Exclusively Breastfed Infant Microbiota Develops over Time and Is Associated with Human Milk Oligosaccharide Intakes
Abstract
:1. Introduction
2. Results
2.1. Participant Characteristics
2.2. PacBio HiFi Sequencing Metrics
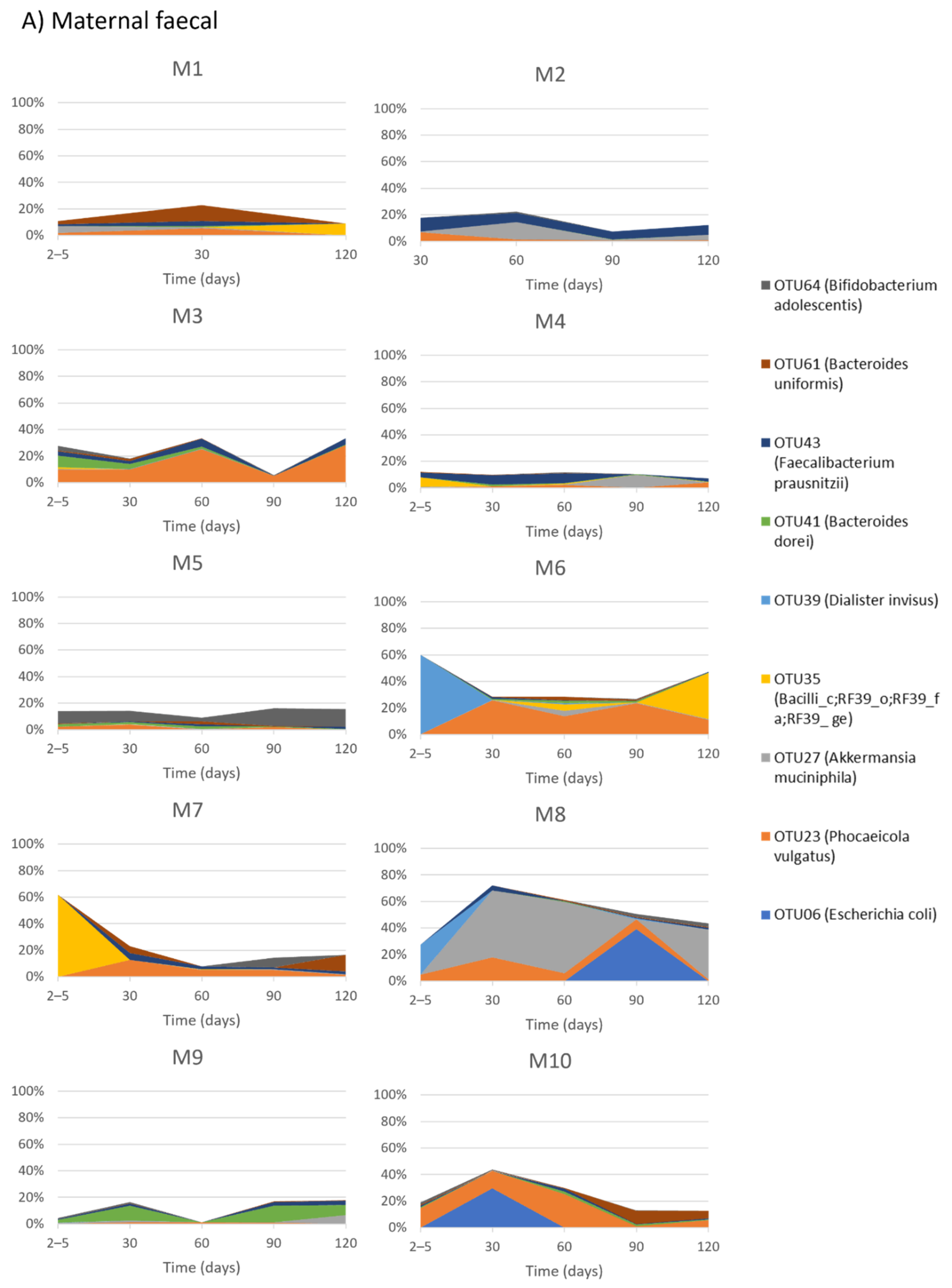
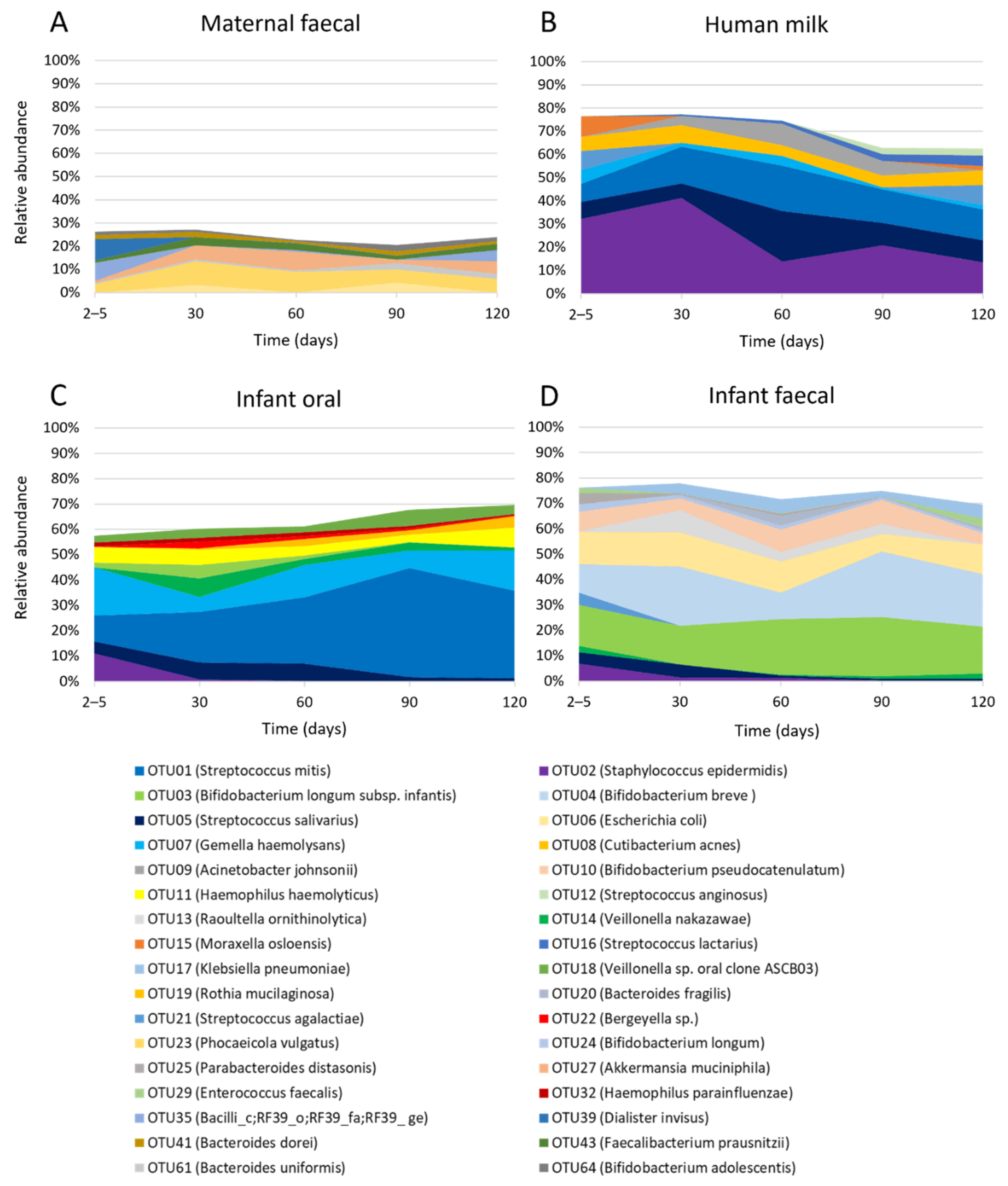
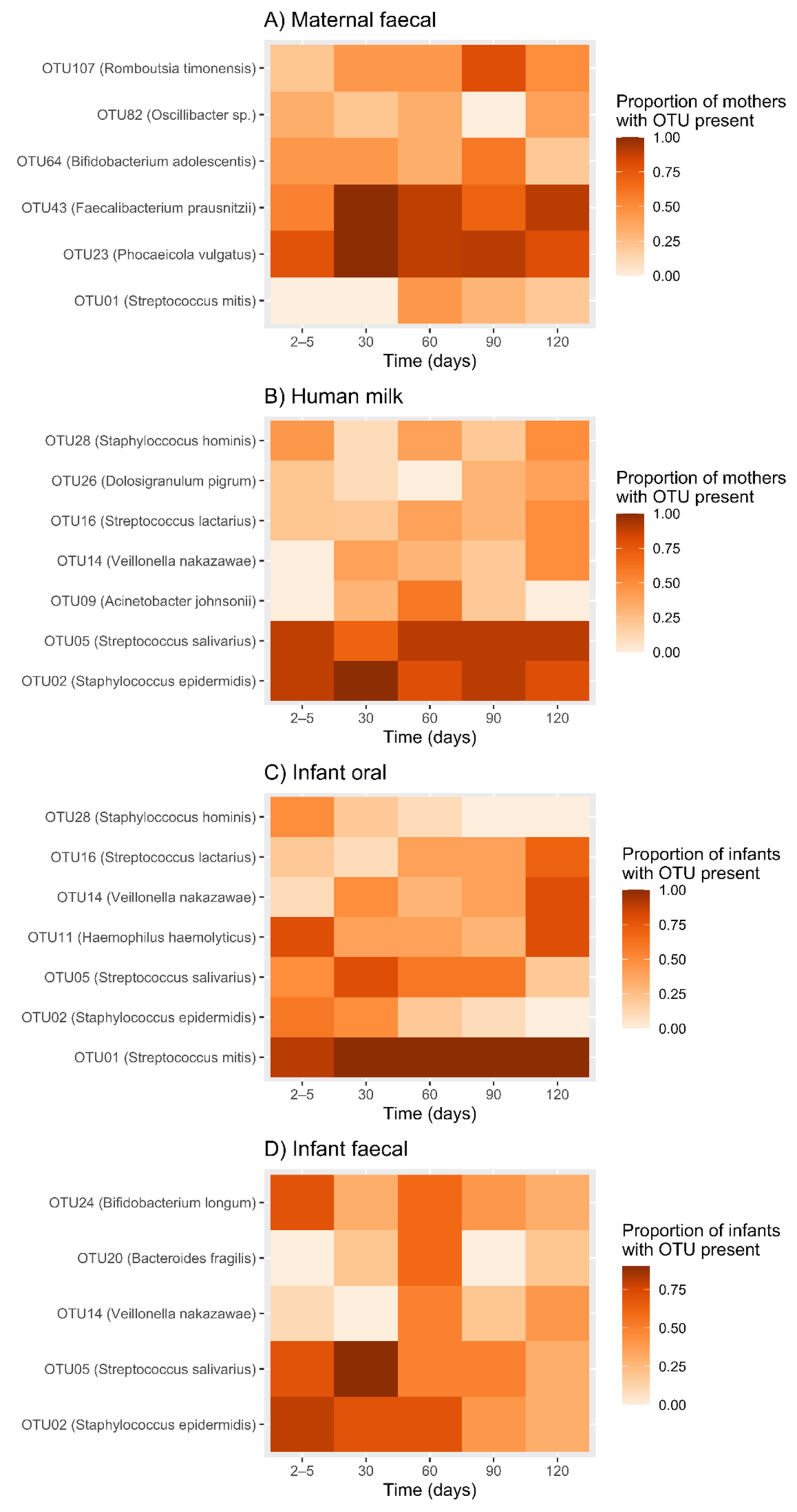
2.3. Temporal Development of Maternal Faecal Bacterial Profiles
2.4. Temporal Development of Human Milk Bacterial Profiles
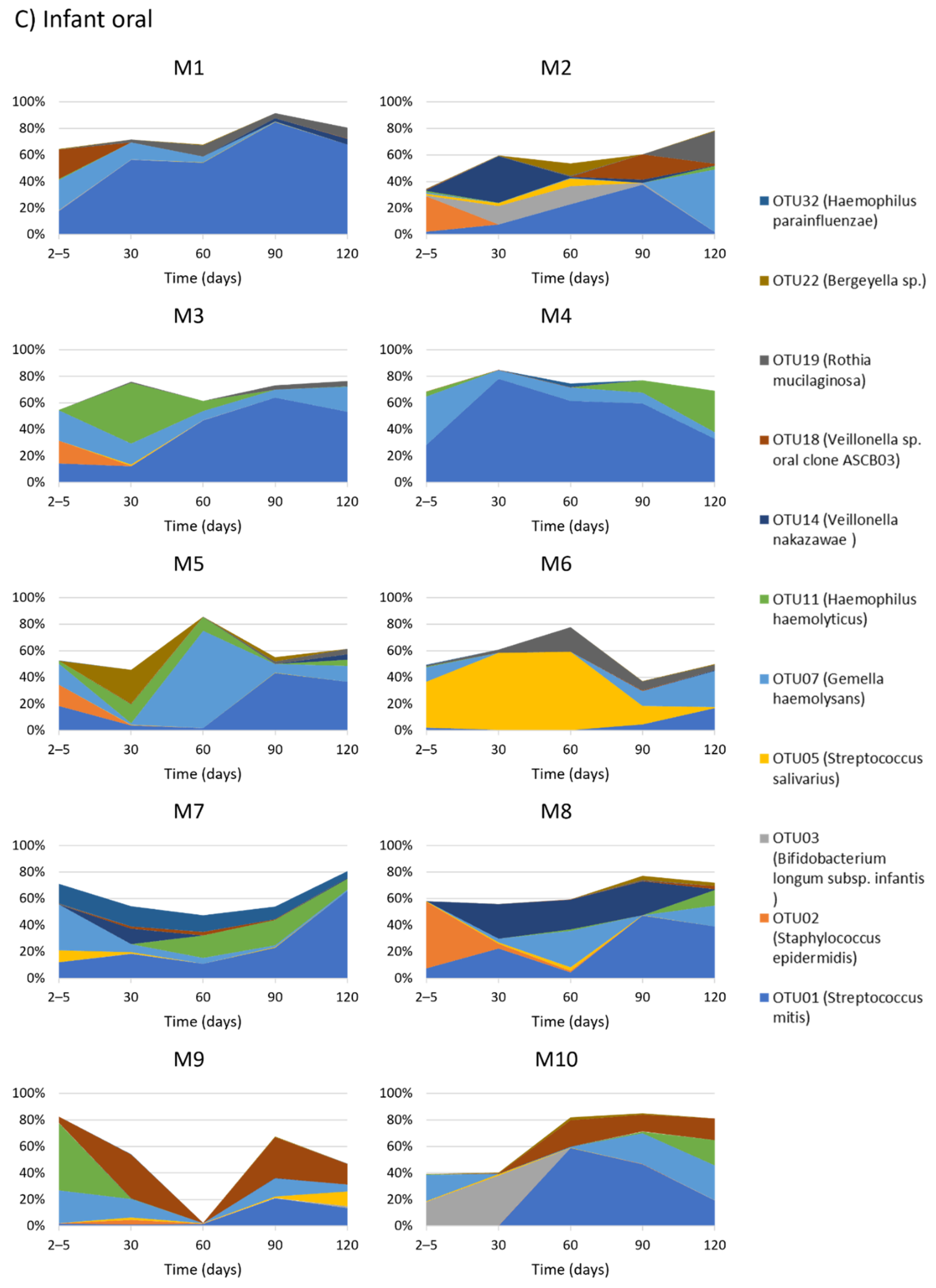
2.5. Temporal Development of Infant Oral Bacterial Profiles
2.6. Temporal Development of Infant Faecal Bacterial Profiles
2.7. Alpha and Beta Diversity between Sample Types
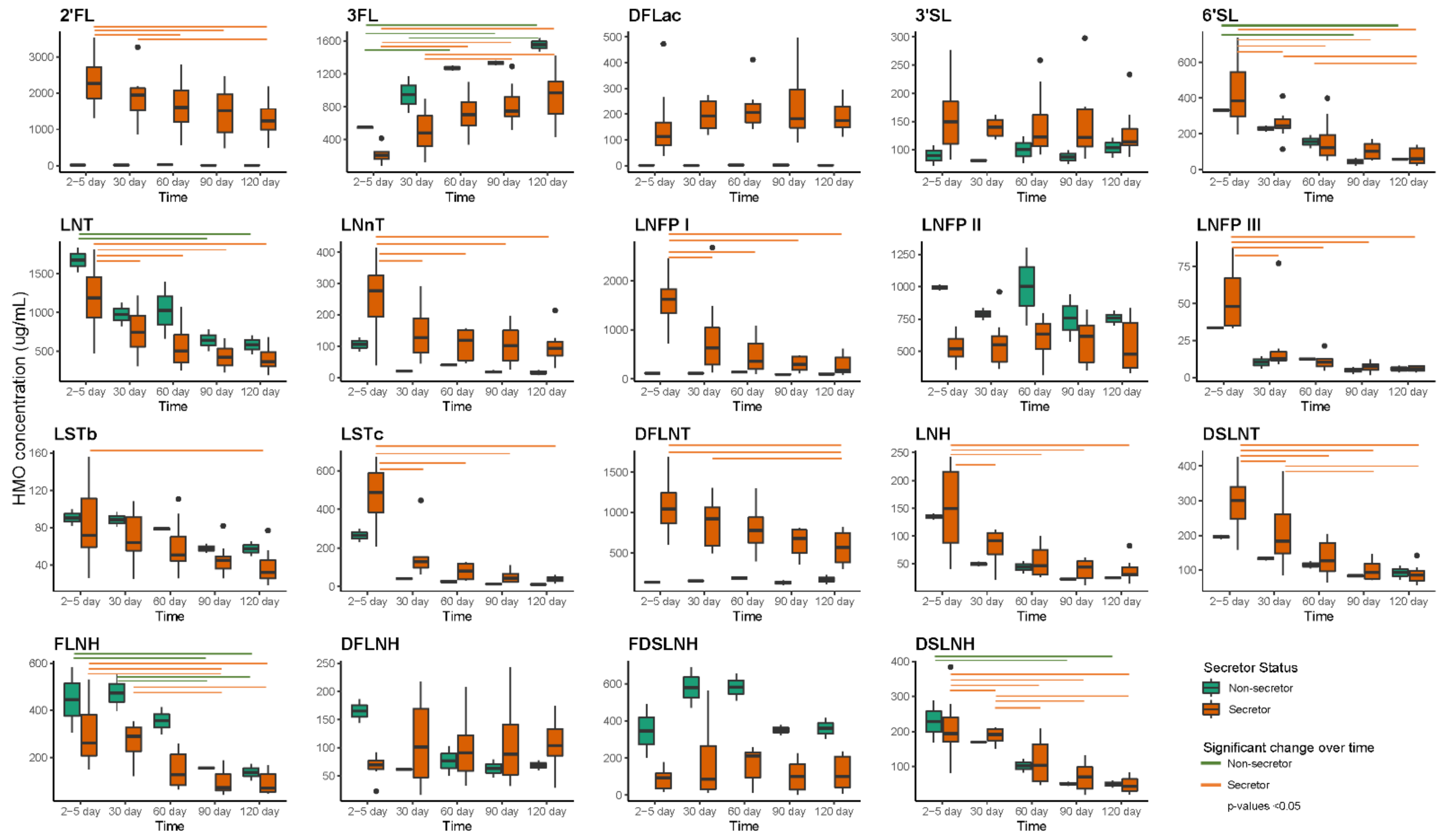
2.8. HMO Concentrations and Intakes over the First Four Months of Lactation
2.9. Associations between HMO Intake and Infant Oral Microbiota
2.10. Associations between HMO Intake and Infant Faecal Microbiota
3. Discussion
4. Materials and Methods
4.1. Study Design
4.2. Sample and Data Collection
4.3. Human Milk Oligosaccharides Analysis
4.4. 24-h Milk Intake
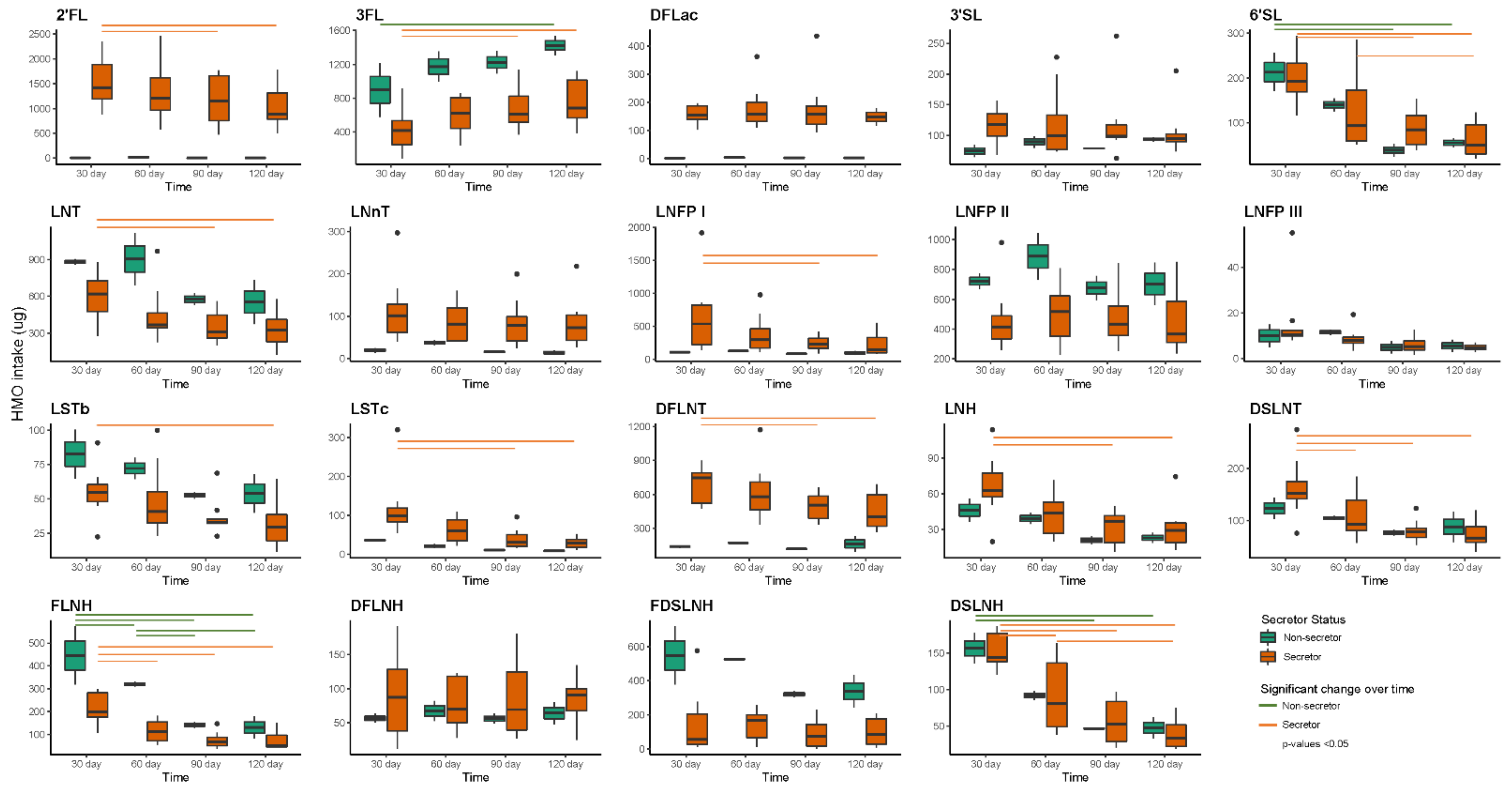
4.5. Daily Intakes of HMOs
4.6. DNA Extraction and Quantification
4.7. 16S rRNA Gene Amplification and Barcoding
4.8. PacBio Sequencing
4.9. Sequencing Data Processing
4.10. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Sample Types | Good’s Coverage (Raw Sequencing Data) | Good’s Coverage (Sequencing Depth 427) |
|---|---|---|
| Maternal faecal | 43.0% (0.0–91.6%) | 38.7% (17.7–89.8%) |
| Human milk | 88.6% (71.3–98.1%) | 82.0% (58.2–97.2%) |
| Infant oral | 86.0% (62.6–93.9%) | 78.3% (53.5–90.3%) |
| Infant faecal | 91.4% (79.1–96.5%) | 85.1% (68.1–93.4%) |
| OTUs | Genus | EC 1 | EC 2 | EC 3 | EC 4 | EC 5 | EC 6 | PCRC 1 | PCRC 2 | PCRC 3 | PCRC 4 | PCRC 5 | PCRC 6 | PCRC 7 | PCRC 8 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OTU01 | Streptococcus | 1815 | 4536 | 1 | 1 | 1 | 1 | 3 | 2 | 2 | 2 | 3 | 0 | 1 | 0 |
| OTU02 | Staphylococcus | 3 | 0 | 24 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 2 | 3 |
| OTU03 | Bifidobacterium | 4 | 3 | 4677 | 2032 | 22 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| OTU04 | Bifidobacterium | 3 | 0 | 1047 | 208 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 2 |
| OTU05 | Streptococcus | 2140 | 470 | 64 | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 1 |
| OTU06 | Escherichia-Shigella | 2 | 0 | 623 | 44 | 217 | 87 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 |
| OTU07 | Gemella | 1925 | 508 | 2 | 1 | 0 | 0 | 1 | 0 | 2 | 1 | 0 | 1 | 0 | 0 |
| OTU09 | Acinetobacter | 165 | 716 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| OTU10 | Bifidobacterium | 0 | 0 | 22 | 1 | 6 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| OTU11 | Haemophilus | 232 | 333 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU13 | Raoultella | 0 | 0 | 947 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU16 | Streptococcus | 7 | 87 | 4 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU18 | Veillonella | 22 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU19 | Rothia | 1347 | 147 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU20 | Bacteroides | 0 | 1 | 53 | 9 | 8 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 4 | 0 |
| OTU23 | Bacteroides | 1 | 14 | 4 | 30 | 836 | 210 | 0 | 3 | 1 | 0 | 1 | 0 | 102 | 0 |
| OTU24 | Bifidobacterium | 0 | 0 | 104 | 5 | 5 | 0 | 0 | 1 | 0 | 0 | 3 | 0 | 1 | 0 |
| OTU25 | Parabacteroides | 1 | 1 | 1087 | 135 | 8 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 21 | 0 |
| OTU26 | Dolosigranulum | 2 | 2 | 0 | 0 | 0 | 1 | 0 | 5704 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU27 | Akkermansia | 2 | 0 | 0 | 5 | 58 | 13 | 0 | 0 | 0 | 0 | 2 | 0 | 417 | 0 |
| OTU29 | Enterococcus | 0 | 0 | 37 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU30 | Bacilli_c;RF39_o;RF39_fa;RF39_ge | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 8991 | 4 |
| OTU31 | Bifidobacterium | 1 | 0 | 343 | 29 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU33 | Phascolarctobacterium | 1 | 1 | 53 | 27 | 27 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 536 | 0 |
| OTU34 | Corynebacterium | 15 | 0 | 4 | 0 | 0 | 0 | 7289 | 856 | 2 | 0 | 0 | 0 | 0 | 0 |
