Aldehyde Dehydrogenase 2 as a Therapeutic Target in Oxidative Stress-Related Diseases: Post-Translational Modifications Deserve More Attention
Abstract
:1. Introduction
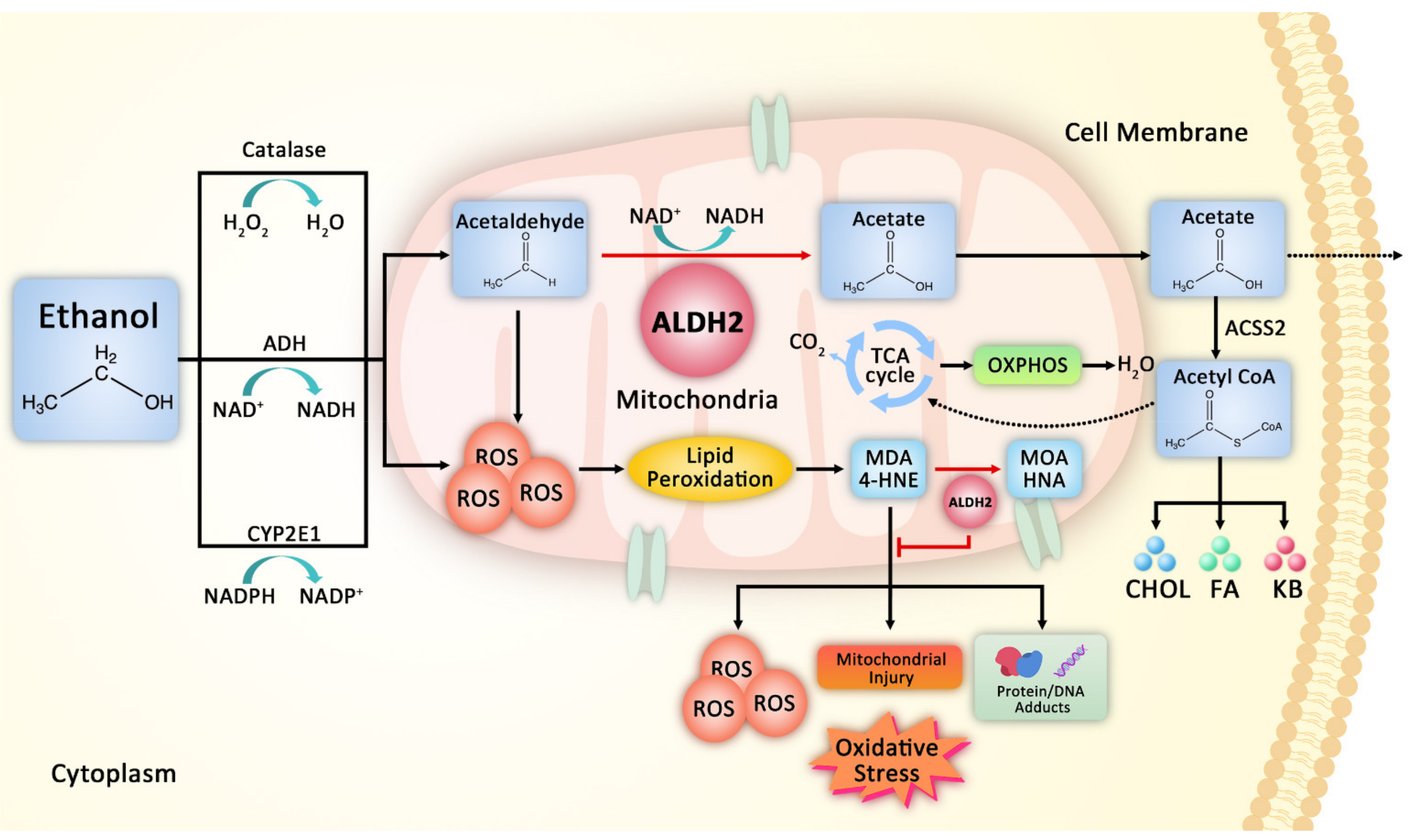
2. ALDH2 in Ethanol and Aldehyde Metabolism
2.1. Human ALDH Protein Family
2.2. Effect of ALDH2 on Detoxification of Reactive Aldehyde
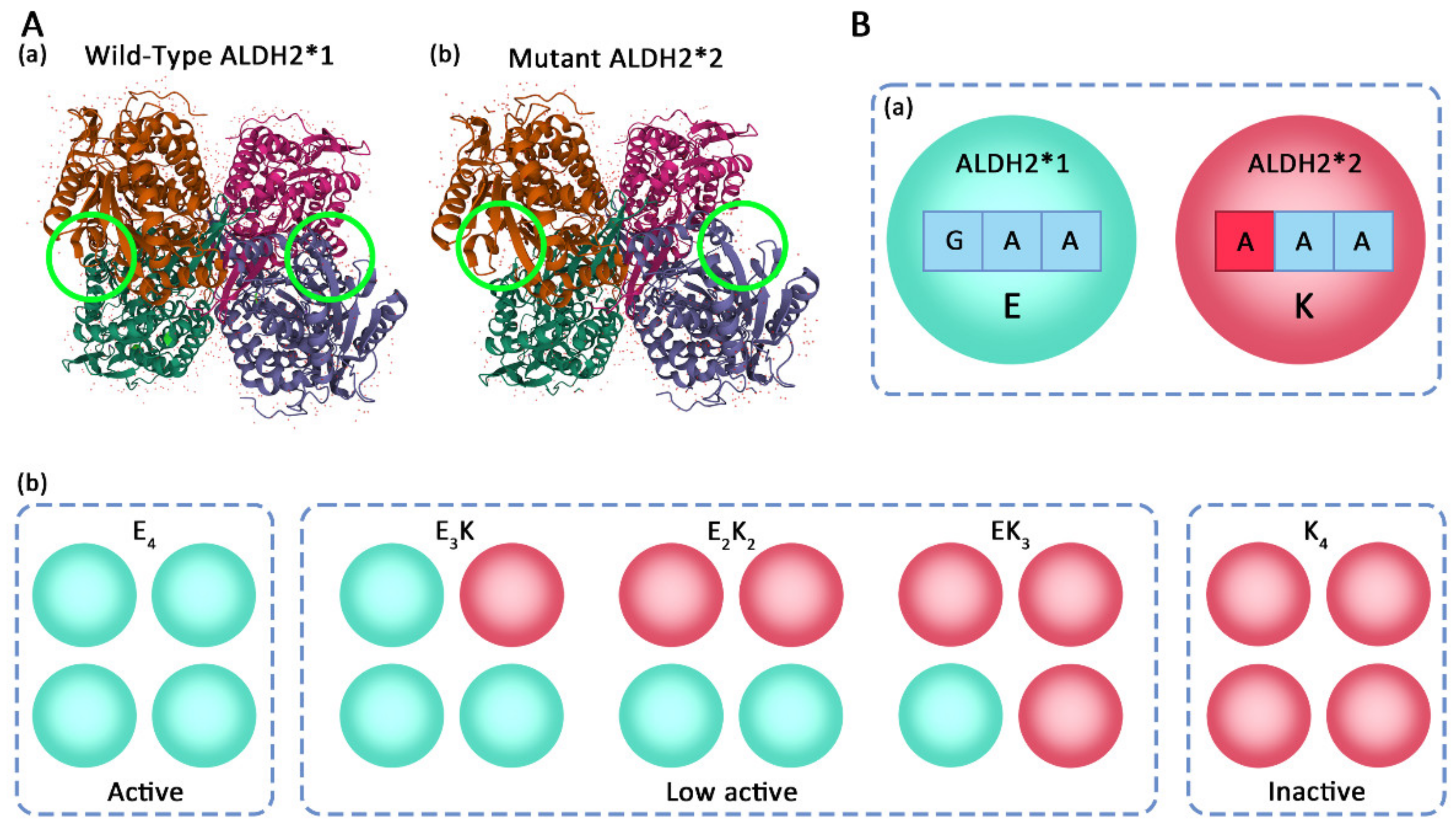
3. Structure and Gene Polymorphism of ALDH2
4. ALDH2 in Oxidative Stress-Related Physiological and Pathological Processes
4.1. Fibrosis
4.2. Apoptosis
4.3. Aging
4.4. Nervous System Injury
4.5. Other Oxidative Stress-Related Diseases
5. Regulation of ALDH2 Activity
5.1. Activators
5.2. Inhibitors
5.3. Post-Translational Modifications (PTMs) of ALDH2
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Seno, T.; Inoue, N.; Gao, D.; Okuda, M.; Sumi, Y.; Matsui, K.; Yamada, S.; Hirata, K.-I.; Kawashima, S.; Tawa, R.; et al. Involvement of NADH/NADPH oxidase in human platelet ROS production. Thromb. Res. 2001, 103, 399–409. [Google Scholar] [CrossRef]
- Breitzig, M.; Bhimineni, C.; Lockey, R.; Kolliputi, N. 4-Hydroxy-2-nonenal: A critical target in oxidative stress? Am. J. Physiol. Cell Physiol. 2016, 311, C537–C543. [Google Scholar] [CrossRef]
- Klaunig, J.E.; Xu, Y.; Isenberg, J.S.; Bachowski, S.; Kolaja, K.L.; Jiang, J.; Stevenson, D.E.; Walborg, E.F., Jr. The role of oxidative stress in chemical carcinogenesis. Environ. Health Perspect. 1998, 106, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Seddon, M.; Looi, Y.H.; Shah, A.M. Oxidative stress and redox signalling in cardiac hypertrophy and heart failure. Heart Off. J. Br. Card. Soc. 2007, 93, 903–907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ayala, A.; Muñoz, M.F.; Argüelles, S. Lipid peroxidation: Production, metabolism, and signaling mechanisms of malondialdehyde and 4-Hydroxy-2-Nonenal. Oxid. Med. Cell. Longev. 2014, 2014, 360438. [Google Scholar] [CrossRef]
- Marchitti, S.A.; Brocker, C.; Stagos, D.; Vasiliou, V. Non-P450 aldehyde oxidizing enzymes: The aldehyde dehydrogenase superfamily. Expert Opin. Drug Metab. Toxicol. 2008, 4, 697–720. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.; Brocker, C.; Koppaka, V.; Ying, C.; Jackson, B.; Matsumoto, A.; Thompson, D.C.; Vasiliou, V. Aldehyde dehydrogenases in cellular responses to oxidative/electrophilicstress. Free Radic. Biol. Med. 2013, 56, 89–101. [Google Scholar] [CrossRef] [Green Version]
- Moazamian, R.; Polhemus, A.; Connaughton, H.; Fraser, B.; Whiting, S.; Gharagozloo, P.; Aitken, R.J. Oxidative stress and human spermatozoa: Diagnostic and functional significance of aldehydes generated as a result of lipid peroxidation. Mol. Hum. Reprod. 2015, 21, 502–515. [Google Scholar] [CrossRef] [Green Version]
- Zhang, R.; Liu, B.; Fan, X.; Wang, W.; Xu, T.; Wei, S.; Zheng, W.; Yuan, Q.; Gao, L.; Yin, X.; et al. Aldehyde Dehydrogenase 2 Protects Against Post-Cardiac Arrest Myocardial Dysfunction Through a Novel Mechanism of Suppressing Mitochondrial Reactive Oxygen Species Production. Front. Pharmacol. 2020, 11, 373. [Google Scholar] [CrossRef]
- Hoshi, H.; Hao, W.; Fujita, Y.; Funayama, A.; Miyauchi, Y.; Hashimoto, K.; Miyamoto, K.; Iwasaki, R.; Sato, Y.; Kobayashi, T.; et al. Aldehyde-stress resulting from Aldh2 mutation promotes osteoporosis due to impaired osteoblastogenesis. J. Bone Miner. Res. 2012, 27, 2015–2023. [Google Scholar] [CrossRef]
- Mittal, M.; Khan, K.; Pal, S.; Porwal, K.; China, S.P.; Barbhuyan, T.K.; Baghe, K.S.; Rawat, T.; Sanyal, S.; Bhadauria, S.; et al. The thiocarbamate disulphide drug, disulfiram induces osteopenia in rats by inhibition of osteoblast function due to suppression of acetaldehyde dehydrogenase activity. Toxicol. Sci. 2014, 139, 257–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Wang, C.C. Glu504Lys single nucleotide polymorphism of aldehyde dehydrogenase 2 gene and the risk of human diseases. Biomed Res. Int. 2015, 2015, 174050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orywal, K.; Szmitkowski, M. Alcohol dehydrogenase and aldehyde dehydrogenase in malignant neoplasms. Clin. Exp. Med. 2017, 17, 131–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Wang, B.L.; Zhang, J.; He, D.Y.; Zhang, Q.; Pan, C.; Yuan, Q.H.; Shi, Y.N.; Tang, H.Y.; Xu, F.; et al. ALDH2 (aldehyde dehydrogenase 2) protects against hypoxia-induced pulmonary hypertension. Arterioscler. Thromb. Vasc. Biol. 2019, 39, 2303–2319. [Google Scholar] [CrossRef]
- Vasiliou, V.; Pappa, A. Polymorphisms of human aldehyde dehydrogenases. Pharmacology 2000, 61, 192–198. [Google Scholar] [CrossRef]
- Wei, S.J.; Xing, J.H.; Wang, B.L.; Xue, L.; Wang, J.L.; Li, R.; Qin, W.D.; Wang, J.; Wang, X.P.; Zhang, M.X.; et al. Poly (ADP-ribose) polymerase inhibition prevents reactive oxygen species induced inhibition of aldehyde dehydrogenase2 activity. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2013, 1833, 479–486. [Google Scholar] [CrossRef] [Green Version]
- Song, W.; Zou, Z.Y.; Xu, F.; Gu, X.X.; Xu, X.F.; Zhao, Q.S. Molecular cloning and expression of a second zebrafish aldehyde dehydrogenase 2 gene (aldh2b). DNA Seq. 2006, 17, 262–269. [Google Scholar] [CrossRef]
- Lee, D.J.; Lee, H.M.; Kim, J.H.; Park, I.S.; Rho, Y.S. Heavy alcohol drinking downregulates ALDH2 gene expression but heavy smoking up-regulates SOD2 gene expression in head and neck squamous cell carcinoma. World J. Surg. Oncol. 2017, 15, 163. [Google Scholar] [CrossRef] [Green Version]
- Molotkov, A.; Duester, G. Genetic evidence that retinaldehyde dehydrogenase Raldh1 (Aldh1a1) functions downstream of alcohol dehydrogenase Adh1 in metabolism of retinol to retinoic acid. J. Biol. Chem. 2003, 278, 36085–36090. [Google Scholar] [CrossRef] [Green Version]
- Vasiliou, V.; Nebert, D.W. Analysis and update of the human aldehyde dehydrogenase (ALDH) gene family. Hum. Genom. 2005, 2, 138–143. [Google Scholar] [CrossRef] [Green Version]
- Paterson, E.K.; Ho, H.; Kapadia, R.; Ganesan, A.K. 9-cis retinoic acid is the ALDH1A1 product that stimulates melanogenesis. Exp. Dermatol. 2013, 22, 202–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, P.H.; Lian, L.; Athanasios, I.; Zavras, A.I. Alcohol intake and folate antagonism via CYP2E1 and ALDH1: Effects on oral carcinogenesis. Med. Hypotheses 2012, 78, 197–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, X.; Molotkov, A.; Manabe, S.-I.; Donmoyer, C.M.; Deltour, L.; Foglio, M.H.; Cuenca, A.E.; Blaner, W.S.; Lipton, S.A.; Duester, G. Targeted disruption of Aldh1a1 (Aaldh1) provides evidence for a complex mechanism of retinoic acid synthesis in the developing retina. Mol. Cell. Biol. 2003, 23, 4637–4648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bchini, R.; Vasiliou, V.; Branlant, G.; Talfournier, F.; Rahuel-Clermont, S. Retinoic acid biosynthesis catalyzed by retinal dehydrogenases relies on a rate-limiting conformational transition associated with substrate recognition. Chem.-Biol. Interact. 2013, 202, 78–84. [Google Scholar] [CrossRef] [Green Version]
- Pai, J.; Haenisch, M.; Muller, C.H.; Goldstein, A.S.; Arnold, S.; Isoherranen, N.; Brabb, T.; Treuting, P.M.; Amory, J.K. Inhibition of retinoic acid biosynthesis by the bisdichloroacetyldiamine WIN 18,446 markedly suppresses spermatogenesis and alters retinoid metabolism in mice. J. Biol. Chem. 2014, 289, 15104–15117. [Google Scholar] [CrossRef] [Green Version]
- Trasion, S.E.; Harrison, E.H.; Wang, T.T.Y. Androgen regulation of aldehyde dehydrogenase 1A3 (ALDH1A3) in the androgen-responsive human prostate cancer cell line LNCaP. Exp. Biol. Med. 2007, 232, 762–771. [Google Scholar] [CrossRef]
- Sullivan, K.E.; Rojas, K.; Cerione, R.A.; Nakano, I.; Wilson, K.F. The stem cell/cancer stem cell marker ALDH1A3 regulates the expression of the survival factor tissue transglutaminase, in mesenchymal glioma stem cells. Oncotarget 2017, 8, 22325–22343. [Google Scholar] [CrossRef] [Green Version]
- Jackson, B.C.; Holmes, R.S.; Backos, D.S.; Reigan, P.; Thompson, D.C.; Vasiliou, V. Comparative genomics, molecular evolution and computational modeling of ALDH1B1 and ALDH2. Chem.-Biol. Interact. 2013, 202, 11–21. [Google Scholar] [CrossRef] [Green Version]
- Jackson, B.C.; Reigan, P.; Miller, B.; Thompson, D.C.; Vasiliou, V. Human ALDH1B1 polymorphisms may affect the metabolism of acetaldehyde and all-trans retinaldehyde—in vitro studies and computational modeling. Pharm. Res. 2015, 32, 1648–1662. [Google Scholar] [CrossRef] [Green Version]
- Hoeferlin, L.A.; Oleinik, N.V.; Krupenko, N.I.; Krupenko, S.A. Activation of p21-dependent G1/G2 arrest in the absence of DNA damage as an antiapoptotic response to metabolic stress. Genes Cancer 2011, 2, 889–899. [Google Scholar] [CrossRef] [Green Version]
