Mycobacterium tuberculosis Acetyltransferase Suppresses Oxidative Stress by Inducing Peroxisome Formation in Macrophages
Abstract
:1. Introduction
2. Results
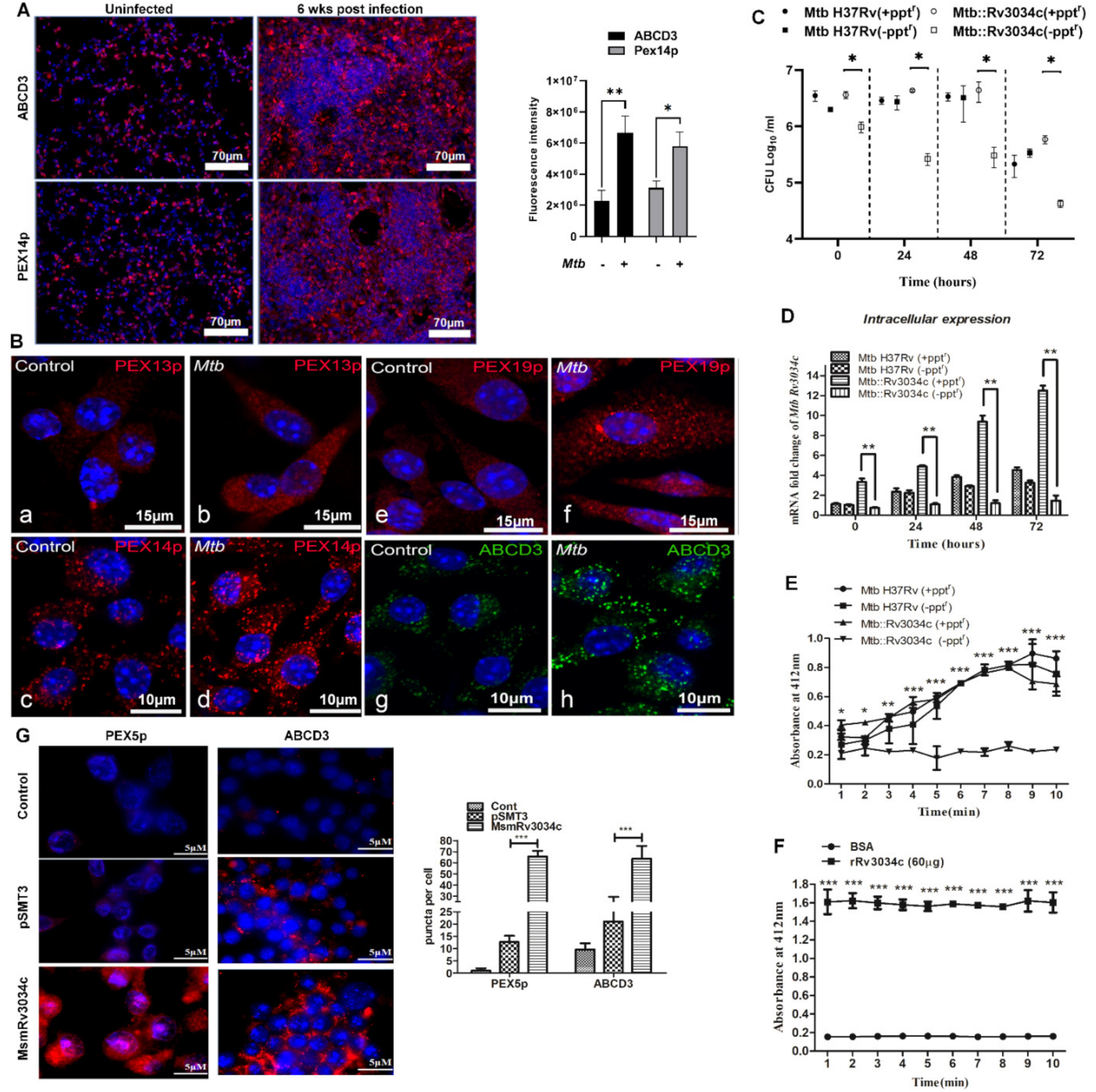
2.1. M. tuberculosis Infection Induces the Expression of Proteins Involved in Peroxisome Biogenesis in Mice Lungs and Bone-Marrow-Derived Macrophages
2.2. Absence of M. tuberculosis Acetyltransferase Reduces Bacterial Growth in Macrophages
2.3. M. tuberculosis Rv3034c Encodes an Acetyltransferase Enzyme
2.4. Recombinant M. smegmatis Expressing Rv3034c and Purified Rv3034c Protein Induced Peroxisome Biogenesis
2.5. M. tuberculosis Rv3034c Induced Peroxisome Biogenesis Scavenges Reactive Oxygen Species Production in Macrophages
2.6. Rv3034c Induces the Peroxisomal β-Oxidation Pathway
2.7. Inhibition of Peroxisomal β-Oxidation Decreases Bacterial Survival Due to Increase in the ROS Production
2.8. M. tuberculosis Rv3034c Induces Peroxisomes through the Macrophage Mannose Receptor (MR)
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Cell Lines and Reagents
4.2. Ethical Statement
4.3. ConA Affinity Purification, Lectin Hybridization, and Mass Spectrometry Analysis
4.4. Construction of Mtb Rv3034c Conditional Mutant
4.5. Cloning and Expression of Rv3034c in Msm
4.6. Rv3034c Protein Purification
4.7. Cellular Localization of Rv3034c and GFP Immunostaining
4.8. Acetyltransferase Assay
4.9. Intracellular Survival Assay
4.10. Isolation of Mouse Peritoneal Macrophages
4.11. Flow Cytometry Analysis
4.12. Mouse Infection
4.13. Immunofluorescence Microscopy
4.14. siRNA Transfection
4.15. Quantitative Real-Time PCR Analysis
4.16. Western Blot Analysis
4.17. Pull-Down Assay
4.18. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Global Tuberculosis Report 2020; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Zammarchi, L.; Bartalesi, F.; Bartoloni, A. Tuberculosis in tropical areas and immigrants. Mediterr. J. Hematol. Infect. Dis. 2014, 6, e2014043. [Google Scholar] [CrossRef]
- Smith, T.; Wolff, K.A.; Nguyen, L. Molecular Biology of Drug Resistance in Mycobacterium tuberculosis. In Pathogenesis of Mycobacterium tuberculosis and Its Interaction with the Host Organism; Pieters, J., McKinney, J.D., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 53–80. [Google Scholar]
- Ehrt, S.; Schnappinger, D. Mycobacterial survival strategies in the phagosome: Defence against host stresses. Cell. Microbiol. 2009, 11, 1170–1178. [Google Scholar] [CrossRef] [Green Version]
