Genome-Wide Analyses of Heat Shock Protein Superfamily Provide New Insights on Adaptation to Sulfide-Rich Environments in Urechis unicinctus (Annelida, Echiura)
Abstract
:1. Introduction
2. Results and Discussion
2.1. Identification and Characterization of the HSPs in U. unicinctus
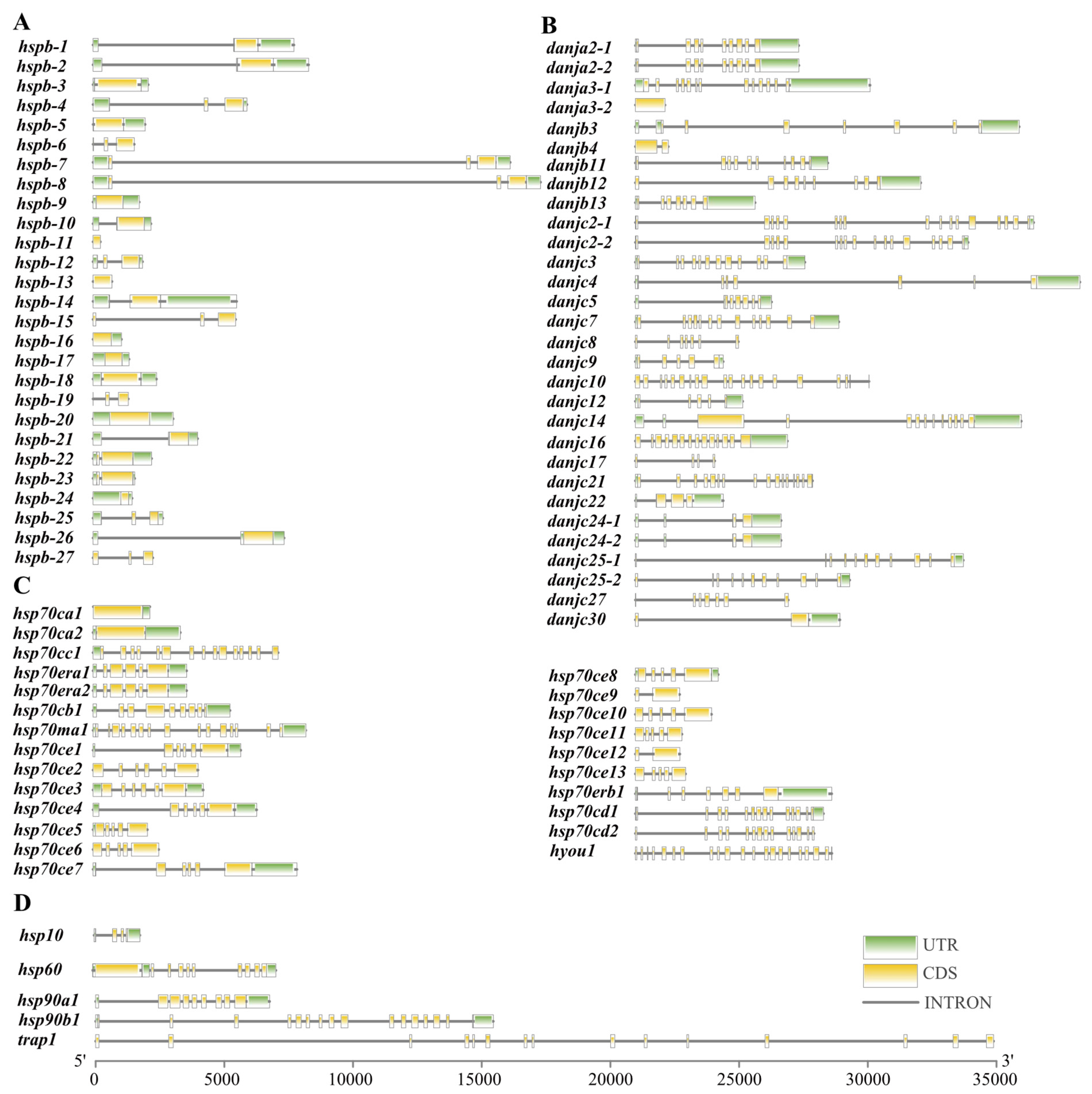
2.2. Genomic Location and Gene Duplication Events in the HSP Gene Superfamily
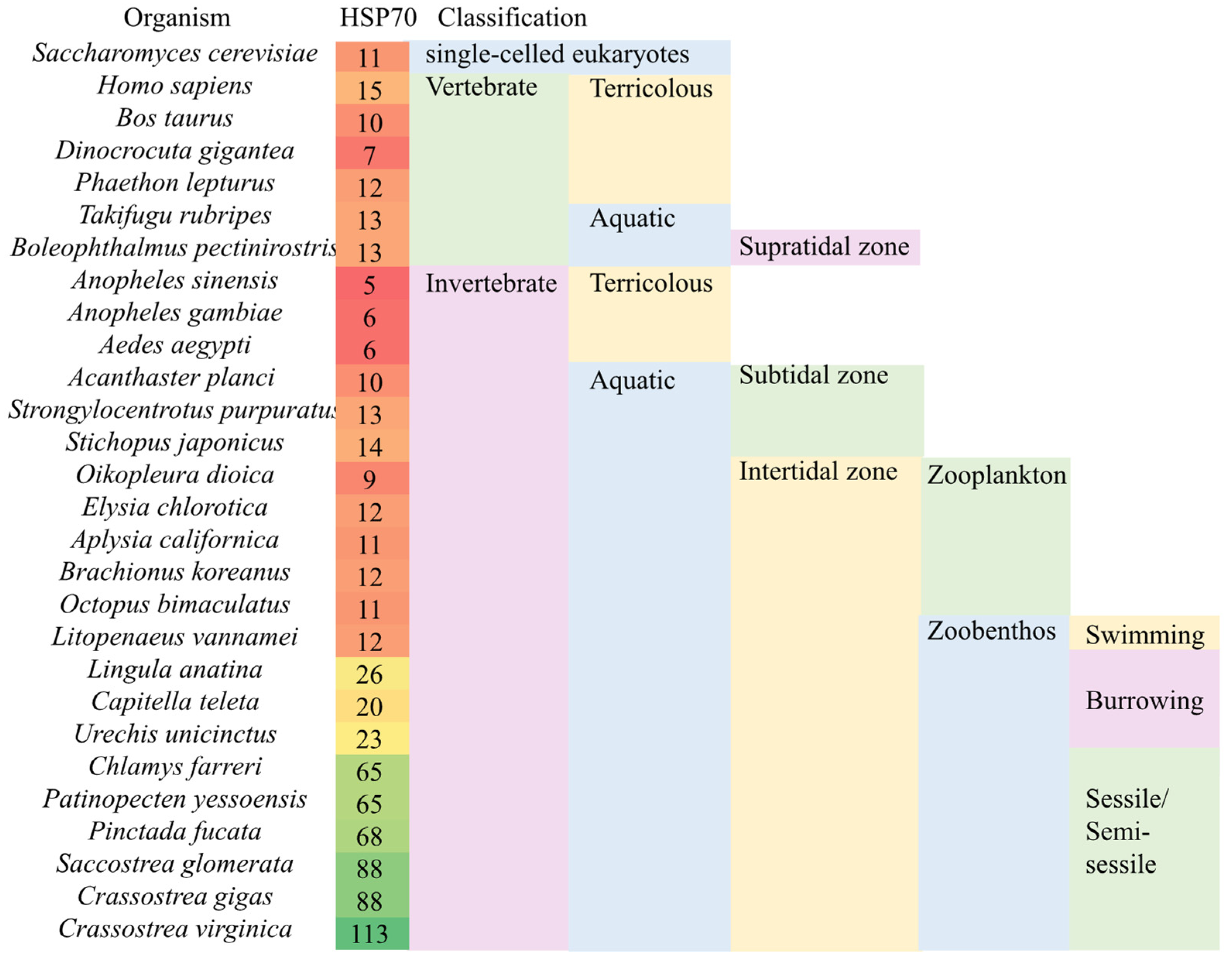
2.3. Expansion of sHSP and HSP70 Family Genes May Be a General Biosignature for Zoobenthos
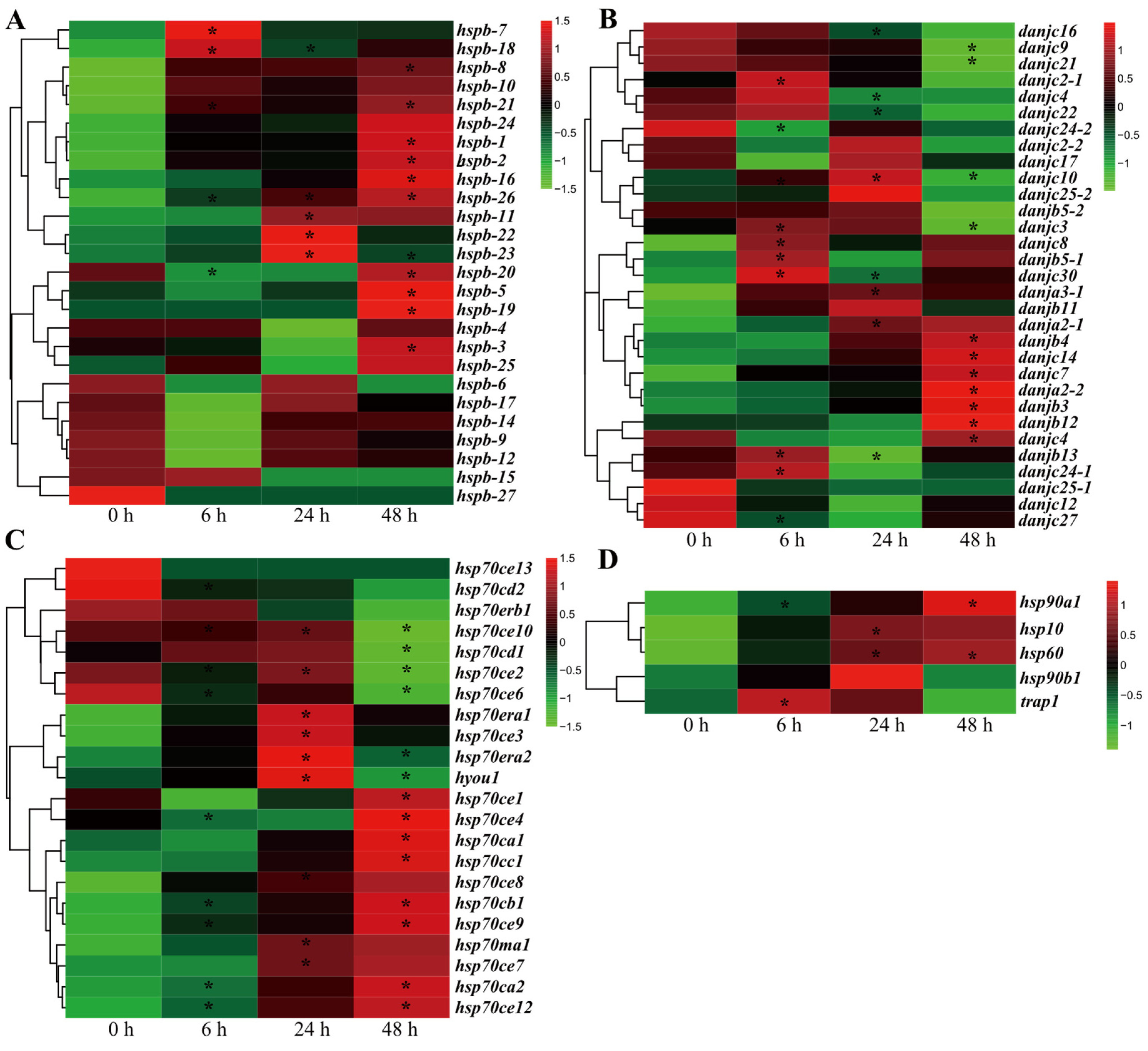
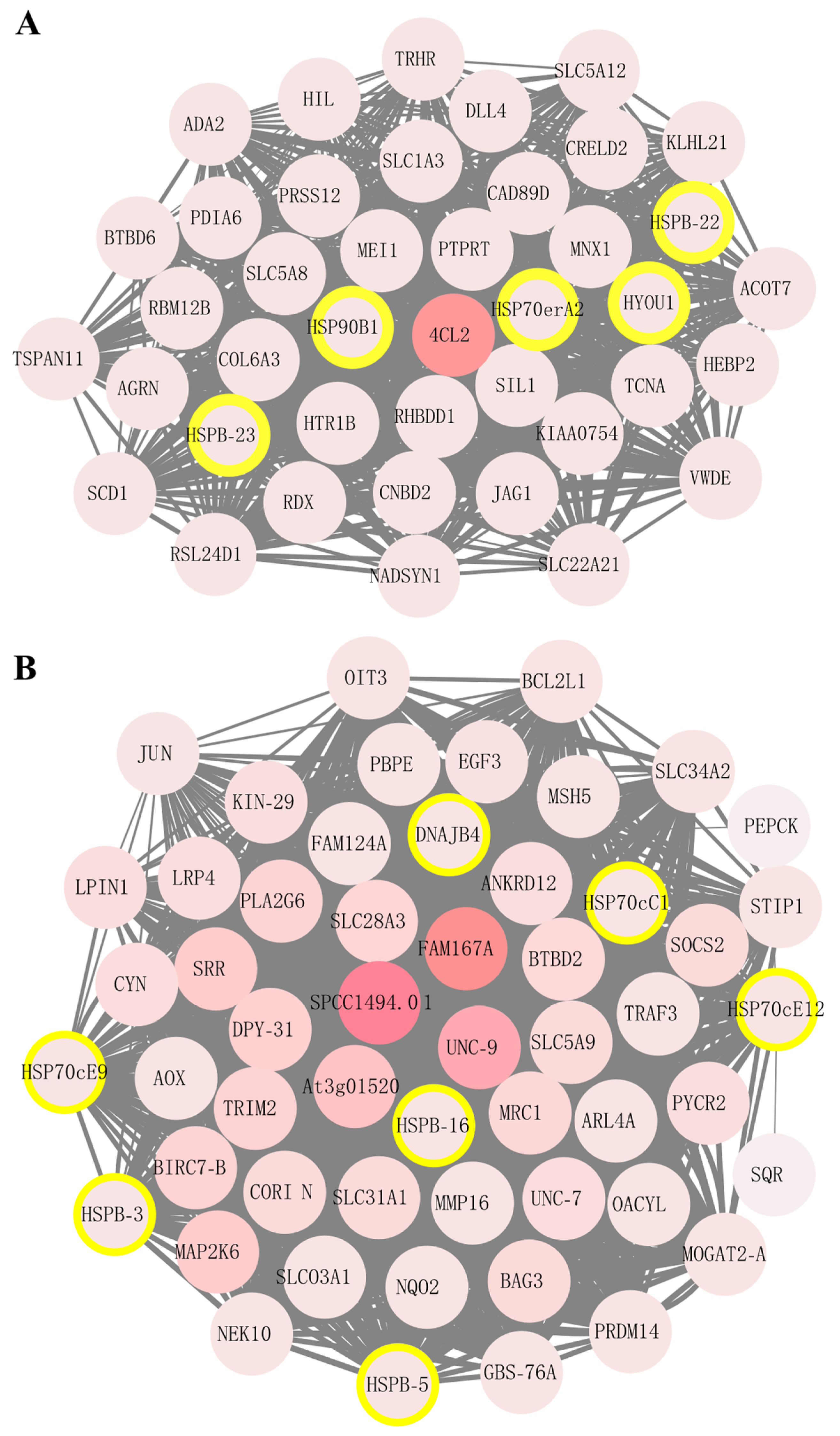
2.4. Most HSP Genes Are Involved in the Sulfide Stress Response in U. unicinctus
2.5. sHSP and HSP70 Genes Are Vital Players in Sulfide Tolerance and Environmental Adaptation in U. unicinctus
3. Materials and Methods
3.1. Animal Materials and Treatment
3.2. RNA Extraction and Illumina Sequencing
3.3. Identification of HSP Members in U. unicinctus
3.4. Multiple Alignment and Phylogenetic Analysis
3.5. Sequence Analysis and Chromosomal Localization
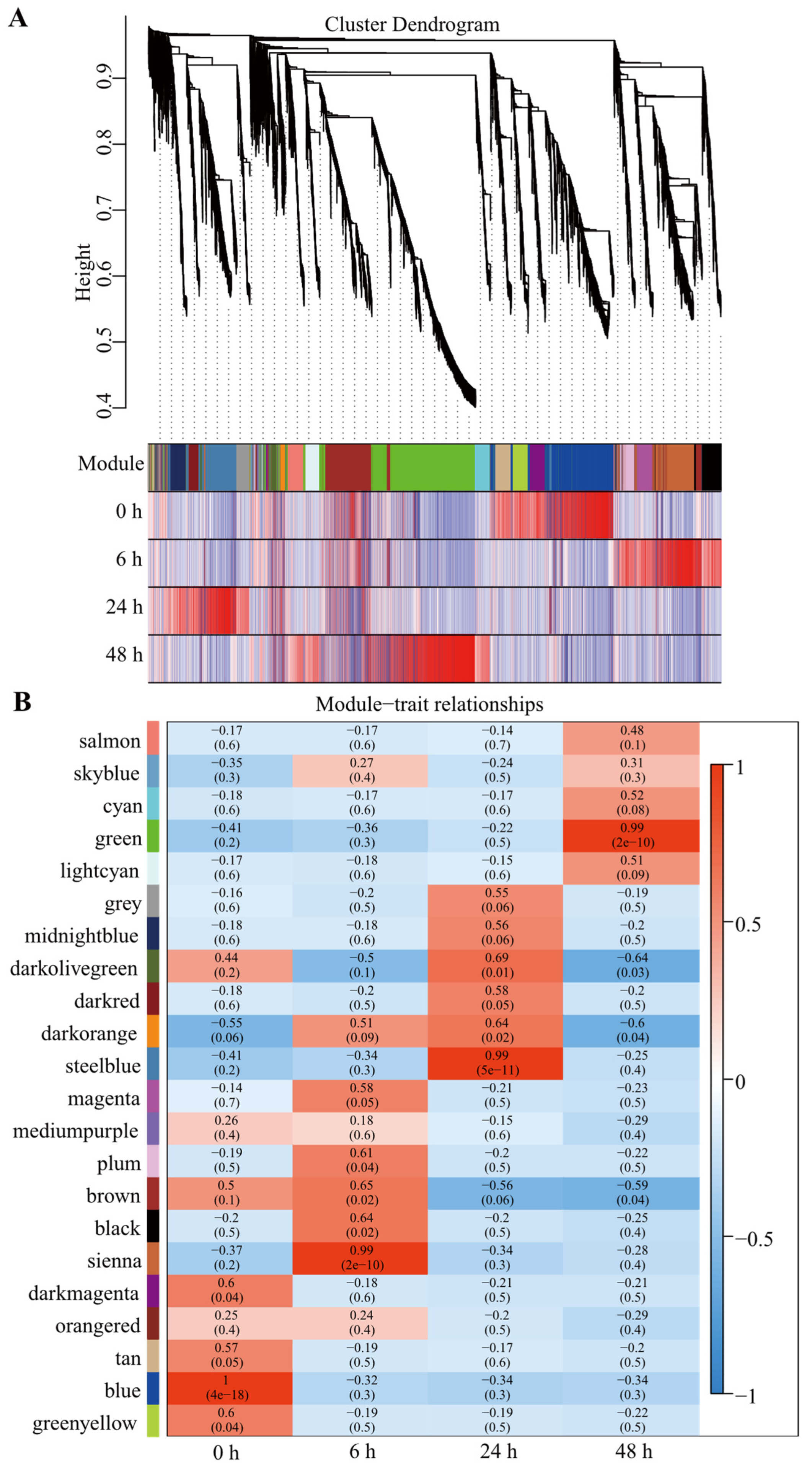
3.6. Expression Profiles and Co-Expression Network Construction
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ritossa, F. A new puffing pattern induced by temperature shock and DNP in Drosophila. Experientia 1962, 18, 571–573. [Google Scholar] [CrossRef]
- De Maio, A.; Hightower, L.E. Heat shock proteins and the biogenesis of cellular membranes. Cell Stress Chaperon. 2021, 26, 15–18. [Google Scholar] [CrossRef] [PubMed]
- Latchman, D.S. Stress proteins: An overview. In Stress Proteins; Latchman, D.S., Ed.; Springer: Berlin/Heidelberg, Germany, 1999; pp. 1–7. [Google Scholar]
- Zhao, L.; Jones, W.A. Expression of heat shock protein genes in insect stress responses. Invertebr. Surviv. J. 2012, 9, 93–101. [Google Scholar]
- Feder, M.E.; Hofmann, G.E. Heat-shock proteins, molecular chaperones, and the stress response: Evolutionary and ecological physiology. Annu. Rev. Physiol. 1999, 61, 243–282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, X.B.; Shao, Y.M.; Miao, S.; Wang, L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell. Mol. Life Sci. 2006, 63, 2560–2570. [Google Scholar] [CrossRef]
- Wong, L.L.; Do, D.T. The role of heat shock proteins in response to extracellular stress in aquatic organisms. In Heat Shock Proteins in Veterinary Medicine and Sciences; Springer: Cham, Switzerland, 2017; pp. 247–274. [Google Scholar]