- Khan, Q.A.; Pediaditakis, P.; Malakhau, Y.; Esmaeilniakooshkghazi, A.; Ashkavand, Z.; Sereda, V.; Krupenko, N.I.; Krupenko, S.A. CHIP E3 ligase mediates proteasomal degradation of the proliferation regulatory protein ALDH1L1 during the transition of NIH3T3 fibroblasts from G0/G1 to S-phase. PLoS ONE 2018, 13, e0199699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krupenko, S.A.; Krupenko, N.I. Loss of ALDH1L1 folate enzyme confers a selective metabolic advantage for tumor progression. Chem.-Biol. Interact. 2019, 302, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Krupenko, N.I.; Dubard, M.E.; Strickland, K.C.; Moxley, K.M.; Oleinik, N.V.; Krupenko, S.A. ALDH1L2 is the mitochondrial homolog of 10-formyltetrahydrofolate dehydrogenase. J. Biol. Chem. 2010, 285, 23056–23063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strickland, K.C.; Krupenko, N.I.; Dubard, M.E.; Hu, C.J.; Tsybovsky, Y.; Krupenko, S.A. Enzymatic properties of ALDH1L2, a mitochondrial 10-formyltetrahydrofolate dehydrogenase. Chem.-Biol. Interact. 2011, 191, 129–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, G.S.; Wang, M.F.; Chen, C.Y.; Luu, S.U.; Chou, H.C.; Li, T.K.; Yin, S.J. Involvement of acetaldehyde for full protection against alcoholism by homozygosity of the variant allele of mitochondrial aldehyde dehydrogenase gene in Asians. Pharmacogenetics 1999, 9, 463–476. [Google Scholar] [CrossRef] [PubMed]
- Dollé, L.; Boulter, L.; Leclercq, I.A.; Grunsven, L.A.v. Next generation of ALDH substrates and their potential to study maturational lineage biology in stem and progenitor cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2015, 308, G573–G578. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.M.; Liu, A.J.; Zang, P.; Dong, W.Z.; Ying, L.; Wang, W.; Xu, P.; Song, X.R.; Cai, J.; Zhang, S.Q.; et al. ALDH2 protects against stroke by clearing 4-HNE. Cell Res. 2013, 23, 915–930. [Google Scholar] [CrossRef]
- Roy, B.; Sundar, K.; Palaniyandi, S.S. 4-hydroxy-2-nonenal decreases coronary endothelial cell migration: Potentiation by aldehyde dehydrogenase 2 inhibition. Vasc. Pharmacol. 2020, 131, 106762. [Google Scholar] [CrossRef]
- Choi, J.-W.; Kim, J.-H.; Cho, S.-C.; Ha, M.-K.; Song, K.-Y.; Youn, H.-D.; Park, S.C. Malondialdehyde inhibits an AMPK-mediated nuclear translocation and repression activity of ALDH2 in transcription. Biochem. Biophys. Res. Commun. 2011, 404, 400–406. [Google Scholar] [CrossRef]
- Townsend, A.J.; Leone-Kabler, S.; Haynes, R.L.; Wu, Y.; Szweda, L.; Bunting, K.D. Selective protection by stably transfected human ALDH3A1 (but not human ALDH1A1) against toxicity of aliphatic aldehydes in V79 cells. Chem.-Biol. Interact. 2001, 130–132, 261–273. [Google Scholar] [CrossRef]
- Wymore, T.; Hempel, J.; Cho, S.S.; Alexander, D.; MacKerell, A.D.; Hugh, B.; Nicholas, H.B., Jr.; Deerfield, W.D. Molecular recognition of aldehydes by aldehyde dehydrogenase and mechanism of nucleophile activation. Proteins Struct. Funct. Bioinform. 2010, 57, 758–771. [Google Scholar] [CrossRef] [PubMed]
- Nakahara, K.; Ohkuni, A.; Kitamura, T.; Abe, K.; Naganuma, T.; Ohno, Y.u.; Zoeller, R.A.; Kihara, A. The Sjögren-Larsson syndrome gene encodes a hexadecenal dehydrogenase of the sphingosine 1-phosphate degradation pathway. Mol. Cell 2012, 46, 461–471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akio, K. Sphingosine 1-phosphate is a key metabolite linking sphingolipids to glycerophospholipids. Biochim. Biophys. Acta 2014, 1841, 766–772. [Google Scholar] [CrossRef]
- Neuber, C.; Schumacher, F.; Gulbins, E.; Kleuser, B. Method to simultaneously determine the sphingosine 1-phosphate breakdown product (2E)-hexadecenal and its fatty acid derivatives using isotope-dilution HPLC–electrospray ionization–quadrupole/time-of-flight mass spectrometry. Anal. Chem. 2014, 86, 9065–9073. [Google Scholar] [CrossRef] [PubMed]
- Marchitti, S.A.; Brocker, C.; Orlicky, D.J.; Vasiliou, V. Molecular characterization, expression analysis, and role of ALDH3B1 in the cellular protection against oxidative stress. Free Radic. Biol. Med. 2010, 49, 1432–1443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marchitti, S.A.; Orlicky, D.J.; Vasiliou, V. Expression and initial characterization of human ALDH3B1. Biochem. Biophys. Res. Commun. 2007, 356, 792–798. [Google Scholar] [CrossRef] [Green Version]
- Kitamura, T.; Takagi, S.; Naganuma, T.; Kihara, A. Mouse aldehyde dehydrogenase ALDH3B2 is localized to lipid droplets via two C-terminal tryptophan residues and lipid modification. Biochem. J. 2015, 465, 79–87. [Google Scholar] [CrossRef] [Green Version]
- Pemberton, T.A.; Srivastava, D.; Sanyal, N.; Henzl, M.T.; Becker, D.F.; Tanner, J.J. Structural studies of yeast Δ1-pyrroline-5-carboxylate dehydrogenase (ALDH4A1): Active site flexibility and oligomeric state. Biochemistry 2014, 53, 1350–1359. [Google Scholar] [CrossRef]
- Tanner, J.J. Structural biology of proline catabolic enzymes. Antioxid. Redox Signal. 2017, 30, 650–673. [Google Scholar] [CrossRef]
- Bogner, A.N.; Stiers, K.M.; McKay, C.M.; Becker, D.F.; Tanner, J.J. Structural basis for the stereospecific inhibition of the dual proline/hydroxyproline catabolic enzyme ALDH4A1 by trans-4-hydroxy-L-proline. Protein Sci. 2021, 30, 1714–1722. [Google Scholar] [CrossRef]
- Campbell, A.C.; Bogner, A.N.; Mao, Y.; Becker, D.F.; Tanner, J.J. Structural analysis of prolines and hydroxyprolines binding to the L-glutamate-γ-semialdehyde dehydrogenase active site of bifunctional proline utilization A. Arch. Biochem. Biophys. 2021, 698, 108727. [Google Scholar] [CrossRef] [PubMed]
- Leo, S.; Capo, C.; Ciminelli, B.M.; Iacovelli, F.; Menduti, G.; Funghini, S.; Donati, M.A.; Falconi, M.; Rossi, L.; Malaspina, P. SSADH deficiency in an Italian family: A novel ALDH5A1 gene mutation affecting the succinic semialdehyde substrate binding site. Metab. Brain Dis. 2017, 32, 1383–1388. [Google Scholar] [CrossRef] [PubMed]
- Vogel, K.R.; Ainslie, G.R.; Jansen, E.E.W.; Salomons, G.S.; Gibson, K.M. Therapeutic relevance of mTOR inhibition in murine succinate semialdehyde dehydrogenase deficiency (SSADHD), a disorder of GABA metabolism. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2017, 1863, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Marcadier, J.L.; Smith, A.M.; Pohl, D.; Schwartzentruber, J.; Al-Dirbashi, O.Y.; Consortium, F.C.; Majewski, J.; Ferdinandusse, S.; Wanders, R.J.; Bulman, D.E.; et al. Mutations in ALDH6A1 encoding methylmalonate semialdehyde dehydrogenase are associated with dysmyelination and transient methylmalonic aciduria. Orphanet J. Rare Dis. 2013, 8, 98. [Google Scholar] [CrossRef] [Green Version]
- Brocker, C.; Cantore, M.; Failli, P.; Vasiliou, V. Aldehyde dehydrogenase 7A1 (ALDH7A1) attenuates reactive aldehyde and oxidative stress induced cytotoxicity. Chem.-Biol. Interact. 2011, 191, 269–277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coulter-Mackie, M.B.; Tiebout, S.; Karnebeek, C.v.; Stockler, S. Overexpression of recombinant human antiquitin in E. coli: Partial enzyme activity in selected ALDH7A1 missense mutations associated with pyridoxine-dependent epilepsy. Mol. Genet. Metab. 2014, 111, 462–466. [Google Scholar] [CrossRef]
- Davis, I.; Yang, Y.; Wherritt, D.; Liu, A. Reassignment of the human aldehyde dehydrogenase ALDH8A1 (ALDH12) to the kynurenine pathway in tryptophan catabolism. J. Biol. Chem. 2018, 293, 9594–9603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Končitíková, R.; Vigouroux, A.; Kopečná, M.; Šebela, M.; Moréra, S.; Kopečný, D. Kinetic and structural analysis of human ALDH9A1. Biosci. Rep. 2019, 39, BSR20190558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wyatt, J.W.; Korasick, D.A.; Qureshi, I.A.; Campbell, A.C.; Gates, K.S.; Tanner, J.J. Inhibition, crystal structures, and in-solution oligomeric structure of aldehyde dehydrogenase 9A1. Arch. Biochem. Biophys. 2020, 691, 108477. [Google Scholar] [CrossRef]
- Vasiliou, V.; Sandoval, M.; Backos, D.S.; Jackson, B.C.; Chen, Y.; Reigan, P.; Lanaspa, M.A.; Johnson, R.J.; Koppaka, V.; Thompson, D.C. ALDH16A1 is a novel non-catalytic enzyme that may be involved in the etiology of gout via protein–protein interactions with HPRT1. Chem.-Biol. Interact. 2013, 202, 22–31. [Google Scholar] [CrossRef] [Green Version]