- Guirado, E.; Schlesinger, L.S.; Kaplan, G. Macrophages in tuberculosis: Friend or foe. Semin. Immunopathol. 2013, 35, 563–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohanty, A.; Tiwari-Pandey, R.; Pandey, N.R. Mitochondria: The indispensable players in innate immunity and guardians of the inflammatory response. J. Cell Commun. Signal. 2019, 13, 303–318. [Google Scholar] [CrossRef] [PubMed]
- Di Cara, F.; Andreoletti, P.; Trompier, D.; Vejux, A.; Bülow, M.H.; Sellin, J.; Lizard, G.; Cherkaoui-Malki, M.; Savary, S. Peroxisomes in Immune Response and Inflammation. Int. J. Mol. Sci. 2019, 20, 3877. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Li, S.; Liu, Y.; Ma, C. Redox regulated peroxisome homeostasis. Redox Biol. 2015, 4, 104–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganguli, G.; Mukherjee, U.; Sonawane, A. Peroxisomes and Oxidative Stress: Their Implications in the Modulation of Cellular Immunity during Mycobacterial Infection. Front. Microbiol. 2019, 10, 1121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Cara, F. Peroxisomes in host defense. PLoS Pathog. 2020, 16, e1008636. [Google Scholar] [CrossRef] [PubMed]
- Del Río, L.A.; López-Huertas, E. ROS Generation in Peroxisomes and its Role in Cell Signaling. Plant Cell Physiol. 2016, 57, 1364–1376. [Google Scholar] [CrossRef] [PubMed]
- Sibirny, A.A. Yeast peroxisomes: Structure, functions and biotechnological opportunities. FEMS Yeast Res. 2016, 16, fow038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karnati, S.; Baumgart-Vogt, E. Peroxisomes in mouse and human lung: Their involvement in pulmonary lipid metabolism. Histochem. Cell Biol. 2008, 130, 719–740. [Google Scholar] [CrossRef] [PubMed]
- Del Río, L.A. Peroxisomes as a cellular source of reactive nitrogen species signal molecules. Arch. Biochem. Biophys. 2011, 506, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Shastri, M.D.; Shukla, S.D.; Chong, W.C.; Dua, K.; Peterson, G.M.; Patel, R.P.; Hansbro, P.M.; Eri, R.; O’Toole, R.F. Role of Oxidative Stress in the Pathology and Management of Human Tuberculosis. Oxid. Med. Cell Longev. 2018, 2018, 7695364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganguli, G.; Pattanaik, K.P.; Jagadeb, M.; Sonawane, A. Mycobacterium tuberculosis Rv3034c regulates mTORC1 and PPAR-γ dependant pexophagy mechanism to control redox levels in macrophages. Cell. Microbiol. 2020, 22, e13214. [Google Scholar] [CrossRef]