- Hartl, F.U. Molecular chaperones in cellular protein folding. Nature 1996, 381, 571–580. [Google Scholar] [CrossRef]
- Kampinga, H.H.; Hageman, J.; Vos, M.J.; Kubota, H.; Tanguay, R.M.; Bruford, E.A.; Cheetham, M.E.; Chen, B.; Hightower, L.E. Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperon. 2009, 14, 105–111. [Google Scholar] [CrossRef] [Green Version]
- Waters, E.R. The evolution, function, structure, and expression of the plant sHSPs. J. Exp. Bot. 2013, 64, 391–403. [Google Scholar] [CrossRef] [Green Version]
- Hageman, J.; Kampinga, H.H. Computational analysis of the human HSPH/HSPA/DNAJ family and cloning of a human HSPH/HSPA/DNAJ expression library. Cell Stress Chaperon. 2009, 14, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Brocchieri, L.; De Macario, E.C.; Macario, A.J. hsp70 genes in the human genome: Conservation and differentiation patterns predict a wide array of overlapping and specialized functions. BMC Evol. Biol. 2008, 8, 19. [Google Scholar] [CrossRef] [Green Version]
- Hu, B.; Li, M.; Yu, X.; Xun, X.; Lu, W.; Li, X.; Li, Y.; Lou, J.; Wang, S.; Zhan, L.L.; et al. Diverse expression regulation of Hsp70 genes in scallops after exposure to toxic Alexandrium dinoflagellates. Chemosphere 2019, 234, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.; Yuan, X.; Zhao, Y.X.; Hu, G.R.; Tu, X.L. Heavy metal pollution in intertidal sediments from Quanzhou Bay, China. J. Environ. Sci. 2008, 20, 664–669. [Google Scholar] [CrossRef]
- Pulgar, J.; Waldisperg, M.; Galbán-Malagón, C.; Maturana, D.; Pulgar, V.M.; Aldana, M. UV radiation impacts body weight, oxygen consumption, and shelter selection in the intertidal vertebrate Girella laevifrons. Sci. Total Environ. 2017, 578, 317–322. [Google Scholar] [CrossRef] [PubMed]
- Bellas, J.; Fernández, N.; Lorenzo, I.; Beiras, R. Integrative assessment of coastal pollution in a Ría coastal system (Galicia, NW Spain): Correspondence between sediment chemistry and toxicity. Chemosphere 2008, 72, 826–835. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, B.B. Mineralization of organic matter in the sea bed—The role of sulphate reduction. Nature 1982, 296, 643–645. [Google Scholar] [CrossRef]
