role of Myosin-II in Cytokinesis: Division of Dictyostelium Cells under Adhesive and Nonadhesive Conditions. Mol. Biol. Cell 1997, 82, 2617. [Google Scholar] [CrossRef] [Green Version]
- Elliott, H.; Fischer, R.S.; Myers, K.A.; Desai, R.A.; Gao, L.; Chen, C.S.; Adelstein, R.S.; Waterman, C.M.; Danuser, G. Myosin II controls cellular branching morphogenesis and migration in three dimensions by minimizing cell-surface curvature. Nat. Cell Biol. 2015, 17, 137–147. [Google Scholar] [CrossRef]
- Clark, A.G.; Wartlick, O.; Salbreux, G.; Paluch, E.K. Stresses at the cell surface during animal cell morphogenesis. Curr. Biol. 2014, 240, R484–R494. [Google Scholar] [CrossRef] [Green Version]
- Franke, W.W.; Hergt, M.; Grund, C. Rearrangement of the vimentin cytoskeleton during adipose conversion: Formation of an intermediate filament cage around lipid globules. Cell 1987, 49, 131–141. [Google Scholar] [CrossRef]
- Audano, M.; Pedretti, S.; Ligorio, S.; Gualdrini, F.; Polletti, S.; Russo, M.; Ghisletti, S.; Bean, C.; Crestani, M.; Caruso, D.; et al. Zc3h10 regulates adipogenesis by controlling translation and F-actin/mitochondria interaction. J. Cell Biol. 2021, 220, e202003173. [Google Scholar] [CrossRef]
- Lo, C.-M.; Buxton, D.B.; Chua, G.C.H.; Dembo, M.; Adelstein, R.S.; Wang, Y.-L. Nonmuscle Myosin IIB Is Involved in the Guidance of Fibroblast Migration. Mol. Biol. Cell 2003, 15, 982–989. [Google Scholar] [CrossRef] [Green Version]
- Beadle, C.; Assanah, M.C.; Monzo, P.; Vallee, R.; Rosenfeld, S.S.; Canoll, P. The Role of Myosin II in Glioma Invasion of the Brain. Mol. Biol. Cell 2008, 19, 3357–3368. [Google Scholar] [CrossRef] [Green Version]
- Sandquist, J.C.; Swenson, K.I.; DeMali, K.A.; Burridge, K.; Means, A.R. Rho Kinase Differentially Regulates Phosphorylation of Nonmuscle Myosin II Isoforms A and B during Cell Rounding and Migration. J. Biol. Chem. 2006, 281, 35873–35883. [Google Scholar] [CrossRef] [Green Version]
- Tullio, A.N.; Accili, D.; Ferrans, V.J.; Yu, Z.X.; Takeda, K.; Grinberg, A.; Westphal, H.; Preston, Y.A.; Adelstein, R.S. Nonmuscle myosin II-B is required for normal development of the mouse heart. Proc. Natl. Acad. Sci. USA 1997, 94, 12407–12412. [Google Scholar] [CrossRef] [Green Version]
- Takeda, K.; Kishi, H.; Ma, X.; Yu, Z.-X.; Adelstein, R.S. Ablation and Mutation of Nonmuscle Myosin Heavy Chain II-B Results in a Defect in Cardiac Myocyte Cytokinesis. Circ. Res. 2003, 93, 330–337. [Google Scholar] [CrossRef]
- Ma, X.; Kawamoto, S.; Hara, Y.; Adelstein, R.S. A point mutation in the motor domain of nonmuscle myosin II-B impairs migration of distinct groups of neurons. Mol. Biol. Cell 2004, 15, 2568–2579. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.T.; Yin, W.; Jin, Y.J.; Panza, P.; Gunawan, F.; Grohmann, B.; Buettner, C.; Sokol, A.M.; Preussner, J.; Guenther, S.; et al. Myh10 deficiency leads to defective extracellular matrix remodeling and pulmonary disease. Nat. Commun. 2018, 9, 4600. [Google Scholar] [CrossRef] [Green Version]