- Terlecky, S.R.; Terlecky, L.J.; Giordano, C.R. Peroxisomes, oxidative stress, and inflammation. World J. Biol. Chem. 2012, 3, 93–97. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Sang, Y.; Lu, J.; Yao, Y.F. Protein Acetylation and Its Role in Bacterial Virulence. Trends Microbiol. 2017, 25, 768–779. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Yang, W.; Fan, X.; Xie, J. Comprehensive analysis of protein acetyltransferases of human pathogen Mycobacterium tuberculosis. Biosci. Rep. 2019, 39, BSR20191661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, W.; VanderVen, B.C.; Walker, S.; Russell, D.G. Novel protein acetyltransferase, Rv2170, modulates carbon and energy metabolism in Mycobacterium tuberculosis. Sci. Rep. 2017, 7, 72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohanty, S.; Jagannathan, L.; Ganguli, G.; Padhi, A.; Roy, D.; Alaridah, N.; Saha, P.; Nongthomba, U.; Godaly, G.; Gopal, R.K.; et al. A mycobacterial phosphoribosyltransferase promotes bacillary survival by inhibiting oxidative stress and autophagy pathways in macrophages and zebrafish. J. Biol. Chem. 2015, 290, 13321–13343. [Google Scholar] [CrossRef] [Green Version]
- Mohanty, S.; Dal Molin, M.; Ganguli, G.; Padhi, A.; Jena, P.; Selchow, P.; Sengupta, S.; Meuli, M.; Sander, P.; Sonawane, A. Mycobacterium tuberculosis EsxO (Rv2346c) promotes bacillary survival by inducing oxidative stress mediated genomic instability in macrophages. Tuberculosis 2016, 96, 44–57. [Google Scholar] [CrossRef]
- Padhi, A.; Naik, S.K.; Sengupta, S.; Ganguli, G.; Sonawane, A. Expression of Mycobacterium tuberculosis NLPC/p60 family protein Rv0024 induce biofilm formation and resistance against cell wall acting anti-tuberculosis drugs in Mycobacterium smegmatis. Microbes Infect. 2016, 18, 224–236. [Google Scholar] [CrossRef]
- Altaf, M.; Miller, C.H.; Bellows, D.S.; O’Toole, R. Evaluation of the Mycobacterium smegmatis and BCG models for the discovery of Mycobacterium tuberculosis inhibitors. Tuberculosis 2010, 90, 333–337. [Google Scholar] [CrossRef] [PubMed]
- Zhai, W.; Wu, F.; Zhang, Y.; Fu, Y.; Liu, Z. The Immune Escape Mechanisms of Mycobacterium tuberculosis. Int. J. Mol. Sci. 2019, 20, 340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dansen, T.B.; Wirtz, K.W. The peroxisome in oxidative stress. IUBMB Life 2001, 51, 223–230. [Google Scholar] [CrossRef]
- Quijano, C.; Trujillo, M.; Castro, L.; Trostchansky, A. Interplay between oxidant species and energy metabolism. Redox Biol. 2016, 8, 28–42. [Google Scholar] [CrossRef] [Green Version]
- Shi, R.; Zhang, Y.; Shi, Y.; Shi, S.; Jiang, L. Inhibition of peroxisomal β-oxidation by thioridazine increases the amount of VLCFAs and Aβ generation in the rat brain. Neurosci. Lett. 2012, 528, 6–10. [Google Scholar] [CrossRef] [PubMed]
- Esparza, M.; Palomares, B.; García, T.; Espinosa, P.; Zenteno, E.; Mancilla, R. PstS-1, the 38-kDa Mycobacterium tuberculosis glycoprotein, is an adhesin, which binds the macrophage mannose receptor and promotes phagocytosis. Scand. J. Immunol. 2015, 81, 46–55. [Google Scholar] [CrossRef] [PubMed]