- Xu, X.H.; Zhu, X.Y.; Xu, G.C.; Xu, J.T.; Tang, Y.; Huo, W. Molecular cloning and expression of the stress gene HSP70 in the marine crab Charybdis Japonica (Decapoda: Brachyura: Portunidae) in response to ammonia-N, Nitrite-N, and sulfide exposure. J. Crustacean Biol. 2016, 36, 675–683. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.N.; Li, S.G.; Gao, Y.C.; Zhan, A.B. Genome-wide identification, characterization and expression analyses of heat shock protein-related genes in a highly invasive ascidian Ciona savignyi. Front. Physiol. 2018, 9, 1043. [Google Scholar] [CrossRef] [Green Version]
- Duan, Y.; Wang, Y.; Liu, Q.; Xiong, D.; Zhang, J. Transcriptomic and microbiota response on Litopenaeus vannamei intestine subjected to acute sulfide exposure. Fish Shellfish Immunol. 2019, 88, 335–343. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, S.; Liu, T.; Chen, M.; Li, W.; Zhang, X. The transcriptomic responses of the ark shell, Anadara broughtonii, to sulfide and hypoxia exposure. Mol. Biol. Rep. 2019, 46, 4245–4257. [Google Scholar] [CrossRef]
- Park, J.C.; Kim, D.-H.; Lee, Y.; Lee, M.-C.; Kim, T.K.; Yim, J.H.; Lee, J.-S. Genome-wide identification and structural analysis of heat shock protein gene families in the marine rotifer Brachionus spp.: Potential application in molecular ecotoxicology. Comp. Biochem. Physiol. Part D 2020, 36, 100749. [Google Scholar] [CrossRef]
- Park, J.C.; Lee, J.S. Genome-wide identification of heat shock proteins in harpacticoid, cyclopoid, and calanoid copepods: Potential application in marine ecotoxicology. Mar. Pollut. Bull. 2021, 169, 112545. [Google Scholar] [CrossRef]
- Knighton, L.E.; Waller, S.J.; Strom, O.; Wolfgeher, D.; Reitzel, A.M.; Truman, A.W. Dynamic remodeling of the interactomes of Nematostella vectensis Hsp70 isoforms under heat shock. J. Proteom. 2019, 206, 103416. [Google Scholar] [CrossRef] [PubMed]
- Arp, A.J.; Hansen, B.M.; Julian, D. The burrow environment and coelomic fluid characteristics of the echiuran worm Urechis caupo from three northern California population sites. Mar. Biol. 1992, 13, 613–623. [Google Scholar] [CrossRef]
- Cooper, C.E.; Brown, G.C. The inhibition of mitochondrial cytochrome oxidase by the gases carbon monoxide, nitric oxide, hydrogen cyanide and hydrogen sulfide: Chemical mechanism and physiological significance. J. Bioen. Biomembr. 2008, 40, 533. [Google Scholar] [CrossRef] [PubMed]
- Pietri, R.; Roman-Morales, E.; Lopez-Garriga, J. Hydrogen sulfide and heme proteins: Knowledge andmysteries. Antioxid. Redox Signal 2011, 15, 393–404. [Google Scholar] [CrossRef] [Green Version]