| OTU35 | Bacilli_c;RF39_o;RF39_fa;RF39_ge | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 2408 | 0 |
| OTU41 | Bacteroides | 1 | 3 | 1 | 4 | 92 | 30 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 |
| OTU43 | Faecalibacterium | 1 | 5 | 0 | 36 | 97 | 33 | 0 | 0 | 1 | 0 | 0 | 0 | 23 | 0 |
| OTU45 | Lactobacillus | 124 | 123 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU48 | Streptococcus | 128 | 53 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU49 | Streptococcus | 108 | 52 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU57 | Lachnospiraceae_ge | 0 | 0 | 5 | 0 | 4 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 51 | 0 |
| OTU61 | Bacteroides | 5 | 1 | 1 | 15 | 181 | 92 | 1 | 2 | 0 | 0 | 0 | 0 | 92 | 0 |
| OTU64 | Bifidobacterium | 2 | 0 | 0 | 0 | 26 | 7 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| OTU68 | Streptococcus | 48 | 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU81 | Streptococcus | 33 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU85 | Streptococcus | 58 | 20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU90 | Gemella | 20 | 15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU97 | Streptococcus | 67 | 44 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU99 | Bifidobacterium | 0 | 0 | 24 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU109 | Blautia | 0 | 0 | 0 | 0 | 40 | 9 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| OTU112 | Anaerostipes | 0 | 0 | 0 | 5 | 29 | 8 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| OTU114 | Fusicatenibacter | 11 | 1 | 0 | 5 | 41 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 |
| OTU118 | Erysipelotrichaceae_UCG-003 | 0 | 0 | 0 | 1 | 29 | 16 | 0 | 0 | 0 | 0 | 0 | 0 | 7 | 0 |
| OTU124 | Bifidobacterium | 0 | 0 | 32 | 4 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU125 | Streptococcus | 37 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU139 | Prevotella | 0 | 43 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU155 | Streptococcus | 18 | 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU156 | Streptococcus | 27 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU167 | Agathobacter | 0 | 0 | 0 | 2 | 23 | 21 | 0 | 0 | 0 | 0 | 1 | 0 | 296 | 0 |
| OTU168 | Enterobacteriaceae_unclassified | 0 | 0 | 52 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU170 | Bacteroides | 0 | 0 | 0 | 2 | 62 | 20 | 0 | 0 | 0 | 0 | 0 | 0 | 24 | 0 |
| OTU192 | Ruminococcus | 0 | 0 | 1 | 0 | 86 | 34 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU195 | Ruminococcus | 0 | 0 | 0 | 1 | 2 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 27 | 0 |
| OTU203 | Bacteroides | 0 | 1 | 0 | 0 | 38 | 31 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU205 | Bifidobacterium | 1 | 0 | 165 | 15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU206 | Bacteroides | 1 | 3 | 0 | 0 | 26 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| OTU207 | Subdoligranulum | 0 | 1 | 0 | 2 | 21 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| OTU213 | Parasutterella | 0 | 3 | 0 | 0 | 112 | 29 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU217 | Gemella | 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU226 | Prevotella | 0 | 112 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU254 | Parabacteroides | 0 | 0 | 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU262 | Enterobacteriaceae_unclassified | 0 | 0 | 28 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU272 | Bifidobacterium | 0 | 1 | 70 | 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU276 | Lachnospiraceae_ge | 1 | 0 | 1 | 4 | 21 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| OTU285 | Enterobacteriaceae_unclassified | 0 | 0 | 28 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU292 | Bifidobacterium | 0 | 0 | 48 | 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU300 | Bifidobacterium | 0 | 0 | 66 | 17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU303 | Bifidobacterium | 0 | 0 | 33 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU306 | Bifidobacterium | 0 | 0 | 38 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU318 | Enterobacteriaceae_unclassified | 0 | 0 | 30 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU322 | Enterobacteriaceae_unclassified | 0 | 0 | 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU357 | Acinetobacter | 2 | 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU367 | Bifidobacterium | 0 | 0 | 41 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU380 | Bifidobacterium | 0 | 0 | 29 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU462 | Campylobacter | 0 | 162 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU558 | Bacilli_c;RF39_o;RF39_fa;RF39_ge | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 126 | 0 |