- Pan, R.; Liu, J.; Wang, S.; Hu, J. Peroxisomes: Versatile organelles with diverse roles in plants. New Phytol. 2020, 225, 1410–1427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maruyama, J.; Kitamoto, K. Expanding functional repertoires of fungal peroxisomes: Contribution to growth and survival processes. Front. Physiol. 2013, 4, 177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, D.M.; Jeon, B.Y.; Lee, H.M.; Jin, H.S.; Yuk, J.M.; Song, C.H.; Lee, S.H.; Lee, Z.W.; Cho, S.N.; Kim, J.M.; et al. Mycobacterium tuberculosis eis regulates autophagy, inflammation, and cell death through redox-dependent signaling. PLoS Pathog. 2010, 6, e1001230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Griffin, J.E.; Gawronski, J.D.; Dejesus, M.A.; Ioerger, T.R.; Akerley, B.J.; Sassetti, C.M. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 2011, 7, e1002251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maranha, A.; Moynihan, P.J.; Miranda, V.; Correia Lourenço, E.; Nunes-Costa, D.; Fraga, J.S.; José Barbosa Pereira, P.; Macedo-Ribeiro, S.; Ventura, M.R.; Clarke, A.J.; et al. Octanoylation of early intermediates of mycobacterial methylglucose lipopolysaccharides. Sci. Rep. 2015, 5, 13610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Cara, F.; Sheshachalam, A.; Braverman, N.E.; Rachubinski, R.A.; Simmonds, A.J. Peroxisome-Mediated Metabolism Is Required for Immune Response to Microbial Infection. Immunity 2017, 47, 93–106.e7. [Google Scholar] [CrossRef] [Green Version]
- Ganaie, A.A.; Lella, R.K.; Solanki, R.; Sharma, C. Thermostable hexameric form of Eis (Rv2416c) protein of M. tuberculosis plays an important role for enhanced intracellular survival within macrophages. PLoS ONE 2011, 6, e27590. [Google Scholar] [CrossRef]
- Abuhammad, A.; Fullam, E.; Lowe, E.D.; Staunton, D.; Kawamura, A.; Westwood, I.M.; Bhakta, S.; Garner, A.C.; Wilson, D.L.; Seden, P.T.; et al. Piperidinols that show anti-tubercular activity as inhibitors of arylamine N-acetyltransferase: An essential enzyme for mycobacterial survival inside macrophages. PLoS ONE 2012, 7, e52790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butler, R.E.; Cihlarova, V.; Stewart, G.R. Effective generation of reactive oxygen species in the mycobacterial phagosome requires K+ efflux from the bacterium. Cell. Microbiol. 2010, 12, 1186–1193. [Google Scholar] [CrossRef] [PubMed]
- Fisher, A.B. Redox signaling across cell membranes. Antioxid. Redox Signal. 2009, 11, 1349–1356. [Google Scholar] [CrossRef] [Green Version]