- Abe, H.; Sato-Okoshi, W.; Tanaka, M.; Okoshi, K.; Teramoto, W.; Kondoh, T.; Nishitani, G.; Endo, Y. Swimming behavior of the spoon worm Urechis unicinctus (Annelida, Echiura). Zoology 2014, 117, 216–223. [Google Scholar] [CrossRef]
- Huang, J.; Zhang, L.; Li, J.; Shi, X.; Zhang, Z. Proposed function of alternative oxidase in mitochondrial sulphide oxidation detoxification in the Echiuran worm, Urechis unicinctus. J. Mar. Biol. Assoc. UK 2013, 93, 2145–2154. [Google Scholar] [CrossRef]
- Li, X.; Liu, X.; Qin, Z.; Wei, M.; Hou, X.; Zhang, T.; Zhang, Z. A novel transcription factor Rwdd1 and its SUMOylation inhibit the expression of sqr, a key gene of mitochondrial sulfide metabolism in Urechis unicinctus. Aquat. Toxicol. 2018, 204, 180–189. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, L.; Zhang, Z.; Ma, X.; Liu, J. Transcriptional response to sulfide in the Echiuran Worm Urechis unicinctus by digital gene expression analysis. BMC Genom. 2015, 16, 829. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Qin, Z.; Li, X.; Ma, X.; Gao, B.; Zhang, Z. NF1, Sp1 and HSF1 are synergistically involved in sulfide-induced sqr activation in echiuran worm Urechis unicinctus. Aquat. Toxicol. 2016, 175, 232–240. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Z.; Ma, X.; Li, X.; Zhou, D.; Gao, B.; Bai, Y. Sulfide exposure results in enhanced sqr transcription through upregulating the expression and activation of HSF1 in echiuran worm Urechis unicinctus. Aquat. Toxicol. 2016, 170, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.B.; Zhang, Z.F.; Shao, M.Y.; Kang, K.H.; Shi, X.L.; Dong, Y.P.; Li, J.L. Response of sulfide: Quinone oxidoreductase to sulfide exposure in the echiuran worm Urechis unicinctus. Mar. Biotechnol. 2012, 14, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, X.; Liu, J.; Zhang, Z. Characteristics and function of sulfur dioxygenase in echiuran worm Urechis unicinctus. PLoS ONE 2013, 8, e81885. [Google Scholar] [CrossRef] [Green Version]
- Shafqat, W.; Jaskani, M.J.; Maqbool, R.; Khan, A.S.; Naqvi, S.A.; Ali, Z.; Khan, I.A. Genome Wide Analysis of Citrus sinensis Heat Shock Proteins. Iran. J. Biotechnol. 2020, 18, e2529. [Google Scholar]
- Evgen’Ev, M.B.; Garbuz, D.G.; Zatsepina, O.G. Heat Shock Proteins and Whole Body Adaptation to Extreme Environments; Springer: Berlin/Heidelberg, Germany, 2014. [Google Scholar]