| OTU624 | Bacilli_c;RF39_o;RF39_fa;RF39_ge | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 111 | 0 |
| OTU694 | Faecalibacterium | 0 | 0 | 0 | 0 | 24 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU726 | Bacilli_c;RF39_o;RF39_fa;RF39_ge | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 84 | 0 |
| OTU795 | Bacteroides | 0 | 0 | 0 | 0 | 32 | 11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| OTU942 | Bacilli_c;RF39_o;RF39_fa;RF39_ge | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 64 | 0 |
| OTU1372 | Bacilli_c;RF39_o;RF39_fa;RF39_ge | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 37 | 0 |
| OTU1827 | Bacilli_c;RF39_o;RF39_fa;RF39_ge | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 28 | 0 |
| OTU2087 | Bacilli_c;RF39_o;RF39_fa;RF39_ge | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 26 | 0 |
| Others | Others | 3283 | 2255 | 3614 | 728 | 4727 | 1681 | 80 | 59 | 15 | 5 | 20 | 7 | 2763 | 5 |
| OTUs | Category | Day 2–5 | Day 30 | Day 60 | Day 90 | Day 120 |
|---|---|---|---|---|---|---|
| Maternal Faecal | ||||||
| OTU01 (Streptococcus mitis) | Absent | 9 (100%) | 9 (100%) | 5 (55.56%) | 7 (70%) | 8 (80%) |
| Low | 0 (0%) | 0 (0%) | 3 (33.33%) | 3 (30%) | 2 (20%) | |
| High | 0 (0%) | 0 (0%) | 1 (11.11%) | 0 (0%) | 0 (0%) | |
| OTU23 (Phocaeicola vulgatus) | Absent | 2 (22.22%) | 0 (0%) | 1 (11.11%) | 1 (10%) | 2 (20%) |
| Low | 5 (55.56%) | 3 (33.33%) | 3 (33.33%) | 5 (50%) | 5 (50%) | |
| Medium | 2 (22.22%) | 6 (66.67%) | 5 (55.56%) | 4 (40%) | 3 (30%) | |
| OTU43 (Faecalibacterium prausnitzii) | Absent | 4 (44.44%) | 0 (0%) | 1 (11.11%) | 3 (30%) | 1 (10%) |
| Low | 5 (55.56%) | 6 (66.67%) | 5 (55.56%) | 6 (60%) | 8 (80%) | |
| Medium | 0 (0%) | 3 (33.33%) | 3 (33.33%) | 1 (10%) | 1 (10%) | |
| OTU64 (Bifidobacterium adolescentis) | Absent | 5 (55.56%) | 5 (55.56%) | 6 (66.67%) | 4 (40%) | 8 (80%) |
| Low | 3 (33.33%) | 3 (33.33%) | 3 (33.33%) | 4 (40%) | 1 (10%) | |
| Medium | 1 (11.11%) | 1 (11.11%) | 0 (0%) | 2 (20%) | 1 (10%) | |
| OTU82 (Oscillibacter sp.) | Absent | 6 (66.67%) | 7 (77.78%) | 6 (66.67%) | 10 (100%) | 6 (60%) |
| Low | 2 (22.22%) | 2 (22.22%) | 3 (33.33%) | 0 (0%) | 3 (30%) | |
| Medium | 1 (11.11%) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (10%) | |
| OTU107 (Romboutsia timonensis) | Absent | 7 (77.78%) | 5 (55.56%) | 5 (55.56%) | 2 (20%) | 5 (50%) |
| Low | 1 (11.11%) | 4 (44.44%) | 4 (44.44%) | 7 (70%) | 5 (50%) | |
| Medium | 1 (11.11%) | 0 (0%) | 0 (0%) | 1 (10%) | 0 (0%) | |
| Human Milk | ||||||
| OTU02 (Staphylococcus epidermidis) | Absent | 1 (11.11%) | 0 (0%) | 2 (20%) | 1 (10%) | 2 (20%) |
| Low | 3 (33.33%) | 1 (10%) | 5 (50%) | 4 (40%) | 2 (20%) | |
| Medium | 2 (22.22%) | 3 (30%) | 0 (0%) | 3 (30%) | 5 (50%) | |
| High | 3 (33.33%) | 6 (60%) | 3 (30%) | 2 (20%) | 1 (10%) | |
| OTU05 (Streptococcus salivarius) | Absent | 1 (11.11%) | 3 (30%) | 1 (10%) | 1 (10%) | 1 (10%) |
| Low | 5 (55.56%) | 5 (50%) | 3 (30%) | 4 (40%) | 6 (60%) | |
| Medium | 2 (22.22%) | 2 (20%) | 3 (30%) | 3 (30%) | 2 (20%) | |
| High | 1 (11.11%) | 0 (0%) | 3 (30%) | 2 (20%) | 1 (10%) | |
| OTU09 (Acinetobacter johnsonii) | Absent | 9 (100%) | 0 (0%) | 4 (40%) | 0 (0%) | 10 (100%) |
| Low | 0 (0%) | 0 (0%) | 4 (40%) | 0 (0%) | 0 (0%) | |
| Medium | 0 (0%) | 0 (0%) | 1 (10%) | 0 (0%) | 0 (0%) | |
| High | 0 (0%) | 0 (0%) | 1 (10%) | 0 (0%) | 0 (0%) | |
| OTU14 (Veillonella nakazawae) | Absent | 9 (100%) | 6 (60%) | 7 (70%) | 8 (80%) | 5 (50%) |
| Low | 0 (0%) | 4 (40%) | 3 (30%) | 2 (20%) | 4 (40%) | |
| Medium | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (10%) | |
| OTU16 (Streptococcus lactarius) | Absent | 7 (77.78%) | 8 (80%) | 6 (60%) | 7 (70%) | 5 (50%) |
| Low | 2 (22.22%) | 1 (10%) | 3 (30%) | 1 (10%) | 2 (20%) | |