- Bicknell, L.S.; Pitt, J.; Aftimos, S.; Ramadas, R.; Maw, M.A.; Robertson, S.P. A missense mutation in ALDH18A1, encoding Δ1-pyrroline-5-carboxylate synthase (P5CS), causes an autosomal recessive neurocutaneous syndrome. Eur. J. Hum. Genet. 2008, 16, 1176–1186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolthuis, D.F.G.J.; Asbeck, E.V.; Mohamed, M.; Gardeitchik, T.; Lim-Melia, E.R.; Wevers, R.A.; Morava, E. Cutis laxa, fat pads and retinopathy due to ALDH18A1 mutation and review of the literature. Eur. J. Paediatr. Neurol. 2014, 18, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Holmes, R.S. Comparative and evolutionary studies of ALDH18A1 genes and proteins. Chem.-Biol. Interact. 2016, 276, 2–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steenhof, M.; Kibæk, M.; Larsen, M.J.; Christensen, M.; Lund, A.M.; Brusgaard, K.; Hertz, J.M. Compound heterozygous mutations in two different domains of ALDH18A1 do not affect the amino acid levels in a patient with hereditary spastic paraplegia. Neurogenetics 2018, 19, 145–149. [Google Scholar] [CrossRef] [PubMed]
- Oota, H.; Pakstis, A.J.; Bonne-Tamir, B.; Goldman, D.; Grigorenko, E.; Kajuna, S.L.B.; Karoma, N.J.; Kungulilo, S.; Lu, R.B.; Odunsi, K.; et al. The evolution and population genetics of the ALDH2 locus: Random genetic drift, selection, and low levels of recombination. Ann. Hum. Genet. 2004, 68, 93–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serio, R.N.; Lu, C.; Gross, S.S.; Gudas, L.J. Different effects of knockouts in ALDH2 and ACSS2 on embryonic stem cell differentiation. Alcohol. Clin. Exp. Res. 2019, 43, 1859–1871. [Google Scholar] [CrossRef] [PubMed]
- Dalleau, S.; Baradat, M.; Guéraud, F.; Huc, L. Cell death and diseases related to oxidative stress: 4-hydroxynonenal (HNE) in the balance. Cell Death Differ. 2013, 20, 1615–1630. [Google Scholar] [CrossRef] [Green Version]
- Tudek, B.; Zdżalik-Bielecka, D.; Tudek, A.; Kosicki, K.; Fabisiewicz, A.; Speina, E. Lipid peroxidation in face of DNA damage, DNA repair and other cellular processes. Free Radic. Biol. Med. 2016, 107, 77–89. [Google Scholar] [CrossRef]
- Adav, S.S.; Sze, S.K. Hypoxia-induced degenerative protein modifications associated with aging and age-associated disorders. Aging Dis. 2020, 11, 341–364. [Google Scholar] [CrossRef] [Green Version]
- Li, S.Y.; Gomelsky, M.; Duan, J.; Zhang, Z.; Gomelsky, L.; Zhang, X.; Epstein, P.N.; Ren, J. Overexpression of aldehyde dehydrogenase-2 (ALDH2) transgene prevents acetaldehyde-induced cell injury in human umbilical vein endothelial cells: Role of ERK and p38 mitogen-activated protein kinase. J. Mol. Cell. Cardiol. 2004, 40, 283–294. [Google Scholar] [CrossRef]
- Lee, J.Y.; Jung, G.Y.; Heo, H.J.; Yun, M.R.; Park, J.Y.; Bae, S.S.; Hong, K.W.; Lee, W.S.; Kim, C.D. 4-Hydroxynonenal induces vascular smooth muscle cell apoptosis through mitochondrial generation of reactive oxygen species. Toxicol. Lett. 2006, 166, 212–221. [Google Scholar] [CrossRef] [PubMed]
- Yalcinkaya, T.; Uzilday, B.; Ozgur, R.; Turkan, I. The roles of reactive carbonyl species in induction of antioxidant defence and ROS signalling in extreme halophytic model Eutrema parvulum and glycophytic model Arabidopsis thaliana. Environ. Exp. Bot. 2019, 160, 81–91. [Google Scholar] [CrossRef]
- Yun, M.R.; Park, H.M.; Seo, K.W.; Lee, S.J.; Im, D.S.; Kim, C.D. 5-Lipoxygenase plays an essential role in 4-HNE-enhanced ROS production in murine macrophages via activation of NADPH oxidase. Free Radic. Res. 2010, 44, 742–750. [Google Scholar] [CrossRef]
- Endo, J.; Sano, M.; Katayama, T.; Hishiki, T.; Shinmur, K.; Morizane, S.; Matsuhash, T.; Katsumata, Y.; Zhang, Y.; Ito, H.; et al. Metabolic remodeling induced by mitochondrial aldehyde stress stimulates tolerance to oxidative stress in the heart. Circ. Res. 2009, 105, 1118–1127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhai, X.X.; Zhang, Z.X.; Liu, W.W.; Liu, B.S.; Zhang, R.; Wang, W.J.; Zheng, W.; Xu, F.; Wang, J.L.; Chen, Y.G. Protective effect of ALDH2 against cyclophosphamide-induced acute hepatotoxicity via attenuating oxidative stress and reactive aldehydes. Biochem. Biophys. Res. Commun. 2018, 499, 93–98. [Google Scholar] [CrossRef]
- Manzo-Avalos, S.; Saavedra-Molina, A. Cellular and mitochondrial effects of alcohol consumption. Int. J. Environ. Res. Public Health 2010, 7, 4281–4304. [Google Scholar] [CrossRef] [Green Version]
- Shi, L.; Tu, B.P. Acetyl-CoA and the regulation of metabolism: Mechanisms and consequences. Curr. Opin. Cell Biol. 2015, 33, 125–131. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.M.; Ren, J. ALDH2 in alcoholic heart diseases: Molecular mechanism and clinical implications. Pharmacol. Ther. 2011, 132, 86–95. [Google Scholar] [CrossRef] [Green Version]
- Siu, G.M.; Draper, H.H. Metabolism of malonaldehyde in vivo and in vitro. Lipids 1982, 17, 349–355. [Google Scholar] [CrossRef]
- Grune, T.; Siems, W.; Kowalewski, J.; Zollner, H.; Esterbauer, H. Identification of metabolic pathways of the lipid peroxidation product 4-hydroxynonenal by enterocytes of rat small intestine. Biochem. Int. 1991, 25, 963–971. [Google Scholar] [CrossRef]
- Zhong, H.Q.; Yin, H.Y. Role of lipid peroxidation derived 4-hydroxynonenal (4-HNE) in cancer: Focusing on mitochondria. Redox Biol. 2015, 4, 193–199. [Google Scholar] [CrossRef] [Green Version]
- Lv, L.Z.; Ye, W.J.; Song, P.Y.; Chen, Y.B.; Yang, J.; Zhang, C.M.; Chen, X.P.; Luo, F.Y. Relationship between ALDH2 genotype and in-stent restenosis in Chinese Han patients after percutaneous coronary intervention. BMC Cardiovasc. Disord. 2019, 19, 176. [Google Scholar] [CrossRef] [PubMed]
- Braun, T.; Bober, E.; Singh, S.; Agarwal, D.P.; Goedde, H.W. Evidence for a signal peptide at the amino-terminal end of human mitochondrial aldehyde dehydrogenase. FEBS Lett. 1987, 215, 233–236. [Google Scholar] [CrossRef] [Green Version]
- Ni, L.; Zhou, J.Z.; Hurley, T.D.; Weiner, H. Human liver mitochondrial aldehyde dehydrogenase: Three-dimensional structure and the restoration of solubility and activity of chimeric forms. Protein Sci. 2010, 8, 2784–2790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, E.D.; Jiao, T.T.; Shen, Y.L.; Xu, Y.J.; Sun, Y.Q.; Cai, Z.C.; Zhang, Q.; Li, J.M. Molecular mechanism of HSF1-upregulated ALDH2 by PKC in ameliorating pressure overload-induced heart failure in mice. BioMed Res. Int. 2020, 2020, 3481623. [Google Scholar] [CrossRef]
- Hsu, L.C.; Bendel, R.E.; Yoshida, A. Genomic structure of the human mitochondrial aldehyde dehydrogenase gene. Genomics 1988, 2, 57–65. [Google Scholar] [CrossRef]
- Weiner, H.; Wei, B.X.; Zhou, J.Z. Subunit communication in tetrameric class 2 human liver aldehyde dehydrogenase as the basis for half-of-the-site reactivity and the dominance of the oriental subunit in a heterotetramer. Chem. Biol. Interact. 2001, 130–132, 47–56. [Google Scholar] [CrossRef]
- Gong, D.X.; Zhang, H.; Hu, S.S. Mitochondrial aldehyde dehydrogenase 2 activation and cardioprotection. J. Mol. Cell. Cardiol. 2013, 55, 58–63. [Google Scholar] [CrossRef]
- Steinmetz, C.G.; Xie, P.G.; Weiner, H.; Hurley, T.D. Structure of mitochondrial aldehyde dehydrogenase: The genetic component of ethanol aversion. Structure 1997, 5, 701–711. [Google Scholar] [CrossRef]
- Larson, H.N.; Zhou, J.Z.; Chen, Z.Q.; Stamler, J.S.; Weiner, H.; Hurley, T.D. Structural and functional consequences of coenzyme binding to the inactive Asian variant of mitochondrial aldehyde dehydrogenase. J. Biol. Chem. 2007, 282, 12940–12950. [Google Scholar] [CrossRef] [Green Version]
- González-Segura, L.; Ho, K.K.; Perez-Miller, S.; Weine, H.; Hurley, T.D. Catalytic contribution of threonine 244 in human ALDH2. Chem. Biol. Interact. 2013, 202, 32–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore, S.A.; Baker, H.M.; Blythe, T.J.; Kitson, K.E.; Kitson, T.M.; Baker, E.N. Sheep liver cytosolic aldehyde dehydrogenase: The structure reveals the basis for the retinal specificity of class 1 aldehyde dehydrogenases. Structure 1998, 6, 1541–1551. [Google Scholar] [CrossRef] [Green Version]
- Perez-Miller, S.J.; Hurley, T.D. Coenzyme isomerization is integral to catalysis in aldehyde dehydrogenase. Biochemistry 2003, 42, 7100–7109. [Google Scholar] [CrossRef]
- Wenzl, M.V.; Beretta, M.; Gorren, A.C.F.; Zeller, A.; Baral, P.K.; Gruber, K.; Russwurm, M.; Koesling, D.; Schmidt, K.; Mayer, B. Role of the general base Glu-268 in nitroglycerin bioactivation and superoxide formation by aldehyde dehydrogenase-2. J. Biol. Chem. 2009, 284, 19878–19886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, R.; Zhao, Z.H.; Sun, M.Y.; Luo, J.C.; Xiao, Y.C. ALDH2 gene polymorphism in different types of cancers and its clinical significance. Life Sci. 2016, 147, 59–66. [Google Scholar] [CrossRef]
- Yang, L.Y.; Li, L.L.; Tang, H.Y.; Ma, T.B.; Li, Y.L.; Zhang, X.W.; Shi, X.L.; Liu, J.Y. Alcohol-aggravated episodic pain in humans with SCN11A mutation and ALDH2 polymorphism. Pain 2020, 161, 1470–1482. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, T.; Nishimoto, Y.; Hirata, T.; Abe, Y.; Takebayashi, T.; Arai, Y. ALDH2 p.E504K variation and sex are major factors associated with current and quitting alcohol drinking in Japanese Oldest Old. Genes 2021, 12, 799. [Google Scholar] [CrossRef]