- Guo, M.; Liu, J.H.; Lu, J.P.; Zhai, Y.F.; Wang, H.; Gong, Z.H.; Wang, S.B.; Lu, M.H. Genome-wide analysis of the CaHsp20 gene family in pepper: Comprehensive sequence and expression profile analysis under heat stress. Front. Plant Sci. 2015, 6, 806. [Google Scholar] [CrossRef] [Green Version]
- Ouyang, S.; Zhu, W.; Hamilton, J.; Lin, H.; Campbell, M.; Childs, K.; Nissen, F.T.; Malek, R.L.; Lee, Y.; Zheng, L.; et al. The TIGR rice genome annotation resource: Improvements and new features. Nucleic Acids Res. 2007, 35 (Suppl. 1), D883–D887. [Google Scholar] [CrossRef] [Green Version]
- Chang, D.; Duda, T.F. Extensive and continuous duplication facilitates rapid evolution and diversification of gene families. Mol. Biol. Evol. 2012, 29, 2019–2029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, X.; He, Y.; Zhang, L.; Lelong, C.; Jouaux, A. Immune and stress responses in oysters with insights on adaptation. Fish Shellfish Immun. 2015, 46, 107–119. [Google Scholar] [CrossRef]
- Dunn, M.J.; Kinney, G.M.; Washington, P.M.; Berman, J.; Anderson, M.Z. Functional diversification accompanies gene family expansion of MED2 homologs in Candida albicans. PLoS Genet. 2018, 14, e1007326. [Google Scholar] [CrossRef] [Green Version]
- Xiao, J.; Liu, Q.Y.; Du, J.H.; Zhu, W.L.; Li, Q.Y.; Chen, X.L.; Chen, X.H.; Liu, H.; Zhou, X.Y.; Zhao, Y.Z.; et al. Integrated analysis of physiological, transcriptomic and metabolomic responses and tolerance mechanism of nitrite exposure in Litopenaeus vannamei. Sci. Total Environ. 2020, 711, 134416. [Google Scholar] [CrossRef]
- Zeng, D.; Yang, C.; Li, Q.; Zhu, W.; Chen, X.; Peng, M.; Chen, X.H.; Lin, Y.; Wang, H.L.; Liu, H.; et al. Identification of a quantitative trait loci (QTL) associated with ammonia tolerance in the Pacific white shrimp (Litopenaeus vannamei). BMC Genom. 2020, 21, 857. [Google Scholar] [CrossRef] [PubMed]
- Blake, J.A.; Grassle, J.P.; Eckelbarger, K.J. Capitella teleta, a new species designation for the opportunistic and experimental Capitella sp. I, with a review of the literature for confirmed records. Zoosymposia 2009, 2, 25–53. [Google Scholar] [CrossRef] [Green Version]
- Hochstein, R.; Zhang, Q.; Sadowsky, M.J.; Forbes, V.E. The deposit feeder Capitella teleta has a unique and relatively complex microbiome likely supporting its ability to degrade pollutants. Sci. Total Environ. 2019, 670, 547–554. [Google Scholar] [CrossRef]