| Medium | 0 (0%) | 1 (10%) | 1 (10%) | 2 (20%) | 3 (30%) | |
| OTU26 (Dolosigranulum pigrum) | Absent | 7 (77.78%) | 9 (90%) | 10 (100%) | 7 (70%) | 6 (60%) |
| Low | 2 (22.22%) | 1 (10%) | 0 (0%) | 1 (10%) | 4 (40%) | |
| Medium | 0 (0%) | 0 (0%) | 0 (0%) | 2 (20%) | 0 (0%) | |
| OTU28 (Staphyloccocus hominis) | Absent | 5 (55.56%) | 9 (90%) | 6 (60%) | 8 (80%) | 5 (50%) |
| Low | 3 (33.33%) | 1 (10%) | 4 (40%) | 2 (20%) | 5 (50%) | |
| Medium | 1 (11.11%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | |
| Infant Oral | ||||||
| OTU01 (Streptococcus mitis) | Absent | 1 (10%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Low | 3 (30%) | 4 (40%) | 4 (40%) | 1 (10%) | 1 (10%) | |
| Medium | 6 (60%) | 4 (40%) | 2 (20%) | 2 (20%) | 3 (30%) | |
| High | 0 (0%) | 2 (20%) | 4 (40%) | 7 (70%) | 6 (60%) | |
| OTU02 (Staphylococcus epidermidis) | Absent | 4 (40%) | 5 (50%) | 8 (80%) | 9 (90%) | 10 (100%) |
| Low | 2 (20%) | 5 (50%) | 2 (20%) | 1 (10%) | 0 (0%) | |
| Medium | 3 (30%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | |
| High | 1 (10%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | |
| OTU05 (Streptococcus salivarius) | Absent | 5 (50%) | 2 (20%) | 4 (40%) | 4 (40%) | 8 (80%) |
| Low | 3 (30%) | 7 (70%) | 4 (40%) | 5 (50%) | 1 (10%) | |
| Medium | 1 (10%) | 0 (0%) | 1 (10%) | 1 (10%) | 1 (10%) | |
| High | 1 (10%) | 1 (10%) | 1 (10%) | 0 (0%) | 0 (0%) | |
| OTU11 (Haemophilus haemolyticus) | Absent | 2 (20%) | 6 (60%) | 6 (60%) | 7 (70%) | 2 (20%) |
| Low | 7 (70%) | 2 (20%) | 1 (10%) | 1 (10%) | 4 (40%) | |
| Medium | 0 (0%) | 1 (10%) | 3 (30%) | 2 (20%) | 3 (30%) | |
| High | 1 (10%) | 1 (10%) | 0 (0%) | 0 (0%) | 1 (10%) | |
| OTU14 (Veillonella nakazawae) | Absent | 9 (90%) | 5 (50%) | 7 (70%) | 6 (60%) | 2 (20%) |
| Low | 1 (10%) | 2 (20%) | 2 (20%) | 3 (30%) | 8 (80%) | |
| Medium | 0 (0%) | 2 (20%) | 1 (10%) | 1 (10%) | 0 (0%) | |
| High | 0 (0%) | 1 (10%) | 0 (0%) | 0 (0%) | 0 (0%) | |
| OTU16 (Streptococcus lactarius) | Absent | 8 (80%) | 9 (90%) | 6 (60%) | 6 (60%) | 3 (30%) |
| Low | 2 (20%) | 1 (10%) | 4 (40%) | 3 (30%) | 6 (60%) | |
| Medium | 0 (0%) | 0 (0%) | 0 (0%) | 1 (10%) | 1 (10%) | |
| OTU28 (Staphyloccocus hominis) | Absent | 5 (50%) | 8 (80%) | 9 (90%) | 10 (100%) | 10 (100%) |
| Low | 4 (40%) | 2 (20%) | 1 (10%) | 0 (0%) | 0 (0%) | |
| Medium | 1 (10%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | |
| Infant Faecal | ||||||
| OTU02 (Staphylococcus epidermidis) | Absent | 2 (20%) | 3 (30%) | 3 (30%) | 6 (60%) | 7 (70%) |
| Low | 5 (50%) | 6 (60%) | 6 (60%) | 4 (40%) | 3 (30%) | |
| Medium | 3 (30%) | 1 (10%) | 1 (10%) | 0 (0%) | 0 (0%) | |
| OTU05 (Streptococcus salivarius) | Absent | 3 (30%) | 1 (10%) | 5 (50%) | 5 (50%) | 7 (70%) |
| Low | 4 (40%) | 7 (70%) | 4 (40%) | 5 (50%) | 3 (30%) | |
| Medium | 3 (30%) | 1 (10%) | 1 (10%) | 0 (0%) | 0 (0%) | |
| High | 0 (0%) | 1 (10%) | 0 (0%) | 0 (0%) | 0 (0%) | |
| OTU14 (Veillonella nakazawae) | Absent | 9 (90%) | 10 (100%) | 5 (50%) | 8 (80%) | 6 (60%) |
| Low | 0 (0%) | 0 (0%) | 5 (50%) | 1 (10%) | 2 (20%) | |
| Medium | 1 (10%) | 0 (0%) | 0 (0%) | 1 (10%) | 2 (20%) | |
| OTU20 (Bacteroides fragilis) | Absent | 10 (100%) | 8 (80%) | 4 (40%) | 10 (100%) | 8 (80%) |
| Low | 0 (0%) | 2 (20%) | 5 (50%) | 0 (0%) | 1 (10%) | |
| Medium | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (10%) | |
| High | 0 (0%) | 0 (0%) | 1 (10%) | 0 (0%) | 0 (0%) | |
| OTU24 (Bifidobacterium longum) | Absent | 3 (30%) | 7 (70%) | 4 (40%) | 6 (60%) | 7 (70%) |
| Low | 3 (30%) | 1 (10%) | 4 (40%) | 4 (40%) | 2 (20%) | |
| Medium | 4 (40%) | 2 (20%) | 2 (20%) | 0 (0%) | 1 (10%) | |
| Bacterial OTUs | Comparison (Parameter—Intercept) | Estimated Difference | SE | p-Value |
|---|---|---|---|---|
| Day 30 | ||||
| LNnT | ||||
| OTU11 (Haemophilus haemolyticus) | Low—Absent | 83.375 | 30.368 | 0.034 |
| OTU11 (Haemophilus haemolyticus) | High—Absent | 240.073 | 40.174 | 0.001 |
| LNFP I | ||||
| OTU18 (Veillonella sp. oral clone ASCB03) | High—Absent | 1541.626 | 351.386 | 0.003 |
| OTU32 (Haemophilus parainfluenzae) | Low—Absent | 1502.522 | 332.410 | 0.003 |
| LNFP III | ||||
| OTU18 (Veillonella sp. oral clone ASCB03) | High—Absent | 44.042 | 3.996 | <0.001 |
| OTU32 (Haemophilus parainfluenzae) | Low—Absent | 44.201 | 3.754 | <0.001 |