- Del Río, L.A.; Sandalio, L.M.; Corpas, F.J.; Palma, J.M.; Barroso, J.B. Reactive oxygen species and reactive nitrogen species in peroxisomes. Production, scavenging, and role in cell signaling. Plant Physiol. 2006, 141, 330–335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Peroxisomes. In Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002. [Google Scholar]
- Kretschmer, M.; Wang, J.; Kronstad, J.W. Peroxisomal and mitochondrial β-oxidation pathways influence the virulence of the pathogenic fungus Cryptococcus neoformans. Eukaryot. Cell 2012, 11, 1042–1054. [Google Scholar] [CrossRef] [Green Version]
- Imanaka, T.; Aihara, K.; Suzuki, Y.; Yokota, S.; Osumi, T. The 70-kDa peroxisomal membrane protein (PMP70), an ATP-binding cassette transporter. Cell Biochem. Biophys. 2000, 32, 131–138. [Google Scholar] [CrossRef]
- Wanders, R.J.; Waterham, H.R.; Ferdinandusse, S. Metabolic Interplay between Peroxisomes and Other Subcellular Organelles Including Mitochondria and the Endoplasmic Reticulum. Front. Cell Dev. Biol. 2015, 3, 83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puckett, S.; Trujillo, C.; Wang, Z.; Eoh, H.; Ioerger, T.R.; Krieger, I.; Sacchettini, J.; Schnappinger, D.; Rhee, K.Y.; Ehrt, S. Glyoxylate detoxification is an essential function of malate synthase required for carbon assimilation in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2017, 114, E2225–E2232. [Google Scholar] [CrossRef] [Green Version]
- Anes, E.; Peyron, P.; Staali, L.; Jordao, L.; Gutierrez, M.G.; Kress, H.; Hagedorn, M.; Maridonneau-Parini, I.; Skinner, M.A.; Wildeman, A.G.; et al. Dynamic life and death interactions between Mycobacterium smegmatis and J774 macrophages. Cell. Microbiol. 2006, 8, 939–960. [Google Scholar] [CrossRef] [PubMed]
- Jena, P.; Mohanty, S.; Mohanty, T.; Kallert, S.; Morgelin, M.; Lindstrøm, T.; Borregaard, N.; Stenger, S.; Sonawane, A.; Sørensen, O.E. Azurophil granule proteins constitute the major mycobactericidal proteins in human neutrophils and enhance the killing of mycobacteria in macrophages. PLoS ONE 2012, 7, e50345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Padhi, A.; Pattnaik, K.; Biswas, M.; Jagadeb, M.; Behera, A.; Sonawane, A. Mycobacterium tuberculosis LprE Suppresses TLR2-Dependent Cathelicidin and Autophagy Expression to Enhance Bacterial Survival in Macrophages. J. Immunol. 2019, 203, 2665–2678. [Google Scholar] [CrossRef]