- Zhang, G.; Fang, X.; Guo, X.; Li, L.; Luo, R.; Xu, F.; Yang, P.; Zhang, L.; Wang, X.; Qi, H.; et al. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 2012, 490, 49–54. [Google Scholar] [CrossRef] [Green Version]
- Takeuchi, T.; Koyanagi, R.; Gyoja, F.; Kanda, M.; Hisata, K.; Fujie, M.; Goto, H.; Yamasaki, S.; Nagai, K.; Morino, Y.; et al. Bivalve-specific gene expansion in the pearl oyster genome: Implications of adaptation to a sessile lifestyle. Zool. Lett. 2016, 2, 3. [Google Scholar] [CrossRef] [Green Version]
- Powell, D.; Subramanian, S.; Suwansa-Ard, S.; Zhao, M.; O’Connor, W.; Raftos, D.; Elizur, A. The genome of the oyster Saccostrea offers insight into the environmental resilience of bivalves. DNA Res. 2018, 25, 655–665. [Google Scholar] [CrossRef] [Green Version]
- Luo, Y.J.; Takeuchi, T.; Koyanagi, R.; Yamada, L.; Kanda, M.; Khalturina, M.; Fujie, M.; Yamasaki, S.I.; Endo, K.; Satoh, N. The Lingula genome provides insights into brachiopod evolution and the origin of phosphate biomineralization. Nat. Commun. 2015, 61, 8301. [Google Scholar] [CrossRef]
- Simakov, O.; Marletaz, F.; Cho, S.J.; Edsinger-Gonzales, E.; Havlak, P.; Hellsten, U.; Kuo, G.H.; Larsson, T.; Lv, J.; Arendt, D.; et al. Insights into bilaterian evolution from three spiralian genomes. Nature 2013, 493, 526–531. [Google Scholar] [CrossRef] [Green Version]
- Albertin, C.B.; Simakov, O.; Mitros, T.; Wang, Z.Y.; Pungor, J.R.; Edsinger-Gonzales, E.; Brenner, S.; Ragsdale, C.W.; Rokhsar, D.S. The octopus genome and the evolution of cephalopod neural and morphological novelties. Nature 2015, 524, 220–224. [Google Scholar] [CrossRef] [Green Version]
- Rumpho, M.E.; Worful, J.M.; Lee, J.; Kannan, K.; Tyler, M.S.; Bhattacharya, D.; Moustafa, A.; Manhart, J.R. Horizontal gene transfer of the algal nuclear gene psbO to the photosynthetic sea slug Elysia chlorotica. Proc. Natl. Acad. Sci. USA 2008, 105, 17867–17871. [Google Scholar] [CrossRef] [Green Version]
- Moroz, L.L.; Kohn, A.B. Do different neurons age differently? Direct genome-wide analysis of aging in single identified cholinergic neurons. Front. Aging Neurosci. 2010, 2, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, Z.; Liu, X.; Wang, L.; Wang, X.; Li, Y.; Xiang, J.; Wang, P. Gene expression profiles of four heat shock proteins in response to different acute stresses in shrimp, Litopenaeus vannamei. Comp. Biochem. Physiol. C 2012, 156, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Lotz, S.K.; Knighton, L.E.; Jones, G.W.; Truman, A.W. Not quite the SSAme: Unique roles for the yeast cytosolic Hsp70s. Curr. Genet. 2019, 65, 1127–1134. [Google Scholar] [CrossRef] [PubMed]