| LSTc | ||||
| OTU18 (Veillonella sp. oral clone ASCB03) | High—Absent | 233.322 | 40.045 | <0.001 |
| OTU32 (Haemophilus parainfluenzae) | Low—Absent | 245.268 | 31.792 | <0.001 |
| LNH | ||||
| OTU11 (Haemophilus haemolyticus) | High—Absent | 64.315 | 17.900 | 0.012 |
| DSLNT | ||||
| OTU11 (Haemophilus haemolyticus) | Low—Absent | 113.917 | 28.901 | 0.008 |
| OTU18 (Veillonella sp. oral clone ASCB03) | Low—Absent | −51.026 | 20.311 | 0.040 |
| OTU18 (Veillonella sp. oral clone ASCB03) | High—Absent | 110.395 | 33.168 | 0.013 |
| OTU32 (Haemophilus parainfluenzae) | Low—Absent | 124.808 | 34.216 | 0.008 |
| FLNH | ||||
| OTU05 (Streptococcus salivarius) | High—Absent | 323.804 | 94.591 | 0.011 |
| Day 60 | ||||
| 3FL | ||||
| OTU02 (Staphylococcus epidermidis) | Low—Absent | −550.146 | 203.242 | 0.027 |
| DFLac | ||||
| OTU32 (Haemophilus parainfluenzae) | Medium—Absent | 224.329 | 70.758 | 0.016 |
| LNnT | ||||
| OTU03 (Bifidobacterium longum subsp. infantis) | Low—Absent | 100.502 | 31.407 | 0.015 |
| LNFP I | ||||
| OTU18 (Veillonella sp. oral clone ASCB03) | Medium—Absent | 788.647 | 204.195 | 0.006 |
| LNFP II | ||||
| OTU02 (Staphylococcus epidermidis) | Low—Absent | −381.626 | 155.937 | 0.040 |
| LNFP III | ||||
| OTU18 (Veillonella sp. oral clone ASCB03) | Medium—Absent | 11.980 | 3.171 | 0.007 |
| LSTb | ||||
| OTU14 (Veillonella nakazawae) | Low—Absent | 42.776 | 15.187 | 0.026 |
| LSTc | ||||
| OTU01 (Streptococcus mitis) | Medium—Low | 69.136 | 17.701 | 0.006 |
| DFLNT | ||||
| OTU14 (Veillonella nakazawae) | Low—Absent | 553.393 | 176.628 | 0.017 |
| DSLNT | ||||
| OTU14 (Veillonella nakazawae) | Low—Absent | 70.404 | 20.949 | 0.012 |
| Day 90 | ||||
| DFLac | ||||
| OTU01 (Streptococcus mitis) | Medium—Low | 326.879 | 98.311 | 0.013 |
| OTU32 (Haemophilus parainfluenzae) | Medium—Absent | 321.837 | 79.181 | 0.004 |
| 3′SL | ||||
| OTU01 (Streptococcus mitis) | Medium—Low | 115.877 | 48.265 | 0.047 |
| OTU32 (Haemophilus parainfluenzae) | Medium—Absent | 167.484 | 20.209 | <0.001 |
| 6′SL | ||||
| OTU19 (Rothia mucilaginosa) | Low—Absent | −60.645 | 18.879 | 0.015 |
| OTU19 (Rothia mucilaginosa) | Medium—Absent | −85.766 | 30.830 | 0.027 |
| LNT | ||||
| OTU05 (Streptococcus salivarius) | Low—Absent | −275.077 | 47.665 | <0.001 |
| LNFP I | ||||
| OTU18 (Veillonella sp. oral clone ASCB03) | Medium—Absent | 242.589 | 62.818 | 0.008 |
| LNFP II | ||||
| OTU22 (Bergeyella sp.) | Low—Absent | −251.985 | 90.486 | 0.024 |
| LSTb | ||||
| OTU07 (Gemella haemolysans) | Low—Absent | −39.158 | 10.458 | 0.007 |
| OTU07 (Gemella haemolysans) | Medium—Absent | −28.050 | 9.783 | 0.024 |
| LSTc | ||||
| OTU32 (Haemophilus parainfluenzae) | Medium—Absent | 68.280 | 18.015 | 0.005 |
| DSLNT | ||||
| OTU07 (Gemella haemolysans) | Low—Absent | −56.769 | 14.608 | 0.006 |
| OTU07 (Gemella haemolysans) | Medium—Absent | −44.494 | 13.664 | 0.014 |
| OTU18 (Veillonella sp. oral clone ASCB03) | Medium—Absent | 45.434 | 9.703 | 0.003 |
| DSLNH | ||||
| OTU19 (Rothia mucilaginosa) | Low—Absent | −46.494 | 11.472 | 0.005 |
| Day 120 | ||||
| 2′FL | ||||
| OTU03 (Bifidobacterium longum subsp. infantis) | Low—Absent | 1044.095 | 324.676 | 0.012 |
| OTU18 (Veillonella sp. oral clone ASCB03) | Medium—Absent | 1143.659 | 348.875 | 0.014 |
| 3FL | ||||
| OTU03 (Bifidobacterium longum subsp. infantis) | Low—Absent | −597.110 | 241.857 | 0.039 |
| OTU18 (Veillonella sp. oral clone ASCB03) | Medium—Absent | −736.135 | 221.592 | 0.013 |
| 3′SL | ||||
| OTU32 (Haemophilus parainfluenzae) | Low—Absent | 112.458 | 11.639 | <0.001 |
| 6′SL | ||||
| OTU01 (Streptococcus mitis) | High—Low | −58.021 | 20.220 | 0.024 |
| OTU03 (Bifidobacterium longum subsp. infantis) | Low—Absent | 63.180 | 18.410 | 0.009 |
| OTU18 (Veillonella sp. oral clone ASCB03) | Medium—Absent | 67.677 | 20.171 | 0.012 |
| LNFP I | ||||
| OTU03 (Bifidobacterium longum subsp. infantis) | Low—Absent | 291.768 | 88.867 | 0.011 |
| OTU18 (Veillonella sp. oral clone ASCB03) | Medium—Absent | 323.706 | 93.534 | 0.011 |
| LNH | ||||
| OTU03 (Bifidobacterium longum subsp. infantis) | Low—Absent | 31.952 | 9.363 | 0.009 |
| OTU18 (Veillonella sp. oral clone ASCB03) | Medium—Absent | 32.123 | 10.591 | 0.019 |
| DSLNT | ||||
| OTU01 (Streptococcus mitis) | High—Low | −58.388 | 21.383 | 0.029 |