- Makni-Maalej, K.; Chiandotto, M.; Hurtado-Nedelec, M.; Bedouhene, S.; Gougerot-Pocidalo, M.A.; Dang, P.M.; El-Benna, J. Zymosan induces NADPH oxidase activation in human neutrophils by inducing the phosphorylation of p47phox and the activation of Rac2: Involvement of protein tyrosine kinases, PI3Kinase, PKC, ERK1/2 and p38MAPkinase. Biochem. Pharmacol. 2013, 85, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Lewendon, A.; Ellis, J.; Shaw, W.V. Structural and mechanistic studies of galactoside acetyltransferase, the Escherichia coli LacA gene product. J. Biol. Chem. 1995, 270, 26326–26331. [Google Scholar] [CrossRef] [Green Version]
- Edmondson, D.G.; Roth, S.Y. Identification of protein interactions by far Western analysis. Curr. Protoc. Mol. Biol. 2001, 55, 20.6.1–20.6.10. [Google Scholar] [CrossRef]
- Bansal, K.; Sinha, A.Y.; Ghorpade, D.S.; Togarsimalemath, S.K.; Patil, S.A.; Kaveri, S.V.; Balaji, K.N.; Bayry, J. Src homology 3-interacting domain of Rv1917c of Mycobacterium tuberculosis induces selective maturation of human dendritic cells by regulating PI3K-MAPK-NF-kappaB signaling and drives Th2 immune responses. J. Biol. Chem. 2010, 285, 36511–36522. [Google Scholar] [CrossRef] [Green Version]




| Gene Name | Sequence (5′>>3′) |
|---|---|
| Rv3034c-Forward | GGA TCC GTG AAC GTC CTC AGT TTG GGC TCG T |
| Rv3034c-Reverse | AAG CTT CTA GCG GGC CGC CTT CTT GC |
| Rv3034c N-ter Reverse | TCACCGCCCCCGCGGGGGCGGGCCGCA |
| Rv3034c C-ter Forward | AAAACTGCAGGTGCCACCGGCGTCACCTCGC |
| Mtb::Rv3034(±pptr) Forward | CACC CCA TGG GTG TAC GTG GTG TAA GTG TCG |
| Mtb::Rv3034(±pptr) Reverse | AGCT GCA TGC AGCACCAGTCGGCCATTAGCA |
| Msm-pSC300:Rv3034c Forward | ATGGATCCCTGCAGGTGAACGTCCTCAGTTTGGGCT |
| Msm-pSC300:Rv3034c Reverse | ATTAAGCTTGATATC GCGGGCCGCCTTCTTGCGTT |
| MsmpSC300:Rv3034cC-ter Forward | ATTAAGCTT GATATCGAATTCCTGCAGCT |
| Start–End | Observed Mass | Mr(expt) | Delta | Sequence |
|---|---|---|---|---|
| 1–16 | 1648.7180 | 1647.7107 | −0.1296 | MNVLSLGSSSGVVWGR.V |
| 17–32 | 1468.4590 | 1467.4517 | −0.3528 | R.VPITAPAGAATGVTSR.A |
| 33–40 | 915.0550 | 914.0477 | −0.3551 | R.ADAHSQMR.R |
| 42–50 | 936.1210 | 935.1137 | −0.3575 | R.YAQTGPTAK.L |
| 51–66 | 1755.8930 | 1754.8857 | 0.0261 | K.LSSAPMTTMWGAPLHR.R |
| 81–88 | 892.5120 | 891.5047 | −0.0382 | K.FLTLASLK.W |
| 95–103 | 1168.1250 | 1167.1177 | −0.4899 | R.AYTPWYLVR.Y |
| 104–106 | 524.2900 | 523.2827 | 0.0284 | R.YWR.L |
| 112–120 | 1034.6180 | 1033.6107 | 0.0075 | K.LANPHIITR.G |
| 121–127 | 751.4140 | 750.4067 | −0.0031 | R.GMVFLGK.G |
| 128–144 | 1833.0130 | 1832.0057 | 0.0265 | K.GVEIHATPELAQLEIGR.W |
| 145–151 | 854.5430 | 853.5357 | 0.0911 | R.WVHIGDK.N |
| 152–155 | 503.7740 | 502.7667 | 0.4804 | K.NTIR.A |
| 156–162 | 769.5000 | 768.4927 | 0.1049 | R.AHEGSLR.F |
| 167–171 | 543.3640 | 542.3567 | 0.0027 | K.VVLGR.D |
| 221–230 | 1085.7020 | 1084.6947 | 0.1030 | R.IGPDTWIGVK.V |