- Wang, X.P.; Sheikh, S.; Saigal, D.; Robinson, L.; Weiner, H. Heterotetramers of human liver mitochondrial (class 2) aldehyde dehydrogenase expressed in Escherichia coli: A model to study the heterotetramers expected to be found in Oriental people. J. Biol. Chem. 1996, 271, 31172–31178. [Google Scholar] [CrossRef] [Green Version]
- Lai, C.L.; Yao, C.T.; Chau, G.Y.; Yang, L.F.; Kuo, T.Y.; Chiang, C.P.; Yin, S.J. Dominance of the inactive Asian variant over activity and protein contents of mitochondrial aldehyde dehydrogenase 2 in human liver. Alcohol. Clin. Exp. Res. 2014, 38, 44–50. [Google Scholar] [CrossRef]
- Zuo, W.; Zhan, Z.Y.; Ma, L.; Bai, W.; Zeng, S.G. Effect of ALDH2 polymorphism on cancer risk in Asians. Medicine 2019, 98, e14855. [Google Scholar] [CrossRef]
- Idewaki, Y.; Iwase, M.; Fujii, H.; Ohkuma, T.; Ide, H.; Kaizu, S.; Jodai, T.; Kikuchi, Y.; Hirano, A.; Nakamura, U.; et al. Association of genetically determined aldehyde dehydrogenase 2 activity with diabetic complications in relation to alcohol consumption in japanese patients with type 2 diabetes mellitus: The fukuoka diabetes registry. PLoS ONE 2015, 10, e0143288. [Google Scholar] [CrossRef]
- Zhong, S.S.; Li, L.X.; Zhang, Y.L.; Zhang, L.L.; Lu, J.H.; Guo, S.Y.; Liang, N.N.; Ge, J.; Zhu, M.J.; Tao, Y.Z.; et al. Acetaldehyde dehydrogenase 2 interactions with LDLR and AMPK regulate foam cell formation. J. Clin. Investig. 2019, 129, 252–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohta, S.; Ohsawa, I.; Kamino, K.; Ando, F.; Shimokata, H. Mitochondrial ALDH2 deficiency as an oxidative stress. Ann. New York Acad. Sci. 2004, 1011, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Gross, E.R.; Zambelli, V.O.; Small, B.A.; Ferreira, J.C.B.; Chen, C.-H.; Mochly-Rosen, D. A personalized medicine approach for Asian Americans with the aldehyde dehydrogenase 2*2 variant. Annu. Rev. Pharmacol. Toxicol. 2015, 55, 107–127. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.H.; Ferreira, J.C.B.; Gross, E.R.; Mochly-Rosen, D. Targeting aldehyde dehydrogenase 2: New therapeutic opportunities. Physiol. Rev. 2014, 94, 1–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vasiliou, V.; Pappa, A.; Petersen, D.R. Role of aldehyde dehydrogenases in endogenous and xenobiotic metabolism. Chem.-Biol. Interact. 2000, 129, 1–19. [Google Scholar] [CrossRef]
- Wen, Y.Q.; Wang, X.P.; Xiao, S.Y.; Wang, Y.J. Ectopic expression of VpALDH2B4, a novel aldehyde dehydrogenase gene from Chinese wild grapevine (Vitis pseudoreticulata), enhances resistance to mildew pathogens and salt stress in Arabidopsis. Planta 2012, 236, 525–539. [Google Scholar] [CrossRef]
- Khan, M.; Qiao, F.; Kumar, P.; Islam, S.M.T.; Singh, A.K.; Won, J.; Sing, I. Neuroprotective effects of Alda-1 mitigate spinal cord injury in mice: Involvement of Alda-1-induced ALDH2 activation-mediated suppression of reactive aldehyde mechanisms. Neural Regen. Res. 2021, 17, 185–193. [Google Scholar] [CrossRef]
- Kim, J.; Chen, C.H.; Yang, J.Y.; Mochly-Rosen, D. Aldehyde dehydrogenase 2*2 knock-in mice show increased reactive oxygen species production in response to cisplatin treatment. J. Biomed. Sci. 2017, 24, 33. [Google Scholar] [CrossRef] [Green Version]
- Yuan Hou, J.; Xiong Zhong, Z.; Ting Deng, Q.; Dong Liu, S.; Fang Lin, L. Association between the polymorphism of aldehyde dehydrogenase 2 gene and cerebral infarction in a Hakka population in southern China. Biochem. Genet. 2020, 58, 322–334. [Google Scholar] [CrossRef]
- Yuan, Q.H.; Cao, S.C.; Dong, Q.Q.; Wang, Z.; Xu, Y.S.; Han, Q.; Ma, J.J.; Wei, S.J.; Pang, J.J.; Yang, F.H.; et al. ALDH2 activation inhibited cardiac fibroblast-to-myofibroblast transformation via the TGF-β1/Smad signaling pathway. J. Cardiovasc. Pharmacol. 2019, 73, 9. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Xu, D.L.; Wang, S.J.; Fu, H.; Wang, K.Q.; Zou, Y.Z.; Sun, A.J.; Ge, J.B. Inhibition of aldehyde dehydrogenase 2 activity enhances antimycin-induced rat cardiomyocytes apoptosis through activation of MAPK signaling pathway. Biomed. Pharmacother. 2011, 65, 590–593. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Jia, X.J.; Zong, Q.F.; Zhang, G.J.; Ye, H.W.; Hu, J.; Gao, Q.; Guan, S.D. Remote ischemic postconditioning protects the heart by upregulating ALDH2 expression levels through the PI3K/Akt signaling pathway. Mol. Med. Rep. 2014, 10, 536–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, H.J.; Won, Y.S.; Park, O.; Chang, B.; Duryee, M.J.; Thiele, G.E.; Matsumoto, A.; Singh, S.; Abdelmegeed, M.A.; Song, B.J.; et al. Aldehyde dehydrogenase 2 deficiency ameliorates alcoholic fatty liver but worsens liver inflammation and fibrosis in mice. Hepatology 2014, 60, 146–157. [Google Scholar] [CrossRef] [Green Version]
- Ma, X.; Luo, Q.; Zhu, H.; Liu, X.; Dong, Z.; Zhang, K.; Zou, Y.; Wu, J.; Ge, J.; Sun, A. Aldehyde dehydrogenase 2 activation ameliorates CCl4-induced chronic liver fibrosis in mice by up-regulating Nrf2/HO-1 antioxidant pathway. J. Cell. Mol. Med. 2017, 22, 3965–3978. [Google Scholar] [CrossRef]
- D’Souza, Y.; Elharram, A.; Soon-Shiong, R.; Andrew, R.D.; Bennett, B.M. Characterization of Aldh2−/− mice as an age-related model of cognitive impairment and Alzheimer’s disease. Mol. Brain 2015, 8, 27. [Google Scholar] [CrossRef]
- Ghoweria, A.O.; Gagolewiczb, P.; Fraziera, H.N.; Ganta, J.C.; Andrewb, R.D.; Bennettb, B.M.; Thibault, O. Neuronal calcium imaging, excitability, and plasticity changes in the aldh2−/− mouse model of sporadic Alzheimer’s disease. J. Alzheimer’s Dis. 2020, 77, 1623–1637. [Google Scholar] [CrossRef]
- Grünblatt, E.; Riederer, P. Aldehyde dehydrogenase (ALDH) in Alzheimer’s and Parkinson’s disease. J. Neural Transm. 2016, 123, 83–90. [Google Scholar] [CrossRef]
- Yu, R.L.; Tu, S.C.; Wu, R.M.; Lu, P.A.; Tan, C.H. Interactions of COMT and ALDH2 genetic polymorphisms on symptoms of parkinson’s disease. Brain Sci. 2021, 11, 361. [Google Scholar] [CrossRef]
- Xue, L.; Xue, L.; Yang, F.H.; Han, Z.Q.; Cui, S.M.; Dai, S.; Xu, F.; Zhang, C.X.; Wang, X.P.; Pang, J.J.; et al. ALDH2 mediates the dose-response protection of chronic ethanol against endothelial senescence through SIRT1/p53 pathway. Biochem. Biophys. Res. Commun. 2018, 504, 777–783. [Google Scholar] [CrossRef]
- Hu, J.F.; Wang, H.X.; Li, H.H.; Hu, J.; Yu, Y.; Gao, Q. Inhibition of ALDH2 expression aggravates renal injury in a rat sepsis syndrome model. Exp. Ther. Med. 2017, 14, 2249–2254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Q.K.; He, G.Z.; Wang, J.; Wang, Y.K.; Chen, W. Pretreatment with the ALDH2 agonist Alda-1 reduces intestinal injury induced by ischaemia and reperfusion in mice. Clin. Sci. 2017, 131, 1123–1136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henderson, N.C.; Rieder, F.; Wynn, T.A. Fibrosis: From mechanisms to medicines. Nature 2020, 587, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Santibañez, J.F.; Quintanilla, M.; Bernabeu, C. TGF-β/TGF-β receptor system and its role in physiological and pathological conditions. Clin. Sci. 2011, 121, 233–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rauchman, M.; Griggs, D. Emerging strategies to disrupt the central TGF-β axis in kidney fibrosis. Transl. Res. 2019, 209, 90–104. [Google Scholar] [CrossRef]
- Akhurst, R.J. Targeting TGF-β signaling for therapeutic gain. Cold Spring Harb. Perspect. Biol. 2017, 9, a022301. [Google Scholar] [CrossRef] [Green Version]
- Fernandez, I.E.; Eickelberg, O. The Impact of TGF-β on lung fibrosis. Proc. Am. Thorac. Soc. 2012, 9, 111–116. [Google Scholar] [CrossRef]
- Gu, X.Y.; Fang, T.T.; Kang, P.F.; Hu, J.F.; Yu, Y.; Li, Z.H.; Cheng, X.Y.; Gao, Q. Effect of ALDH2 on high glucose-induced cardiac fibroblast oxidative stress, apoptosis, and fibrosis. Oxid. Med. Cell. Longev. 2017, 2017, 9257967. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.J.; Hua, Y.; Chen, H.M.; Yang, H.Y.; Zhang, T.; Huang, G.Q.; Fan, H.J.; Tan, Z.B.; Huang, X.F.; Liu, B.; et al. Aldehyde dehydrogenase-2 protects against myocardial infarction-related cardiac fibrosis through modulation of the Wnt/β-catenin signaling pathway. Ther. Clin. Risk Manag. 2015, 11, 1371–1381. [Google Scholar] [CrossRef] [Green Version]
- Gu, L.; Zhu, Y.J.; Yang, X.; Guo, Z.J.; Xu, W.B.; Tian, X.L. Effect of TGF-β/Smad signaling pathway on lung myofibroblast differentiation. Acta Pharmacol. Sin. 2007, 28, 382–391. [Google Scholar] [CrossRef] [Green Version]
- Yu, H.Y.; Königshoff, M.; Jayachandran, A.; Handley, D.; Seeger, W.; Kaminski, N.; Eickelberg, O. Transgelin is a direct target of TGF-β/Smad3-dependent epithelial cell migration in lung fibrosis. FASEB J. 2008, 22, 1778–1789. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.Y.; Gao, W.K.; Dang, Y.Y.; Liu, X.; Li, Y.J.; Peng, X.; Ye, X.Y. Both ERK/MAPK and TGF-Beta/Smad signaling pathways play a role in the kidney fbrosis of diabetic mice accelerated by blood glucose fluctuation. J. Diabetes Res. 2013, 2013, 463740. [Google Scholar] [CrossRef] [Green Version]