| FDSLNH | ||||
| OTU05 (Streptococcus salivarius), | Low—Absent | 307.528 | 96.080 | 0.015 |
| DSLNH | ||||
| OTU01 (Streptococcus mitis) | High—Low | −22.305 | 8.279 | 0.031 |
| OTU03 (Bifidobacterium longum subsp. infantis) | Low—Absent | 32.307 | 11.696 | 0.025 |
| Bacterial OTUs | Comparison (Parameter—Intercept) | Estimated Difference | SE | p-Value |
|---|---|---|---|---|
| Day 30 | ||||
| 2′FL | ||||
| OTU04 (Bifidobacterium breve) | Low—Absent | −1666.940 | 425.648 | 0.008 |
| 3FL | ||||
| OTU13 (Raoultella ornithinolytica) | High—Absent | 735.702 | 258.385 | 0.025 |
| OTU24 (Bifidobacterium longum) | Low—Absent | 723.455 | 265.973 | 0.030 |
| DFLac | ||||
| OTU04 (Bifidobacterium breve) | Low—Absent | −138.030 | 28.935 | 0.003 |
| 3′SL | ||||
| OTU03 (Bifidobacterium longum subsp. infantis) | Medium—Absent | −41.128 | 14.211 | 0.028 |
| OTU20 (Bacteroides fragilis) | Low—Absent | 48.548 | 19.523 | 0.038 |
| LNnT | ||||
| OTU03 (Bifidobacterium longum subsp. infantis) | Low—Absent | 205.125 | 55.873 | 0.010 |
| OTU17 (Klebsiella pneumoniae) | Low—Absent | 232.141 | 40.266 | <0.001 |
| OTU20 (Bacteroides fragilis) | Low—Absent | 126.620 | 53.970 | 0.047 |
| LNFP I | ||||
| OTU05 (Streptococcus salivarius) | Medium—Absent | 1813.872 | 453.887 | 0.007 |
| OTU10 (Bifidobacterium pseudocatenulatum) | High—Absent | 1475.480 | 350.940 | 0.006 |
| OTU17 (Klebsiella pneumoniae) | High—Absent | 1492.947 | 323.246 | 0.002 |
| OTU25 (Parabacteroides distasonis) | Low—Absent | 1524.848 | 316.978 | 0.001 |
| LNFP II | ||||
| OTU17 (Klebsiella pneumoniae) | Low—Absent | 484.536 | 170.968 | 0.025 |
| LNFP III | ||||
| OTU05 (Streptococcus salivarius) | Low—Absent | 5.726 | 2.302 | 0.047 |
| OTU05 (Streptococcus salivarius) | Medium—Absent | 50.291 | 3.045 | <0.001 |
| OTU05 (Streptococcus salivarius) | High—Absent | 11.672 | 3.045 | 0.009 |
| OTU10 (Bifidobacterium pseudocatenulatum) | High—Absent | 44.116 | 4.077 | <0.001 |
| OTU17 (Klebsiella pneumoniae) | High—Absent | 44.575 | 3.925 | <0.001 |
| OTU25 (Parabacteroides distasonis) | Low—Absent | 44.540 | 3.651 | <0.001 |
| LSTb | ||||
| OTU24 (Bifidobacterium longum) | Low—Absent | 49.510 | 17.349 | 0.025 |
| LSTc | ||||
| OTU05 (Streptococcus salivarius) | Medium—Absent | 281.536 | 46.123 | <0.001 |
| OTU10 (Bifidobacterium pseudocatenulatum) | High—Absent | 247.574 | 33.782 | <0.001 |
| OTU17 (Klebsiella pneumoniae) | Low—Absent | 70.097 | 19.648 | 0.009 |
| OTU17 (Klebsiella pneumoniae) | High—Absent | 241.556 | 37.923 | <0.001 |
| OTU25 (Parabacteroides distasonis) | Low—Absent | 238.437 | 36.608 | <0.001 |
| DFLNT | ||||
| OTU04 (Bifidobacterium breve) | Low—Absent | −597.364 | 136.154 | 0.005 |
| DSLNT | ||||
| OTU10 (Bifidobacterium pseudocatenulatum) | High—Absent | 124.563 | 37.241 | 0.016 |
| OTU25 (Parabacteroides distasonis) | Low—Absent | 133.074 | 41.169 | 0.012 |
| FLNH | ||||
| OTU13 (Raoultella ornithinolytica) | High—Absent | 342.258 | 81.920 | 0.004 |
| OTU24 (Bifidobacterium longum) | Low—Absent | 351.494 | 83.120 | 0.004 |
| DFLNH | ||||
| OTU20 (Bacteroides fragilis) | Low—Absent | 88.416 | 37.035 | 0.044 |
| DSLNH | ||||
| OTU20 (Bacteroides fragilis) | Low—Absent | 35.677 | 15.099 | 0.046 |
| Day 60 | ||||
| 3FL | ||||
| OTU05 (Streptococcus salivarius) | Low—Absent | −457.444 | 140.352 | 0.014 |
| OTU05 (Streptococcus salivarius) | Medium—Absent | −721.382 | 229.193 | 0.016 |
| LNT | ||||
| OTU06 (Escherichia coli) | Medium—Absent | 498.920 | 145.761 | 0.014 |
| OTU29 (Enterococcus faecalis) | Low—Absent | 636.978 | 249.903 | 0.034 |
| LNnT | ||||
| OTU17 (Klebsiella pneumoniae) | Low—Absent | 76.183 | 23.105 | 0.013 |
| OTU17 (Klebsiella pneumoniae) | Medium—Absent | 70.373 | 23.105 | 0.019 |
| LNFP I | ||||
| OTU25 (Parabacteroides distasonis) | Low—Absent | 700.864 | 210.358 | 0.013 |
| LNFP II | ||||
| OTU29 (Enterococcus faecalis) | Low—Absent | 520.197 | 204.372 | 0.034 |
| LNFP III | ||||
| OTU25 (Parabacteroides distasonis) | Low—Absent | 11.677 | 2.660 | 0.003 |
| LSTb | ||||
| OTU06 (Escherichia coli) | Medium—Absent | 39.951 | 11.096 | 0.011 |
| OTU25 (Parabacteroides distasonis) | Low—Absent | 55.080 | 20.155 | 0.029 |
| DFLNT | ||||
| OTU25 (Parabacteroides distasonis) | Low—Absent | 666.649 | 211.012 | 0.016 |