- Recuenco, M.C.; Ohmori, T.; Tanigawa, S.; Taguchi, A.; Fujimura, S.; Conti, M.A.; Wei, Q.; Kiyonari, H.; Abe, T.; Adelstein, R.S.; et al. Nonmuscle Myosin II Regulates the Morphogenesis of Metanephric Mesenchyme–Derived Immature Nephrons. J. Am. Soc. Nephrol. 2015, 26, 1081. [Google Scholar] [CrossRef] [Green Version]
- Ma, X.; Adelstein, R.S. A Point Mutation in Myh10 Causes Major Defects in Heart Development and Body Wall Closure. Circ. Cardiovasc. Genet. 2014, 7, 257–265. [Google Scholar] [CrossRef] [Green Version]
- Cheung, S.Y.; Sayeed, M.; Nakuluri, K.; Li, L.; Feldman, B.J. MYH9 facilitates autoregulation of adipose tissue depot development. JCI Insight 2021, 6. [Google Scholar] [CrossRef]
- Wasik, A.A.; Dumont, V.; Tienari, J.; Nyman, T.A.; Fogarty, C.L.; Forsblom, C.; Lehto, M.; Lehtonen, E.; Groop, P.H.; Lehtonen, S. Septin 7 reduces nonmuscle myosin IIA activity in the SNAP23 complex and hinders GLUT4 storage vesicle docking and fusion. Exp. Cell Res. 2017, 350, 336–348. [Google Scholar] [CrossRef]
- Stall, R.; Ramos, J.; Kent Fulcher, F.; Patel, Y.M. Regulation of myosin IIA and filamentous actin during insulin-stimulated glucose uptake in 3T3-L1 adipocytes. Exp. Cell Res. 2014, 322, 81–88. [Google Scholar] [CrossRef] [Green Version]
- Kislev, N.; Egozi, M.; Benayahu, D. Imaging analysis to quantitate the Interplay of membrane and cytoplasm protein dynamics. bioRxiv 2021. [Google Scholar] [CrossRef]
- Kim, S.; Moustaid-Moussa, N. Secretory.; Endocrine and Autocrine/Paracrine Function of the Adipocyte. J. Nutr. 2000, 1302, 3110S–3115S. [Google Scholar] [CrossRef] [PubMed]
- Hemmingsen, M.; Vedel, S.; Skafte-Pedersen, P.; Sabourin, D.; Collas, P.; Bruus, H.; Dufva, M. The Role of Paracrine and Autocrine Signaling in the Early Phase of Adipogenic Differentiation of Adipose-derived Stem Cells. PLoS ONE 2013, 8, e63638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia, N.A.; Moncayo-Arlandi, J.; Sepulveda, P.; Diez-Juan, A. Cardiomyocyte exosomes regulate glycolytic flux in endothelium by direct transfer of GLUT transporters and glycolytic enzymes. Cardiovasc. Res. 2016, 109, 397–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Even-Faitelson, L.; Ravid, S. PAK1 and aPKCζ Regulate Myosin II-B Phosphorylation: A Novel Signaling Pathway Regulating Filament Assembly. Mol. Biol. Cell 2006, 17, 2869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Leeuwen, F.N.; van Delft, S.; Kain, H.E.; van der Kammen, R.A.; Collard, J.G. Rac regulates phosphorylation of the myosin-II heavy chain.; actinomyosin disassembly and cell spreading. Nat. Cell Biol. 1999, 1, 242–248. [Google Scholar] [CrossRef]
- Straussman, R.; Even, L.; Ravid, S. Myosin II heavy chain isoforms are phosphorylated in an EGF-dependent manner: Involvement of protein kinase C. J. Cell Sci. 2001, 114, 3047–3057. [Google Scholar] [CrossRef]
- Dulyaninova, N.G.; House, R.P.; Betapudi, V.; Bresnick, A.R. Myosin-IIA Heavy-Chain Phosphorylation Regulates the Motility of MDA-MB-231 Carcinoma Cells. Mol. Biol. Cell 2007, 18, 3144–3155. [Google Scholar] [CrossRef] [Green Version]