- Meng, X.M.; Huang, X.R.; Xiao, J.; Chung, A.C.K.; Qin, W.; Chen, H.Y.; Lan, H.Y. Disruption of Smad4 impairs TGF-β/Smad3 and Smad7 transcriptional regulation during renal inflammation and fibrosis in vivo and in vitro. Kidney Int. 2012, 81, 266–279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harris, W.T.; Kelly, D.R.; Zhou, Y.; Wang, D.; Macewen, M.; Hagood, J.S.; Clancy, J.P.; Ambalavanan, N.; Sorscher, E.J. Myofibroblast differentiation and enhanced Tgf-B signaling in cystic fibrosis lung disease. PLoS ONE 2013, 8, e70196. [Google Scholar] [CrossRef]
- Xiao, Z.C.; Zhang, J.; Peng, X.G.; Dong, Y.J.; Jia, L.X.; Li, H.H.; Du, J. The Notch γ-secretase inhibitor ameliorates kidney fibrosis via inhibition of TGF-β/Smad2/3 signaling pathway activation. Int. J. Biochem. Cell Biol. 2014, 55, 65–71. [Google Scholar] [CrossRef]
- Chu, H.Y.; Shi, Y.; Jiang, S.; Zhong, Q.C.; Zhao, Y.Q.; Liu, Q.M.; Ma, Y.Y.; Shi, X.G.; Ding, W.F.; Zhou, X.D.; et al. Treatment effects of the traditional Chinese medicine Shenks in bleomycin-induced lung fibrosis through regulation of TGF-beta/Smad3 signaling and oxidative stress. Sci. Rep. 2017, 7, 2252. [Google Scholar] [CrossRef] [PubMed]
- Louzada, R.A.; Corre, R.; Hassani, R.A.E.; Meziani, L.; Jaillet, M.; Cazes, A.; Crestani, B.; Deutsch, E.; Dupuy, C. NADPH oxidase DUOX1 sustains TGF-β1 signalling and promotes lung fibrosis. Eur. Respir. J. 2020, 57, 1901949. [Google Scholar] [CrossRef] [PubMed]
- Donepudi, M.; Grutter, M.G. Structure and zymogen activation of caspases. Biophys. Chem. 2002, 101–102, 145–153. [Google Scholar] [CrossRef]
- Chakrabarty, S.; Verhelst, S.H.L. Controlled inhibition of apoptosis by photoactivatable caspase inhibitors. Cell Chem. Biol. 2020, 27, 1434–1440.e10. [Google Scholar] [CrossRef]
- Li, D.R.; Chen, J.; Ai, Y.W.; Gu, X.Q.; Li, L.; Che, D.; Jiang, Z.D.; Li, L.; Chen, S.; Huang, H.W.; et al. Estrogen-related hormones induce apoptosis by stabilizing schlafen-12 protein turnover. Mol. Cell 2019, 75, 1103–1116. [Google Scholar] [CrossRef]
- Torcia, M.; Chiara, G.D.; Nencioni, L.; Ammendola, S.; Labardi, D.; Lucibello, M.; Rosini, P.; Marlier, L.N.J.L.; Bonini, P.; Sbarba, P.D.; et al. Nerve growth factor inhibits apoptosis in memory B lymphocytes via inactivation of p38 MAPK, prevention of Bcl-2 phosphorylation, and cytochrome c release. J. Biol. Chem. 2001, 276, 39027–39036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.D.; Weng, Q.J.; Zhang, L.; He, Q.J.; Yang, B. VEGF and Bcl-2 interact via MAPKs signaling pathway in the response to hypoxia in neuroblastoma. Cell. Mol. Neurobiol. 2009, 29, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Y.; Wang, R.; Wang, Y.J.; Peng, R.Q.; Wu, Y.; Yuan, Y.F. Ginkgo biloba extract mitigates liver fibrosis and apoptosis by regulating p38 MAPK, NF-κB/IκBα, and Bcl-2/Bax signaling. Drug Des. Dev. Ther. 2015, 9, 6303–6317. [Google Scholar] [CrossRef] [Green Version]
- Andreka, G.; Vertesaljai, M.; Szantho, G.; Font, G.; Piroth, Z.; Fontos, G.; Juhasz, E.D.; Szekely, L.; Szelid, Z.; Turner, M.S.; et al. Remote ischaemic postconditioning protects the heart during acute myocardial infarction in pigs. Heart 2007, 93, 749–752. [Google Scholar] [CrossRef] [PubMed]
- Li, S.Y.; Gilbert, S.A.B.; Li, Q.; Ren, J. Aldehyde dehydrogenase-2 (ALDH2) ameliorates chronic alcohol ingestion-induced myocardial insulin resistance and endoplasmic reticulum stress. J. Mol. Cell. Cardiol. 2009, 47, 247–255. [Google Scholar] [CrossRef] [Green Version]
- Zhong, Z.B.; Hu, Q.C.; Fu, Z.; Wang, R.; Xiong, Y.; Zhang, Y.; Liu, Z.Z.; Wang, Y.F.; Ye, Q.F. Increased expression of aldehyde dehydrogenase 2 reduces renal cell apoptosis during ischemia/reperfusion injury after hypothermic machine perfusion. Artif. Organs 2016, 40, 596–603. [Google Scholar] [CrossRef]
- Cao, S.C.; Bian, Y.; Zhou, X.; Yuan, Q.H.; Wei, S.J.; Xue, L.; Yang, F.H.; Dong, Q.Q.; Wang, W.J.; Zheng, B.Y.; et al. A small-molecule activator of mitochondrial aldehyde dehydrogenase 2 reduces the severity of cerulein-induced acute pancreatitis. Biochem. Biophys. Res. Commun. 2019, 522, 518–524. [Google Scholar] [CrossRef]
- Shen, C.; Wang, C.; Han, S.S.; Wang, Z.J.; Dong, Z.; Zhao, X.N.; Wang, P.; Zhu, H.; Sun, X.L.; Ma, X.; et al. Aldehyde dehydrogenase 2 deficiency negates chronic low-to-moderate alcohol consumption-induced cardioprotecion possibly via ROS-dependent apoptosis and RIP1/RIP3/MLKL-mediated necroptosis. Biochim. Biophys. Acta 2017, 1863, 1912–1918. [Google Scholar] [CrossRef]
- Xu, D.; Guthrie, J.R.; Mabry, S.; Sack, T.M.; Truog, W.E. Mitochondrial aldehyde dehydrogenase attenuates hyperoxia-induced cell death through activation of ERK/MAPK and PI3K-Akt pathways in lung epithelial cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 2016, 291, L966–L975. [Google Scholar] [CrossRef]
- Kim, G.-J.; Song, D.H.; Yoo, H.S.; Chung, K.-H.; Lee, K.J.; An, J.H. Hederagenin supplementation alleviates the pro-inflammatory and apoptotic response to alcohol in rats. Nutrients 2017, 9, 41. [Google Scholar] [CrossRef] [Green Version]
- Tian, X.L.; Li, Y. Endothelial cell senescence and age-related vascular diseases. J. Genet. Genom. 2014, 41, 485–495. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.H.; Wang, J.L.; Huang, H.J.; Huang, Y.; Mi, T.; Zhang, C.T.; Zhang, L. Fibroblast growth factor 21 delayed endothelial replicative senescence and protected cells from H2O2-induced premature senescence through SIRT1. Am. J. Transl. Res. 2017, 9, 4492–4501. [Google Scholar] [PubMed]
- Koc, A.; Gasch, A.P.; Rutherford, J.C.; Kim, H.Y.; Gladyshev, V.N. Methionine sulfoxide reductase regulation of yeast lifespan reveals reactive oxygen species-dependent and -independent components of aging. Proc. Natl. Acad. Sci. USA 2004, 101, 7999–8004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nannelli, G.; Ziche, M.; Donnini, S.; Morbidelli, L. Endothelial aldehyde dehydrogenase 2 as a target to maintain vascular wellness and function in ageing. Biomedicines 2020, 8, 4. [Google Scholar] [CrossRef] [Green Version]
- Gu, C.H.; Xing, Y.; Jiang, L.; Chen, M.; Xu, M.; Yin, Y.; Li, C.; Yang, Z.; Yu, L.; Ma, H. Impaired cardiac SIRT1 activity by carbonyl stress contributes to aging-related ischemic intolerance. PLoS ONE 2013, 8, e74050. [Google Scholar] [CrossRef] [Green Version]
- Haigis, M.C.; Sinclair, D.A. Mammalian sirtuins: Biological insights and disease relevance. Annu. Rev. Pathol. Mech. Dis. 2010, 5, 253–295. [Google Scholar] [CrossRef] [Green Version]
- Prola, A.; Silva, J.P.D.; Guilbert, A.; Lecru, L.; Piquereau, J.; Ribeiro, M.; Mateo, P.; Gressette, M.; Fortin, D.; Boursier, C.; et al. SIRT1 protects the heart from ER stress-induced cell death through eIF2α deacetylation. Cell Death Differ. 2016, 24, 343–356. [Google Scholar] [CrossRef] [Green Version]
- Xue, L.; Cui, S.M.; Cui, Z.Q.; Yang, F.H.; Pang, J.J.; Xu, F.; Chen, Y.G. ALDH2: A new protector against age-independent myocardial senescence. Int. J. Cardiol. 2016, 210, 38–40. [Google Scholar] [CrossRef]
- Nannelli, G.; Terzuoli, E.; Giorgio, V.; Donnini, S.; Lupetti, P.; Giachetti, A.; Bernardi, P.; Ziche, M. ALDH2 activity reduces mitochondrial oxygen reserve capacity in endothelial cells and induces senescence properties. Oxid. Med. Cell. Longev. 2018, 2018, 9765027. [Google Scholar] [CrossRef] [Green Version]
- Zhu, H.; Wang, Z.; Dong, Z.; Wang, C.; Cao, Q.; Fan, F.; Zhao, J.J.; Liu, X.W.; Yuan, M.; Sun, X.L.; et al. Aldehyde dehydrogenase 2 deficiency promotes atherosclerotic plaque instability through accelerating mitochondrial ROS-mediated vascular smooth muscle cell senescence. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2018, 1865, 1782–1792. [Google Scholar] [CrossRef]
- Xue, L.; Zhu, W.Y.; Yang, F.H.; Dai, S.; Han, Z.Q.; Xu, F.; Guo, P.; Chen, Y.G. Appropriate dose of ethanol exerts anti-senescence and anti-atherosclerosis protective effects by activating ALDH2. Biochem. Biophys. Res. Commun. 2019, 512, 319–325. [Google Scholar] [CrossRef] [PubMed]
- Bartoli-Leonard, F.; Saddic, L.; Aikawa, E. Double-edged sword of ALDH2 mutations: One polymorphism can both benefit and harm the cardiovascular system. Eur. Heart J. 2020, 41, 2453–2455. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.H.; Joshi, A.U.; Mochly-Rosen, D. The role of mitochondrial aldehyde dehydrogenase 2 (ALDH2) in neuropathology and neurodegeneration. Acta Neurol. Taiwanica 2016, 25, 111–123. [Google Scholar]