| 231–235 | 573.3350 | 572.3277 | −0.0369 | K.VSVLR.G |
| 236–241 | 604.3450 | 603.3377 | 0.0037 | R.GTTIGR.G |
| 242–252 | 1154.6410 | 1153.6337 | 0.0311 | R.GCVLGSHAVVR.G |
| 253–267 | 1429.8940 | 1428.8867 | 0.1255 | R.GAIPDYSIAVGAPAK.V |
| 284–295 | 1242.6740 | 1241.6667 | 0.0052 | R.AELAAALADIER.K |
| Gene Name | Sequence (5′>>3′) |
|---|---|
| Rv3034c-Forward | ATT CTC AGA TGC GCC GAT AC |
| Rv3034c-Reverse | AGT AGC GCA CCA GGT ACC AC |
| Mfp2-Forward | GCATTGATGTGGTGGTGAAC |
| Mfp2-Reverse | GAATGCGGCCATAGTTCTGT |
| Acaa1-Forward | GGCCTTCTTTCAAGGGAAAC |
| Acaa1-Reverse | CTAAGCCCTGACGACGAGAC |
| Catalase-Forward | ACATGGTCTGGGACTTCTGG |
| Catalase-Reverse | CAAGTTTTTGATGCCCTGGT |
| Acox1-Forward | GCTGAGGAACCTGTGTCTCT |
| Acox1-Reverse | TCAAAGGCATCCACCAAAGC |
| Icl1-Forward | GTTTAGCGAAGCGGTGAAAG |
| Icl1-Reverse | CCGCCAGGGTAATAAACTGA |
| Mas-Forward | GCACCGGCAGCATTTATATT |
| Mas-Reverse | GATCCAGAAAGCCGGTGTTA |
| SigA-Forward | CCAAGGGCTACAAGTTCTCG |
| SigA-Reverse | TGGATCTCCAGCACCTTCTC |
| GAPDH-Forward | AGGGCCCTGACAACTCTTTT |
| GAPDH-Reverse | AGGGGCTACATGGCAACTG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Behera, A.; Jain, P.; Ganguli, G.; Biswas, M.; Padhi, A.; Pattanaik, K.P.; Nayak, B.; Ergün, S.; Hagens, K.; Redinger, N.; et al. Mycobacterium tuberculosis Acetyltransferase Suppresses Oxidative Stress by Inducing Peroxisome Formation in Macrophages. Int. J. Mol. Sci. 2022, 23, 2584. https://doi.org/10.3390/ijms23052584
Behera A, Jain P, Ganguli G, Biswas M, Padhi A, Pattanaik KP, Nayak B, Ergün S, Hagens K, Redinger N, et al. Mycobacterium tuberculosis Acetyltransferase Suppresses Oxidative Stress by Inducing Peroxisome Formation in Macrophages. International Journal of Molecular Sciences. 2022; 23(5):2584. https://doi.org/10.3390/ijms23052584
Chicago/Turabian StyleBehera, Ananyaashree, Preeti Jain, Geetanjali Ganguli, Mainak Biswas, Avinash Padhi, Kali Prasad Pattanaik, Barsa Nayak, Süleyman Ergün, Kristine Hagens, Natalja Redinger, and et al. 2022. "Mycobacterium tuberculosis Acetyltransferase Suppresses Oxidative Stress by Inducing Peroxisome Formation in Macrophages" International Journal of Molecular Sciences 23, no. 5: 2584. https://doi.org/10.3390/ijms23052584
APA StyleBehera, A., Jain, P., Ganguli, G., Biswas, M., Padhi, A., Pattanaik, K. P., Nayak, B., Ergün, S., Hagens, K., Redinger, N., Saqib, M., Mishra, B. B., Schaible, U. E., Karnati, S., & Sonawane, A. (2022). Mycobacterium tuberculosis Acetyltransferase Suppresses Oxidative Stress by Inducing Peroxisome Formation in Macrophages. International Journal of Molecular Sciences, 23(5), 2584. https://doi.org/10.3390/ijms23052584