- Sabry, J.H.; Moores, S.L.; Ryan, S.; Zang, J.H.; Spudich, J.A. Myosin heavy chain phosphorylation sites regulate myosin localization during cytokinesis in live cells. Mol. Biol. Cell. 1997, 82, 2605–2615. [Google Scholar] [CrossRef] [Green Version]
- Dulyaninova, N.G.; Bresnick, A.R. The heavy chain has its day: Regulation of myosin-II assembly. Bioarchitecture 2013, 3, 77–85. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.J.; Yang, C.; Gupta, N.; Zuo, J.; Chang, Y.S.; Fang, F.D. Protein kinase C-zeta regulation of GLUT4 translocation through actin remodeling in CHO cells. J. Mol. Med. 2007, 85, 851–861. [Google Scholar] [CrossRef]
- Uberall, F.; Hellbert, K.; Kampfer, S.; Maly, K.; Villunger, A.; Spitaler, M.; Mwanjewe, J.; Baier-Bitterlich, G.; Baier, G.; Grunicke, H.H. Evidence That Atypical Protein Kinase C-λ and Atypical Protein Kinase C-ζ Participate in Ras-mediated Reorganization of the F-actin Cytoskeleton. J. Cell Biol. 1999, 144, 413–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.Z.; Zhao, H.L.; Zuo, J.; Ho, S.K.; Chan, J.C.; Meng, Y.; Fang, F.D.; Tong, P.C. Protein Kinase Cζ Mediates Insulin-induced Glucose Transport through Actin Remodeling in L6 Muscle Cells. Mol. Biol. Cell 2006, 17, 2322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Juanes-Garcia, A.; Chapman, J.R.; Aguilar-Cuenca, R.; Delgado-Arevalo, C.; Hodges, J.; Whitmore, L.A.; Shabanowitz, J.; Hunt, D.F.; Horwitz, A.R.; Vicente-Manzanares, M. A regulatory motif in nonmuscle myosin II-B regulates its role in migratory front–back polarity. J. Cell Biol. 2015, 209, 23–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomes, E.R.; Jani, S.; Gundersen, G.G. Nuclear Movement Regulated by Cdc42, MRCK, Myosin, and Actin Flow Establishes MTOC Polarization in Migrating Cells. Cell 2005, 121, 451–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schiffhauer, E.S.; Ren, Y.; Iglesias, V.A.; Kothari, P.; Iglesias, P.A.; Robinson, D.N. Myosin IIB assembly state determines its mechanosensitive dynamics. J. Cell Biol. 2019, 218, 895–908. [Google Scholar] [CrossRef] [Green Version]
- Shoham, N.; Gottlieb, R.; Sharabani-Yosef, O.; Zaretsky, U.; Benayahu, D.; Gefen, A. Static mechanical stretching accelerates lipid production in 3T3-L1 adipocytes by activating the MEK signaling pathway. Am. J. Physiol. Physiol. 2012, 302, 429–441. [Google Scholar] [CrossRef] [Green Version]
- Cohen, J.; Raviv, S.; Adir, O.; Padmanabhan, K.; Soffer, A.; Luxenburg, C. The Wave complex controls epidermal morphogenesis and proliferation by suppressing Wnt–Sox9 signaling. J. Cell Biol. 2019, 218, 1390–1406. [Google Scholar] [CrossRef] [Green Version]
- Lim, C.Y.; Bi, X.; Wu, D.; Kim, J.B.; Gunning, P.W.; Hong, W.; Han, W. Tropomodulin3 is a novel Akt2 effector regulating insulin-stimulated GLUT4 exocytosis through cortical actin remodeling. Nat. Commun. 2015, 6, 5951. [Google Scholar] [CrossRef] [Green Version]