| LNH | ||||
| OTU10 (Bifidobacterium pseudocatenulatum) | Low—Absent | 26.025 | 6.182 | 0.006 |
| OTU10 (Bifidobacterium pseudocatenulatum) | Medium—Absent | 15.904 | 6.182 | 0.042 |
| OTU10 (Bifidobacterium pseudocatenulatum) | High—Absent | 42.040 | 8.094 | 0.002 |
| FLNH | ||||
| OTU20 (Bacteroides fragilis) | Low—Absent | −145.411 | 41.110 | 0.010 |
| OTU20 (Bacteroides fragilis) | High—Absent | −191.839 | 68.517 | 0.027 |
| OTU29 (Enterococcus faecalis) | Low—Absent | 194.456 | 83.620 | 0.049 |
| DFLNH | ||||
| OTU04 (Bifidobacterium breve) | Medium—Absent | 66.004 | 21.207 | 0.021 |
| OTU04 (Bifidobacterium breve) | High—Absent | 65.513 | 16.197 | 0.007 |
| Day 90 | ||||
| 6′SL | ||||
| OTU24 (Bifidobacterium longum) | Low—Absent | −53.183 | 22.870 | 0.049 |
| LNnT | ||||
| OTU13 (Raoultella ornithinolytica) | Low—Absent | 139.726 | 45.827 | 0.019 |
| OTU29 (Enterococcus faecalis) | Low—Absent | 141.980 | 42.530 | 0.010 |
| LNFP III | ||||
| OTU25 (Parabacteroides distasonis) | Low—Absent | 5.314 | 2.284 | 0.048 |
| OTU29 (Enterococcus faecalis) | Low—Absent | 7.553 | 2.902 | 0.032 |
| LSTc | ||||
| OTU03 (Bifidobacterium longum subsp. infantis) | Low—Absent | 56.415 | 14.472 | 0.008 |
| DFLNT | ||||
| OTU24 (Bifidobacterium longum) | Low—Absent | −259.858 | 97.869 | 0.029 |
| FLNH | ||||
| OTU04 (Bifidobacterium breve) | High—Absent | −60.398 | 20.607 | 0.019 |
| FDSLNH | ||||
| OTU25 (Parabacteroides distasonis) | Low—Absent | 187.335 | 78.648 | 0.044 |
| Day 120 | ||||
| 2′FL | ||||
| OTU05 (Streptococcus salivarius) | Low—Absent | 794.461 | 324.411 | 0.040 |
| 3FL | ||||
| OTU05 (Streptococcus salivarius) | Low—Absent | −579.838 | 191.043 | 0.016 |
| DFLac | ||||
| OTU24 (Bifidobacterium longum) | Low—Absent | −143.516 | 16.277 | <0.001 |
| OTU25 (Parabacteroides distasonis) | Low—Absent | −130.102 | 56.234 | 0.049 |
| 3′SL | ||||
| OTU29 (Enterococcus faecalis) | Low—Absent | 113.907 | 12.184 | <0.001 |
| LNT | ||||
| OTU06 (Escherichia coli) | High—Absent | 425.170 | 137.772 | 0.022 |
| OTU25 (Parabacteroides distasonis) | Low—Absent | 402.129 | 149.964 | 0.028 |
| LNFP I | ||||
| OTU20 (Bacteroides fragilis) | Low—Absent | 394.347 | 125.257 | 0.016 |
| LNFP II | ||||
| OTU05 (Streptococcus salivarius) | Low—Absent | −307.140 | 122.055 | 0.036 |
| LSTb | ||||
| OTU06 (Escherichia coli) | High—Absent | 37.797 | 14.733 | 0.043 |
| DFLNT | ||||
| OTU06 (Escherichia coli) | Medium—Absent | 358.632 | 107.565 | 0.016 |
| LNH | ||||
| OTU20 (Bacteroides fragilis) | Low—Absent | 47.773 | 7.970 | <0.001 |
| FLNH | ||||
| OTU06 (Escherichia coli) | High—Absent | 91.548 | 35.911 | 0.044 |
| OTU25 (Parabacteroides distasonis) | Low—Absent | 105.044 | 39.804 | 0.030 |
| DFLNH | ||||
| OTU14 (Veillonella nakazawae) | Low—Absent | −43.460 | 16.298 | 0.032 |
| FDSLNH | ||||
| OTU24 (Bifidobacterium longum) | Low—Absent | 226.449 | 71.658 | 0.016 |
| OTU25 (Parabacteroides distasonis) | Low—Absent | 317.661 | 94.893 | 0.010 |


References
- Stinson, L.F. Establishment of the early-life microbiome: A DOHaD perspective. J. Dev. Orig. Health Dis. 2020, 11, 201–210. [Google Scholar] [CrossRef]
- Stiemsma, L.T.; Michels, K.B. The role of the microbiome in the developmental origins of health and disease. Pediatrics 2018, 141, e20172437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stanislawski, M.A.; Dabelea, D.; Wagner, B.D.; Iszatt, N.; Dahl, C.; Sontag, M.K.; Knight, R.; Lozupone, C.A.; Eggesbo, M. Gut microbiota in the first 2 years of life and the association with body mass index at age 12 in a norwegian birth cohort. mBio 2018, 9, e01751-18. [Google Scholar] [CrossRef] [Green Version]
- Kostic, A.D.; Gevers, D.; Siljander, H.; Vatanen, T.; Hyotylainen, T.; Hamalainen, A.M.; Peet, A.; Tillmann, V.; Poho, P.; Mattila, I.; et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe 2015, 17, 260–273. [Google Scholar] [CrossRef] [Green Version]
- Azad, M.B.; Konya, T.; Guttman, D.S.; Field, C.J.; Sears, M.R.; HayGlass, K.T.; Mandhane, P.J.; Turvey, S.E.; Subbarao, P.; Becker, A.B.; et al. Infant gut microbiota and food sensitization: Associations in the first year of life. Clin. Exp. Allergy 2015, 45, 632–643. [Google Scholar] [CrossRef] [PubMed]