- Gęgotek, A.; Skrzydlewska, E. Biological effect of protein modifications by lipid peroxidation products. Chem. Phys. Lipids 2019, 221, 46–52. [Google Scholar] [CrossRef]
- Burke, W.J.; Kumar, V.B.; Pandey, N.; Panneton, W.M.; Gan, Q.; Franko, M.W.; O’Dell, M.; Li, S.W.; Pan, Y.; Chung, H.D.; et al. Aggregation of α-synuclein by DOPAL, the monoamine oxidase metabolite of dopamine. Acta Neuropathol. 2008, 115, 193–203. [Google Scholar] [CrossRef]
- Plotegher, N.; Berti, G.; Ferrari, E.; Tessari, I.; Zanetti, M.; Lunelli, L.; Greggio, E.; Bisaglia, M.; Veronesi, M.; Girotto, S. DOPAL derived alpha-synuclein oligomers impair synaptic vesicles physiological function. Sci. Rep. 2017, 7, 40699. [Google Scholar] [CrossRef]
- Doorn, J.A.; Florang, V.R.; Schamp, J.H.; Vanle, B.C. Aldehyde dehydrogenase inhibition generates a reactive dopamine metabolite autotoxic to dopamine neurons. Parkinsonism Relat. Disord. 2014, 20, S73–S75. [Google Scholar] [CrossRef] [Green Version]
- Goldsteina, D.S.; Kopin, I.J.; Sharabi, Y. Catecholamine autotoxicity. Implications for pharmacology and therapeutics of Parkinson disease and related disorders. Pharmacol. Ther. 2014, 144, 268–282. [Google Scholar] [CrossRef] [Green Version]
- Hou, G.J.; Chen, L.; Liu, G.; Li, L.; Yang, Y.; Yan, H.X.; Zhang, H.L.; Tang, J.; Yang, Y.C.; Lin, X.M.; et al. Aldehyde dehydrogenase-2 (ALDH2) opposes hepatocellular carcinoma progression by regulating AMP-activated protein kinase signaling in mice. Hepatology 2017, 65, 1628–1644. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.Y.; Abel, J.; Neuhaus, T.; Ko, Y.; Harth, V.; Hamajima, N.; Tajima, K.; Yoo, K.Y.; Park, S.K.; Noh, D.Y.; et al. Role of alcohol and genetic polymorphisms of CYP2E1 and ALDH2 in breast cancer development. Pharmacogenetics 2003, 13, 67–72. [Google Scholar] [CrossRef]
- Eom, S.Y.; Zhang, Y.W.; Kim, S.H.; Choe, K.H.; Lee, K.Y.; Park, J.D.; Hong, Y.C.; Kim, Y.D.; Kang, J.W.; Kim, H. Influence of NQO1, ALDH2, and CYP2E1 genetic polymorphisms, smoking, and alcohol drinking on the risk of lung cancer in Koreans. Cancer Causes Control. 2009, 20, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, J.R.; Lee, W.T.; Ou, C.Y.; Huang, C.C.; Chang, C.C.; Tsai, S.T.; Chen, K.C.; Huang, J.S.; Wong, T.Y.; Lai, Y.H.; et al. Validation of alcohol flushing questionnaire to identify ALDH 2 status in a case–control study of head and neck cancer. Alcohol. Clin. Exp. Res. 2019, 43, 1225–1233. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Legrand, A.J.; Cunniffe, S.; Hume, S.; Poletto, M.; Vaz, B.; Ramadan, K.; Yao, D.; Dianov, G.L. Interplay between base excision repair protein XRCC1 and ALDH2 predicts overall survival in lung and liver cancer patients. Cell. Oncol. 2018, 41, 527–539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, R.; Kamatani, Y.; Takahashi, A.; Usami, M.; Hosono, N.; Kawaguchi, T.; Tsunoda, T.; Kamatani, N.; Kubo, M.; Nakamura, Y.; et al. Functional variants in ADH1B and ALDH2 coupled with alcohol and smoking synergistically enhance esophageal cancer risk. Gastroenterology 2009, 137, 1768–1775. [Google Scholar] [CrossRef] [PubMed]
- Miyasaka, K.; Hosoya, H.; Tanaka, Y.; Uegaki, S.; Kino, K.; Shimokata, H.; Kawanami, T.; Funakoshi, A. Association of aldehyde dehydrogenase 2 gene polymorphism with pancreatic cancer but not colon cancer. Geriatr. Gerontol. Int. 2010, 10, S120–S126. [Google Scholar] [CrossRef]
- Shin, C.M.; Kim, N.; Cho, S.-I.; Kim, J.S.; Jung, H.C.; Song, I.S. Association between alcohol intake and risk for gastric cancer with regard to ALDH2 genotype in the Korean population. Int. J. Epidemiol. 2011, 40, 1047–1055. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Huycke, M.; Herman, T.; Wang, X. Glutathione S-transferase alpha 4 induction by activator protein 1 in colorectal cancer. Oncogene 2016, 35, 5795–5806. [Google Scholar] [CrossRef]
- Chen, L.; Wu, M.; Ji, C.; Yuan, M.; Liu, C.; Yin, Q. Silencing transcription factor FOXM1 represses proliferation, migration, and invasion while inducing apoptosis of liver cancer stem cells by regulating the expression of ALDH2. IUBMB Life 2020, 72, 285–295. [Google Scholar] [CrossRef]
- Chen, C.H.; Budas, G.R.; Churchill, E.N.; Disatnik, M.H.; Hurley, T.D.; Mochly-Rosen, D. Activation of aldehyde dehydrogenase-2 reduces ischemic damage to the heart. Science 2008, 321, 1493–1495. [Google Scholar] [CrossRef] [Green Version]
- Budas, G.R.; Disatnik, M.H.; Chen, C.H.; Mochly-Rosen, D. Activation of aldehyde dehydrogenase 2 (ALDH2) confers cardioprotection in protein kinase C epsilon (PKCε) knockout mice. J. Mol. Cell. Cardiol. 2010, 48, 757–764. [Google Scholar] [CrossRef] [Green Version]
- Perez-Miller, S.; Younus, H.; Vanam, R.; Chen, C.H.; Mochly-Rosen, D.; Hurley, T.D. Alda-1 is an agonist and chemical chaperone for the common human aldehyde dehydrogenase 2 variant. Nat. Struct. Mol. Biol. 2010, 17, 159–165. [Google Scholar] [CrossRef] [Green Version]
- Yan, W.; Long, P.; Wei, D.; Yan, W.; Zheng, X.; Chen, G.; Wang, J.; Zhang, Z.; Chen, T.; Chen, M. Protection of retinal function and morphology in MNU-induced retinitis pigmentosa rats by ALDH2: An in-vivo study. BMC Ophthalmol. 2020, 20, 55. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.H.; Sun, L.; Mochly-Rosen, D. Mitochondrial aldehyde dehydrogenase and cardiac diseases. Cardiovasc. Res. 2010, 88, 51–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, S.H.; Zhang, H.F.; Yang, Z.B.; Li, T.B.; Liu, B.; Lou, Z.; Ma, Q.L.; Luo, X.J.; Peng, J. Alda-1 reduces cerebral ischemia/reperfusion injury in rat through clearance of reactive aldehydes. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2014, 387, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhao, Q.; Ye, F.; Huang, C.Y.; Chen, W.M.; Huang, W.Q. Alda-1, an ALDH2 activator, protects against hepatic ischemia/reperfusion injury in rats via inhibition of oxidative stress. Free Radic Res. 2018, 52, 629–638. [Google Scholar] [CrossRef] [PubMed]
- He, M.S.; Long, P.; Yan, W.M.; Chen, T.; Guo, L.F.; Zhang, Z.M.; Wang, S.W. ALDH2 attenuates early-stage STZ-induced aged diabetic rats retinas damage via Sirt1/Nrf2 pathway. Life Sci. 2018, 215, 227–235. [Google Scholar] [CrossRef]
- Hua, T.F.; Yang, M.; Zhou, Y.Y.; Chen, L.M.; Wu, H.M.; Liu, R.Y. Alda-1 prevents pulmonary epithelial barrier dysfunction following severe hemorrhagic shock through clearance of reactive aldehydes. BioMed Res. Int. 2019, 2019, 2476252. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.Y.; Wang, L.; Qin, X.; Turdi, S.; Sun, D.D.; Culver, B.; Reiter, R.J.; Wang, X.M.; Zhou, H.; Ren, J. ALDH2 contributes to melatonin-induced protection against APP/PS1 mutation-prompted cardiac anomalies through cGAS-STING-TBK1-mediated regulation of mitophagy. Signal Transduct. Target. Ther. 2020, 5, 1335–1347. [Google Scholar] [CrossRef]
- Chen, C.H.; Gray, M.O.; Mochly-Rosen, D. Cardioprotection from ischemia by a brief exposure to physiological levels of ethanol: Role of epsilon protein kinase C. Proc. Natl. Acad. Sci. USA 1999, 96, 12784–12789. [Google Scholar] [CrossRef] [Green Version]
- Lang, X.E.; Wang, X.; Zhang, K.R.; Lv, J.Y.; Jin, J.H.; Li, Q.S. Isoflurane preconditioning confers cardioprotection by cctivation of ALDH2. PLoS ONE 2013, 8, e52469. [Google Scholar] [CrossRef] [Green Version]
- Li, W.J.; Yin, L.; Sun, X.L.; Wu, J.; Dong, Z.; Hu, K.; Sun, A.J.; Ge, J.B. Alpha-lipoic acid protects against pressure overload-induced heart failure via ALDH2-dependent Nrf1-FUNDC1 signaling. Cell Death Dis. 2020, 11, 599. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Yang, J.Y.; Mou, Y.H.; Wang, L.H.; Zhou, Y.N.; Wu, C.F. Differences in the activities of resveratrol and ascorbic acid in protection of ethanol-induced oxidative DNA damage in human peripheral lymphocytes. Food Chem. Toxicol. 2012, 50, 168–174. [Google Scholar] [CrossRef] [PubMed]
- Wenzel, P.; Hink, U.; Oelze, M.; Schuppan, S.; Schaeuble, K.; Schildknecht, S.; Ho, K.K.; Weiner, H.; Bachschmid, M.; Münzel, T.; et al. Role of reduced lipoic acid in the redox regulation of mitochondrial aldehyde dehydrogenase (ALDH-2) activity. Implications for mitochondrial oxidative stress and nitrate tolerance. J. Biol. Chem. 2007, 282, 792–799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Wang, H.; Hao, P.; Xue, L.; Chen, Y. Inhibition of aldehyde dehydrogenase 2 by oxidative stress is associated with cardiac dysfunction in diabetic rats. Mol. Med. 2011, 17, 172–179. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Liu, B.; Dai, Z.; Zhang, H.F.; Zhang, Y.S.; Luo, X.J.; Ma, Q.L.; Peng, J. Alpha lipoic acid protects heart against myocardial ischemia-reperfusion injury through a mechanism involving aldehyde dehydrogenase 2 activation. Eur. J. Pharmacol. 2012, 678, 32–38. [Google Scholar] [CrossRef]