- Schramek, D.; Sendoel, A.; Segal, J.P.; Beronja, S.; Heller, E.; Oristian, D.; Reva, B.; Fuchs, E. Direct in vivo RNAi screen unveils myosin IIa as a tumor suppressor of squamous cell carcinomas. Science 2014, 343, 309–313. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Yeung, J.L.; Lee, J.H.; An, J.; Steadman, P.E.; Kim, J.R.; Sung, H.K. Visualization of 3D White Adipose Tissue Structure Using Whole-mount Staining. J. Vis. Exp. 2018, e58683. [Google Scholar] [CrossRef] [Green Version]
- Kislev, N.; Izgilov, R.; Adler, R.; Benayahu, D. Exploring the Cell Stemness and the Complexity of the Adipose Tissue Niche. Biomolecules 2021, 112, 1906. [Google Scholar] [CrossRef]
- Püspöki, Z.; Storath, M.K.; Sage, D.; Unser, M. Transforms and Operators for Directional Bioimage Analysis: A Survey. Adv. Anat. Embryol. Cell Biol. 2016, 219, 69–93. [Google Scholar] [CrossRef] [PubMed]
- Steger, G. An unbiased detector of curvilinear structures. IEEE Trans. Pattern Anal. Mach. Intell. 1998, 20, 113–125. [Google Scholar] [CrossRef] [Green Version]
- Wagner, T.; Hiner, M. Xraynaud. thorstenwagner/ij-ridgedetection: Ridge Detection 1.4.0. Zenodo 2017. [Google Scholar] [CrossRef]
- Arganda-Carreras, I.; Kaynig, V.; Rueden, C.; Eliceiri, K.W.; Schindelin, J.; Cardona, A.; Sebastian Seung, H. Trainable Weka Segmentation: A machine learning tool for microscopy pixel classification. Bioinformatics 2017, 335, 2424–2426. [Google Scholar] [CrossRef] [PubMed]
- Lustig, M.; Feng, Q.; Payan, Y.; Gefen, A.; Benayahu, D. Noninvasive continuous monitoring of adipocyte differentiation: From macro to micro scales. Microsc. Microanal. 2019, 25, 119–128. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Park, C.Y.; Krishnan, A.; Zhu, Q.; Wong, A.K.; Lee, Y.S.; Troyanskaya, O.G. Tissue-aware data integration approach for the inference of pathway interactions in metazoan organisms. Bioinformatics 2015, 31, 1093–1101. [Google Scholar] [CrossRef] [Green Version]
- Carbon, S.; Ireland, A.; Mungall, C.J.; Shu, S.; Marshall, B.; Lewis, S. AmiGO: Online access to ontology and annotation data. Bioinformatics 2009, 25, 288–289. [Google Scholar] [CrossRef]
- Shburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25. [Google Scholar] [CrossRef] [Green Version]
- Gene Ontology Consortium. The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kislev, N.; Mor-Yossef Moldovan, L.; Barak, R.; Egozi, M.; Benayahu, D. MYH10 Governs Adipocyte Function and Adipogenesis through Its Interaction with GLUT4. Int. J. Mol. Sci. 2022, 23, 2367. https://doi.org/10.3390/ijms23042367
Kislev N, Mor-Yossef Moldovan L, Barak R, Egozi M, Benayahu D. MYH10 Governs Adipocyte Function and Adipogenesis through Its Interaction with GLUT4. International Journal of Molecular Sciences. 2022; 23(4):2367. https://doi.org/10.3390/ijms23042367
Chicago/Turabian StyleKislev, Nadav, Lisa Mor-Yossef Moldovan, Rakefet Barak, Michael Egozi, and Dafna Benayahu. 2022. "MYH10 Governs Adipocyte Function and Adipogenesis through Its Interaction with GLUT4" International Journal of Molecular Sciences 23, no. 4: 2367. https://doi.org/10.3390/ijms23042367