- Joyner-Matos, J.; Downs, C.A.; Julian, D. Increased expression of stress proteins in the surf clam Donax variabilis following hydrogen sulfide exposure. Comp. Biochem. Phys. A 2006, 145, 245–257. [Google Scholar] [CrossRef]
- Li, H.; Wang, Y.; Wei, C.; Bai, S.; Zhao, Y.; Li, H.; Wu, B.; Wang, R.; Wu, L.; Xu, C. Mediation of exogenous hydrogen sulfide in recovery of ischemic post-conditioning-induced cardioprotection via down-regulating oxidative stress and up-regulating PI3K/Akt/GSK-3β pathway in isolated aging rat hearts. Cell Biosci. 2015, 5, 11. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.S.; Chen, Y.H.; Chen, N.; Wang, L.J.; Chen, D.X.; Weng, H.L.; Dooley, S.; Ding, H.G. Hydrogen sulfide promotes autophagy of hepatocellular carcinoma cells through the PI3K/Akt/mTOR signaling pathway. Cell Death Dis. 2017, 8, e2688. [Google Scholar] [CrossRef] [Green Version]
- Chi, Q.; Wang, D.; Hu, X.; Li, S.; Li, S. Hydrogen sulfide gas exposure induces necroptosis and promotes inflammation through the MAPK/NF-κB pathway in broiler spleen. Oxid. Med. Cell. Longev. 2019, 2019, 8061823. [Google Scholar] [CrossRef] [Green Version]
- Du, X.; Jin, Z.; Liu, D.; Yang, G.; Pei, Y. Hydrogen sulfide alleviates the cold stress through MPK4 in Arabidopsis thaliana. Plant Physiol. Biochem. 2017, 120, 112–119. [Google Scholar] [CrossRef]
- Chen, X.; Su, W.; Zhang, H.; Zhan, Y.; Zeng, F. Fraxinus mandshurica 4-coumarate-CoA ligase 2 enhances drought and osmotic stress tolerance of tobacco by increasing coniferyl alcohol content. Plant Physiol. Bioch. 2020, 155, 697–708. [Google Scholar] [CrossRef]
- Gu, H.; Hughes, B.T.; Espenshade, P.J. Identification of substrates for the conserved prolyl hydroxylase Ofd1 using quantitative proteomics in fission yeast. bioRxiv 2018, 363747. [Google Scholar] [CrossRef]
- Jin, E.J.; Park, S.; Lyu, X.; Jin, Y. Gap junctions: Historical discoveries and new findings in the Caenorhabditis elegans nervous system. Biol. Open 2020, 9, bio053983. [Google Scholar] [CrossRef]