- Zhang, H.; Xue, L.; Li, B.Y.; Zhang, Z.L.; Tao, S.S. Vitamin D protects against alcohol-induced liver cell injury within an NRF2-ALDH2 feedback loop. Mol. Nutr. Food Res. 2019, 63, 1801014. [Google Scholar] [CrossRef]
- Kobayashi, H.; Nakamura, S.; Sato, Y.; Kobayashi, T.; Miyamoto, K.; Oya, A.; Matsumoto, M.; Nakamura, M.; Kanaji, A.; Miyamoto, T. ALDH2 mutation promotes skeletal muscle atrophy in mice via accumulation of oxidative stress. Bone 2020, 142, 115739. [Google Scholar] [CrossRef]
- Gao, G.Y.; Li, D.J.; Keung, W.M. Synthesis of potential antidipsotropic isoflavones: Inhibitors of the mitochondrial monoamine oxidase-aldehyde dehydrogenase pathway. J. Med. Chem. 2001, 44, 3320–3328. [Google Scholar] [CrossRef]
- Lowe, E.D.; Gao, G.Y.; Johnson, L.N.; Keung, W.M. Structure of daidzin, a naturally occurring anti-alcohol-addiction agent, in complex with human mitochondrial aldehyde dehydrogenase. J. Med. Chem. 2008, 51, 4482–4487. [Google Scholar] [CrossRef]
- Choi, H.; Tostes, R.C.; Webb, R.C. Mitochondrial aldehyde dehydrogenase prevents ROS-induced vascular contraction in angiotensin II hypertensive mice. J. Am. Soc. Hypertens. 2011, 5, 154–160. [Google Scholar] [CrossRef] [Green Version]
- Rivera-Meza, M.; Vásquez, D.; Quintanilla, M.E.; Lagos, D.; Rojas, B.; Herrera-Marschitz, M.; Israel, Y. Activation of mitochondrial aldehyde dehydrogenase (ALDH2) by ALDA-1 reduces both the acquisition and maintenance of ethanol intake in rats: A dual mechanism? Neuropharmacology 2019, 146, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, K.; Kawamoto, T.; Kunugita, N.; Tsukiyama, T.; Okamoto, K.; Yoshida, A.; Nakayama, K.; Nakayama, K.-i. Aldehyde dehydrogenase (ALDH) 2 associates with oxidation of methoxyacetaldehyde; in vitro analysis with liver subcellular fraction derived from human and Aldh2 gene targeting mouse. FEBS Lett. 2000, 476, 306–311. [Google Scholar] [CrossRef] [Green Version]
- Omran, Z.; Sheikh, R.; Baothman, O.A.; Zamzami, M.A.; Alarjah, M. Repurposing disulfiram as an anti-obesity drug: Treating and preventing obesity in high-fat-fed rats. Diabetes Metab. Syndr. Obes. Targets Ther. 2020, 13, 1473–1480. [Google Scholar] [CrossRef] [PubMed]
- Roy, B.; Palaniyandi, S.S. A role for aldehyde dehydrogenase (ALDH) 2 in angiotensin II-mediated decrease in angiogenesis of coronary endothelial cells. Microvasc. Res. 2021, 135, 104133. [Google Scholar] [CrossRef]
- Song, B.J.; Abdelmegeed, M.A.; Yoo, S.H.; Kim, B.J.; Jo, S.A.; Jo, I.; Moon, K.H. Post-translational modifications of mitochondrial aldehyde dehydrogenase and biomedical implications. J. Proteom. 2011, 74, 2691–2702. [Google Scholar] [CrossRef] [Green Version]
- Churchill, E.N.; Disatnik, M.H.; Mochly-Rosen, D. Time-dependent and ethanol-induced cardiac protection from ischemia mediated by mitochondrial translocation of εPKC and activation of aldehyde dehydrogenase 2. J. Mol. Cell. Cardiol. 2009, 46, 278–284. [Google Scholar] [CrossRef] [Green Version]
- Lagranha, C.J.; Deschamps, A.; Aponte, A.; Steenbergen, C.; Murphy, E. Sex differences in the phosphorylation of mitochondrial proteins result in reduced production of reactive oxygen species and cardioprotection in females. Circ. Res. 2010, 106, 1681–1691. [Google Scholar] [CrossRef] [Green Version]
- Budas, G.R.; Churchill, E.N.; Disatnik, M.H.; Sun, L.; Mochly-Rosen, D. Mitochondrial import of PKCε is mediated by HSP90: A role in cardioprotection from ischaemia and reperfusion injury. Cardiovasc. Res. 2010, 88, 83–92. [Google Scholar] [CrossRef] [Green Version]
- Moon, K.H.; Lee, Y.M.; Song, B.J. Inhibition of hepatic mitochondrial aldehyde dehydrogenase by carbon tetrachloride through JNK-mediated phosphorylation. Free Radic. Biol. Med. 2010, 48, 391–398. [Google Scholar] [CrossRef] [Green Version]
- Harris, P.S.; Gomez, J.D.; Backos, D.S.; Fritz, K.S. Characterizing sirtuin 3 deacetylase affinity for aldehyde dehydrogenase 2. Chem. Res. Toxicol. 2017, 30, 785–793. [Google Scholar] [CrossRef] [Green Version]
- Xue, L.; Xu, F.; Meng, L.J.; Wei, S.J.; Wang, J.L.; Hao, P.P.; Bian, Y.; Zhang, Y.; Chen, Y.G. Acetylation-dependent regulation of mitochondrial ALDH2 activation by SIRT3 mediates acute ethanol-induced eNOS activation. FEBS Lett. 2011, 586, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.P.; Bourdi, M.; Li, J.H.; Aponte, A.M.; Chen, Y.; Lombard, D.B.; Gucek, M.; Pohl, L.R.; Sack, M.N. SIRT3-dependent deacetylation exacerbates acetaminophen hepatotoxicity. EMBO Rep. 2011, 12, 840–846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moon, K.H.; Hood, B.L.; Kim, B.J.; Hardwick, J.P.; Conrads, T.P.; Veenstra, T.D.; Song, B.J. Inactivation of oxidized and S-nitrosylated mitochondrial proteins in alcoholic fatty liver of rats. Hepatology 2006, 44, 1218–1230. [Google Scholar] [CrossRef] [PubMed]
- Hao, L.Y.; Sun, Q.; Zhong, W.; Zhang, W.L.; Sun, X.G.; Zhou, Z.X. Mitochondria-targeted ubiquinone (MitoQ) enhances acetaldehyde clearance by reversing alcohol-induced posttranslational modification of aldehyde dehydrogenase 2: A molecular mechanism of protection against alcoholic liver disease. Redox Biol. 2018, 14, 626–636. [Google Scholar] [CrossRef]
- Moon, K.H.; Kim, B.J.; Song, B.J. Inhibition of mitochondrial aldehyde dehydrogenase by nitric oxide-mediated S-nitrosylation. FEBS Lett. 2005, 579, 6115–6120. [Google Scholar] [CrossRef] [Green Version]
- Moon, K.H.; Hood, B.L.; Mukhopadhyay, P.; Mohanraj, R.; Abdelmegeed, M.A.; Kwon, Y.I.; Conrads, T.P.; Veenstra, T.D.; Song, B.J.; Pacher, P. Oxidative inactivation of key mitochondrial proteins leads to dysfunction and injury in hepatic ischemia reperfusion. Gastroenterology 2008, 135, 1344–1357. [Google Scholar] [CrossRef] [Green Version]
- Oelz, M.; Knor, M.; Schell, R.; Kamuf, J.; Pautz, A.; Art, J.; Wenzel, P.; Münzel, T.; Kleinert, H.; Daiber, A. Regulation of human mitochondrial aldehyde dehydrogenase (ALDH-2) activity by electrophiles in vitro. J. Biol. Chem. 2011, 286, 8893–8900. [Google Scholar] [CrossRef] [Green Version]
- Latorre-Muro, P.; Baeza, J.; Armstrong, E.A.; Hurtado-Guerrero, R.; Corzana, F.; Wu, L.E.; Sinclair, D.A.; López-Buesa, P.; Carrodeguas, J.A.; Denu, J.M. Dynamic acetylation of phosphoenolpyruvate carboxykinase toggles enzyme activity between gluconeogenic and anaplerotic reactions. Mol. Cell 2018, 71, 718–732. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.M.; Yang, W.H.; Liu, R.; Wang, L.; Yang, W.H. FOXP3 activates SUMO-conjugating UBC9 gene in MCF7 breast cancer cells. Int. J. Mol. Sci. 2018, 19, 2036. [Google Scholar] [CrossRef] [Green Version]
- Sun, F.X.; Suttapitugsakul, S.; Xiao, H.P.; Wu, R.H. Comprehensive analysis of protein glycation reveals its potential impacts on protein degradation and gene expression in human cells. J. Am. Soc. Mass Spectrom. 2019, 30, 2480–2490. [Google Scholar] [CrossRef]
- Valencia-Sánchez, M.I.; Ioannes, P.D.; Wang, M.; Truong, D.M.; Lee, R.; Armache, J.-P.; Boeke, J.D.; Armache, K.-J. Regulation of the Dot1 histone H3K79 methyltransferase by histone H4K16 acetylation. Science 2021, 371, eabc6663. [Google Scholar] [CrossRef]
- Chi, Z.X.; Byeon, H.E.; Seo, E.; Nguyen, Q.A.T.; Lee, W.; Jeong, Y.; Choi, J.; Pandey, D.; Berkowitz, D.E.; Kim, J.H.; et al. Histone deacetylase 6 inhibitor tubastatin A attenuates angiotensin II-induced hypertension by preventing cystathionine γ-lyase protein degradation. Pharmacol. Res. 2019, 146, 104281. [Google Scholar] [CrossRef]
- Patel, N.; Wang, J.H.; Shiozawa, K.; Jones, K.B.; Zhang, Y.F.; Prokop, J.W.; Davenport, G.G.; Nihira, N.T.; Hao, Z.Y.; Wong, D.; et al. HDAC2 regulates site-specific acetylation of MDM2 and its ubiquitination signaling in tumor suppression. Iscience 2019, 13, 43–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Li, D.Q. MORC2 regulates DNA damage response through a PARP1-dependent pathway. Nucleic Acids Res. 2019, 47, 8502–8520. [Google Scholar] [CrossRef] [PubMed]
- Chi, Z.X.; Byeon, H.E.; Seo, E.; Nguyen, Q.A.T.; Lee, W.; Jeong, Y.Y.; Choi, J.Y.; Pandey, D.; Berkowitz, D.E.; Kim, J.H.; et al. Post-translational modification analysis of Saccharomyces cerevisiae histone methylation enzymes reveals phosphorylation sites of regulatory potential. J. Biol. Chem. 2020, 296, 100192. [Google Scholar] [CrossRef]
- Zhou, F.; Liu, Q.Q.; Zhang, L.L.; Zhu, Q.; Wang, S.S.; Zhu, K.C.; Deng, R.Y.; Liu, Y.; Yuan, G.Y.; Wang, X.; et al. Selective inhibition of CBP/p300 HAT by A-485 results in suppression of lipogenesis and hepatic gluconeogenesis. Cell Death Dis. 2020, 11, 745. [Google Scholar] [CrossRef] [PubMed]
- Karaarslan, U.; İşgüder, R.; Bağ, Ö.; Kışla, M.; Ağın, H.; Ünal, N. Alpha lipoic acid intoxication, treatment and outcome. Clin. Toxicol. 2013, 51, 522. [Google Scholar] [CrossRef]
- Sarezky, D.; Raquib, A.R.; Dunaief, J.L.; Kim, B.J. Tolerability in the elderly population of high-dose alpha lipoic acid: A potential antioxidant therapy for the eye. Clin. Ophthalmol. 2016, 10, 1899–1903. [Google Scholar] [CrossRef] [Green Version]


| Gene Name | Chromosomal Location | Protein Subcellular Location | Expression | Substrate/Ref. |
|---|---|---|---|---|
| ALDH1A1 | 9q21.13 | Cytoplasm, cytosol | Liver, duodenum, 18 other tissues | Retinaldehyde [19,20,21]; Acetaldehyde [22] |
| ALDH1A2 | 15q21.3 | Cytoplasm, cytosol | Endometrium, testes, 8 other tissues | Retinaldehyde [23,24,25] |
| ALDH1A3 | 15q26.3 | Cytoplasm, cytosol | Prostate, bladder, 14 other tissues | Retinaldehyde [26,27] |
| ALDH1B1 | 9p13.1 | Mitochondrion | Liver, kidneys, 19 other tissues | Acetaldehyde [28,29] |
| ALDH1L1 | 3q21.3 | Cytoplasm, cytosol | Liver, kidneys, 8 other tissues | 10-Formyltetrahydrofolate [30,31,32] |
| ALDH1L2 | 12q23.3 | Mitochondrion | Pancreas, salivary gland, 22 other tissues | 10-Formyltetrahydrofolate [33,34] |
| ALDH2 | 12q24.12 | Mitochondrion | Fat, liver, 19 other tissues | Acetaldehyde [35,36]; 4-Hydroxy-2-nonenal (4-HNE) [37,38]; Malondialdehyde (MDA) [39] |
| ALDH3A1 | 17p11.2 | Cytoplasm, cytosol | Esophagus, stomach, skin | Benzaldehyde [36,40,41] |
| ALDH3A2 | 17p11.2 | Endoplasmic reticulum | Skin, adrenal glands, 22 other tissues | Hexadecenal [42,43,44] |
| ALDH3B1 | 11q13.2 | Plasma membrane | Lungs, bone marrow, 17 other tissues | Octaldehyde [36]; Benzaldehyde [45]; 4-HNE [45,46]; Hexanal [45,46]; Nonanal [46] |
| ALDH3B2 | 11q13.2 | Lipid droplet | Skin, esophagus, 3 other tissues | Medium-chain to long-chain aldehydes [47] |
| ALDH4A1 | 1p36.13 | Mitochondrion | Kidneys, liver, 8 other tissues | Glutamate γ-semialdehyde [48,49,50,51] |
| ALDH5A1 | 6p22.3 | Mitochondrion | Liver, brain, 23 other tissues | Succinic semialdehyde [52,53]; 4-HNE [53] |
| ALDH6A1 | 14q24.3 | Mitochondrion | Kidneys, liver, 11 other tissues | Methylmalonate-semialdehyde [54] |
| ALDH7A1 | 5q23.2 | Cytoplasm, cytosol, mitochondrion, nucleus | Kidneys, liver, 24 other tissues | α-Aminoadipic-semialdehyde [55,56] |
| ALDH8A1 | 6q23.3 | Cytoplasm, cytosol | Liver, kidneys | Retinaldehyde [36,57] |
| ALDH9A1 | 1q24.1 | Cytoplasm, cytosol | Fat, thyroid gland, 25 other tissues | γ-Trimethylaminobutyraldehyde [58,59] |
| ALDH16A1 | 19q13.33 | Membrane | Spleen, duodenum, 25 other tissues | Unknown, lacks measurable catalytic activity [60] |
| ALDH18A1 | 10q24.1 | Mitochondrion | Duodenum, small intestine, 25 other tissues | Glutamate; γ-Glutamyl phosphate [61,62,63,64] |
| Organ | Treatment | Tissue/Cell, Species | Disease/ Injury | Molecular Mechanism | ALDH2 Function | Notes | Ref. |
|---|---|---|---|---|---|---|---|
| Heart | ALDH2 (indirectly activated) | Human cardiac fibroblasts (HCFs), human | Myocardial fibrosis | (−) TGF-β1, Smad → (+) ALDH2 → (−) α-SMA, myofibroblastic proliferation, collagen | Inhibit myocardial fibrosis induced by TGF-β1 | TGF-β1: transforming growth factor-β1 ALDH: aldehyde dehydrogenase α-SMA: α-smooth muscle actin Smad: Drosophila mothers against decapentaplegic protein | [111] |
| ALDH2 (inhibited by daidzin) | Heart, rat | Cardiomyocytic apoptosis | Daidzin → (−) ALDH2 → (+) ERK1/2, JNK, p38 MAPK → (+) ROS, 4-HNE → (+) cardiomyocytic apoptosis | Inhibit cardiomyocytic apoptosis by downregulating the MAPK pathway | ERK1/2: extracellular signal–regulated kinase 1/2 JNK: c-Jun NH2-terminal kinase p38 MAPK: p38 mitogen-activated protein kinase 4-HNE: 4-hydroxy-2-nonenal ROS: reactive oxygen species | [112] | |
| ALDH2 (activated by RIPostC) | Heart, rat | Myocardial ischemia/reperfusion (I/R) injury | RIPostC → (+) ALDH2, Bcl-2/Bax ↑ → p-Akt/Akt ↑ → (−) cleaved caspase-3 → (−) cardiomyocytic apoptosis | Help mediate the cardio-protection of RIPostC via the PI3K—Akt pathway | RIPostC: remote ischemic postconditioning Bcl-2: B-cell lymphoma-2 Bax: Bcl-2–associated X protein Akt: protein kinase B PI3K: phosphatidylinositol 3-kinase caspase-3: cysteinyl aspartate–specific proteinase 3 | [113] | |
| Liver | ALDH2−/− (KO) | Liver, mouse | Alcoholic liver injury | Alcohol, CCl4, ALDH2−/− → (+) MAAs, acetaldehyde → (+) IL-6 → (+) ERK1/2, p38 MAPK, STAT3, TGF-β, α-SMA, COLIA1, TIMP-1 → liver inflammation, fibrosis | ALDH2 deficiency accelerates alcohol-induced liver inflammatory response and fibrosis | CCl4: carbon tetrachloride MAAs: malondialdehyde–acetaldehyde adducts IL-6: interleukin-6 STAT3: signal transducer and activator of transcription 3 COLIA1: collagen type I alpha 1 TIMP-1: tissue inhibitor of metalloproteinase-1 | [114] |
| ALDH2−/− (KO) | Liver, mouse | Chronic liver fibrosis | CCl4, ALDH2−/− → (+) TGF-β1, ROS → Bcl-2/Bax ↓, Nrf2/HO-1 ↓; (+) TIMP-1, p62, α-SMA, COLIA1; (−) Parkin → (+) fibrosis | Alleviate CCl4-induced hepatic fibrosis via Nrf2/HO-1 pathway | Nrf2: nuclear factor erythroid 2-related factor 2 HO-1: heme oxygenase-1 | [115] | |
| Nerve | ALDH2−/− (KO) | Brain, mouse | Alzheimer disease (AD) | ALDH2−/− → (+) 4-HNE → (+) p-tau, Aβ, activated caspases → (+) NFTs → synaptic and mitochondrial dysfunction → neurodegeneration | Defects in ALDH2 activity kill neurons by stimulating the accumulation of 4-HNE due to OS | tau: microtubule-associated protein tau NFTs: neurofibrillary tangles Aβ: β-amyloid | [116,117] |
| ALDH2 | Pheochromocytoma (PC12), rat | Parkinson disease (PD) | DA, NE, EPI → DOPAL, DOPEGAL + ALDH2 → DOPAC, DOMA → HVA, VMA, (−) α-Syn, (−) LBs → (−) neurotoxicity | Promote removal of neurotoxic metabolites produced in the metabolism of monoamine neurotransmitters and reduce neurotoxicity | DA: dopamine NE: norepinephrine EPI: epinephrine DOPAL: 3,4-dihydroxyphenylacetaldehyde DOPEGAL: 3,4-dihydroxyphenylglycolaldehyde DOPAC: 3,4-dihydroxyphenylacetic acid DOMA: 3,4-dihydroxymandelic acid HVA: homovanillic acid VMA: vanillylmandelic acid α-Syn: α-synuclein LBs: Lewy bodies | [118,119] | |
| Blood vessel | ALDH2 (transgenic over-expression) | Human umbilical-vein endothelial cells (HUVECs), human | Apoptosis in HUVECs | Alcohol, (+) ALDH2 → (−) ROS, ERK1/2, p38 MAPK, caspase-3 → (−) HUVEC apoptosis | Alleviate OS and apoptosis of HUVECs under acetaldehyde exposure via MAPK pathway | − | [70] |
| ALDH2−/− (KO) | Human aorta epithelial cells (HAECs), human | Endothelial senescence | ALDH2−/− → (+) 4-HNE → (−) SIRT1 → (−) p53 deacetylation → (+) endothelial senescence | Promote SIRT1 nuclear translocation by inhibiting 4-HNE accumulation to alleviate senescence | SIRT1: sirtuin 1 | [120] | |
| Others | ALDH2 (indirectly activated) | Kidney, rat | Acute kidney injury (AKI) | CYA → (−) ALDH2, SOD; (+) plasma CRE, BUN, MDA, p65, NF-κB → aggravate glomerular atrophy | Inhibition of ALDH2 aggravated the renal injury | CYA: cyanamide CRE: creatinine BUN: blood urea nitrogen MDA: malondialdehyde SOD: superoxide dismutase | [121] |
| ALDH2 (activated by Alda-1) | Intestine, mouse | Intestinal I/R injury | Alda-1 → (+) ALDH2 → (−) 4-HNE, MDA, MPO, NO, iNOS, H2O2, caspase-3, NF-κBα, TNF-α, IL-6, IL-1β; (+) IκBα, Bcl-2/Bax ↑ → alleviate inflammatory response, OS | ALDH2 activation can alleviate intestinal I/R injury by relieving inflammatory response and OS | Alda-1: N-(1,3-benzodioxol-5-ylmethyl)-2,6-dichlorobenzamide MPO: myeloperoxidase iNOS: inducible NO synthase H2O2: hydrogen peroxide NF-κB: nuclear factor κ-light-chain-enhancer of activated B cells TNF-α: tumor necrosis factor-alpha IκBα: inhibitor of NF-κB IL-1β: interleukin-1β | [122] | |
| ALDH2*2 (transfected with the ALDH2*2 gene) | Osteoblasts, mouse | Osteoporosis | ALDH2*2 → (+) 4-HNE, acetaldehyde → (+) PPARγ → (−) osteoblastic differentiation → (+) osteoblastic apoptosis → osteoporosis | ALDH2*2 as a dominant-negative form of ALDH2 promotes osteoporosis by impairing osteoblastogenesis | PPARγ: peroxisome proliferator–activated receptor gamma | [10] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, J.; Hao, Y.; Piao, X.; Gu, X. Aldehyde Dehydrogenase 2 as a Therapeutic Target in Oxidative Stress-Related Diseases: Post-Translational Modifications Deserve More Attention. Int. J. Mol. Sci. 2022, 23, 2682. https://doi.org/10.3390/ijms23052682
Gao J, Hao Y, Piao X, Gu X. Aldehyde Dehydrogenase 2 as a Therapeutic Target in Oxidative Stress-Related Diseases: Post-Translational Modifications Deserve More Attention. International Journal of Molecular Sciences. 2022; 23(5):2682. https://doi.org/10.3390/ijms23052682
Chicago/Turabian StyleGao, Jie, Yue Hao, Xiangshu Piao, and Xianhong Gu. 2022. "Aldehyde Dehydrogenase 2 as a Therapeutic Target in Oxidative Stress-Related Diseases: Post-Translational Modifications Deserve More Attention" International Journal of Molecular Sciences 23, no. 5: 2682. https://doi.org/10.3390/ijms23052682