- Mentlein, L.; Thorlacius, G.E.; Meneghel, L.; Sepulveda, J.R.; Brauner, S.; Espinosa, A.; Wahren-Herlenius, M. FAM167A/BLK is a susceptibility locus in autoimmune diseases: Characterisation of the FAM167 gene family. Ann. Rheum. Dis. 2018, 77, A19. [Google Scholar]
- Ma, X.; Liu, X.; Zhou, D.; Bai, Y.; Gao, B.; Zhang, Z.; Qin, Z. The NF-κB pathway participates in the response to sulfide stress in Urechis unicinctus. Fish Shellfish Immunol. 2016, 58, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Xuan, F.; Fu, H.; Zhu, J.; Ge, X.; Gu, Z. Transciptomic and histological analysis of hepatopancreas, muscle and gill tissues of oriental river prawn (Macrobrachium nipponense) in response to chronic hypoxia. BMC Genom. 2015, 16, 491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Liu, X.; Qin, Z.; Liu, J.; Zhang, Z. Expression characteristics of sulfur dioxygenase and its function adaption to sulfide in echiuran worm Urechis unicinctus. Gene 2016, 593, 334–341. [Google Scholar] [CrossRef] [PubMed]
- Yu, E.M.; Yoshinaga, T.; Jalufka, F.L.; Ehsan, H.; Mark Welch, D.B.; Kaneko, G. The complex evolution of the metazoan HSP70 gene family. Sci. Rep. 2021, 11, 17794. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [Green Version]
- Langfelder, P.; Horvath, S. Fast R functions for robust correlations and hierarchical clustering. J. Stat. Softw. 2012, 46, i11. [Google Scholar] [CrossRef] [Green Version]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, D.; Qin, Z.; Wei, M.; Kong, D.; Zheng, Q.; Bai, S.; Lin, S.; Zhang, Z.; Ma, Y. Genome-Wide Analyses of Heat Shock Protein Superfamily Provide New Insights on Adaptation to Sulfide-Rich Environments in Urechis unicinctus (Annelida, Echiura). Int. J. Mol. Sci. 2022, 23, 2715. https://doi.org/10.3390/ijms23052715
Liu D, Qin Z, Wei M, Kong D, Zheng Q, Bai S, Lin S, Zhang Z, Ma Y. Genome-Wide Analyses of Heat Shock Protein Superfamily Provide New Insights on Adaptation to Sulfide-Rich Environments in Urechis unicinctus (Annelida, Echiura). International Journal of Molecular Sciences. 2022; 23(5):2715. https://doi.org/10.3390/ijms23052715
Chicago/Turabian StyleLiu, Danwen, Zhenkui Qin, Maokai Wei, Dexu Kong, Qiaojun Zheng, Shumiao Bai, Siyu Lin, Zhifeng Zhang, and Yubin Ma. 2022. "Genome-Wide Analyses of Heat Shock Protein Superfamily Provide New Insights on Adaptation to Sulfide-Rich Environments in Urechis unicinctus (Annelida, Echiura)" International Journal of Molecular Sciences 23, no. 5: 2715. https://doi.org/10.3390/ijms23052715