- Arrieta, M.C.; Stiemsma, L.T.; Dimitriu, P.A.; Thorson, L.; Russell, S.; Yurist-Doutsch, S.; Kuzeljevic, B.; Gold, M.J.; Britton, H.M.; Lefebvre, D.L.; et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 2015, 7, 307ra152. [Google Scholar] [CrossRef] [PubMed]
- Mohajeri, M.H.; La Fata, G.; Steinert, R.E.; Weber, P. Relationship between the gut microbiome and brain function. Nutr. Rev. 2018, 76, 481–496. [Google Scholar] [CrossRef] [PubMed]
- Van den Elsen, L.W.J.; Garssen, J.; Burcelin, R.; Verhasselt, V. Shaping the gut microbiota by breastfeeding: The gateway to allergy prevention? Front. Pediatr. 2019, 7, 47. [Google Scholar] [CrossRef]
- Stewart, C.J.; Ajami, N.J.; O’Brien, J.L.; Hutchinson, D.S.; Smith, D.P.; Wong, M.C.; Ross, M.C.; Lloyd, R.E.; Doddapaneni, H.; Metcalf, G.A.; et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 2018, 562, 583–588. [Google Scholar] [CrossRef]
- Roswall, J.; Olsson, L.M.; Kovatcheva-Datchary, P.; Nilsson, S.; Tremaroli, V.; Simon, M.-C.; Kiilerich, P.; Akrami, R.; Krämer, M.; Uhlén, M.; et al. Developmental trajectory of the healthy human gut microbiota during the first 5 years of life. Cell Host Microbe 2021, 29, 765–776.e763. [Google Scholar] [CrossRef]
- Nagpal, R.; Tsuji, H.; Takahashi, T.; Nomoto, K.; Kawashima, K.; Nagata, S.; Yamashiro, Y. Ontogenesis of the gut microbiota composition in healthy, full-term, vaginally born and breast-fed infants over the first 3 years of life: A quantitative bird’s-eye view. Front. Microbiol. 2017, 8, 1388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yassour, M.; Jason, E.; Hogstrom, L.J.; Arthur, T.D.; Tripathi, S.; Siljander, H.; Selvenius, J.; Oikarinen, S.; Hyoty, H.; Virtanen, S.M.; et al. Strain-level analysis of mother-to-child bacterial transmission during the first few months of life. Cell Host Microbe 2018, 24, 146–154.e144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferretti, P.; Pasolli, E.; Tett, A.; Asnicar, F.; Gorfer, V.; Fedi, S.; Armanini, F.; Truong, D.T.; Manara, S.; Zolfo, M.; et al. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe 2018, 24, 133–145.e135. [Google Scholar] [CrossRef] [PubMed]
- Murphy, K.; Curley, D.; O’Callaghan, T.F.; O’Shea, C.A.; Dempsey, E.M.; O’Toole, P.W.; Ross, R.P.; Ryan, C.A.; Stanton, C. The composition of human milk and infant faecal microbiota over the first three months of life: A pilot study. Sci. Rep. 2017, 7, 40597. [Google Scholar] [CrossRef] [Green Version]
- Drell, T.; Stsepetova, J.; Simm, J.; Rull, K.; Aleksejeva, A.; Antson, A.; Tillmann, V.; Metsis, M.; Sepp, E.; Salumets, A.; et al. The Influence of different maternal microbial communities on the development of infant gut and oral microbiota. Sci. Rep. 2017, 7, 9940. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, M.; Nakayama, J. Development of the gut microbiota in infancy and its impact on health in later life. Allergol. Int. 2017, 66, 515–522. [Google Scholar] [CrossRef]
- Valles, Y.; Artacho, A.; Pascual-Garcia, A.; Ferrus, M.L.; Gosalbes, M.J.; Abellan, J.J.; Francino, M.P. Microbial succession in the gut: Directional trends of taxonomic and functional change in a birth cohort of Spanish infants. PLoS Genet. 2014, 10, e1004406. [Google Scholar] [CrossRef] [Green Version]
- Lackey, K.A.; Williams, J.E.; Meehan, C.L.; Zachek, J.A.; Benda, E.D.; Price, W.J.; Foster, J.A.; Sellen, D.W.; Kamau-Mbuthia, E.W.; Kamundia, E.W.; et al. What’s normal? microbiomes in human milk and infant feces are related to each other but vary geographically: The INSPIRE study. Front. Nutr. 2019, 6, 45. [Google Scholar] [CrossRef] [Green Version]
- Drago, L.; Toscano, M.; De Grandi, R.; Grossi, E.; Padovani, E.M.; Peroni, D.G. Microbiota network and mathematic microbe mutualism in colostrum and mature milk collected in two different geographic areas: Italy versus Burundi. ISME J. 2017, 11, 875–884. [Google Scholar] [CrossRef]
- McGuire, M.K.; Meehan, C.L.; McGuire, M.A.; Williams, J.E.; Foster, J.; Sellen, D.W.; Kamau-Mbuthia, E.W.; Kamundia, E.W.; Mbugua, S.; Moore, S.E.; et al. What’s normal? oligosaccharide concentrations and profiles in milk produced by healthy women vary geographically. Am. J. Clin. Nutr. 2017, 105, 1086–1100. [Google Scholar] [CrossRef]
- Ding, M.; Qi, C.; Yang, Z.; Jiang, S.; Bi, Y.; Lai, J.; Sun, J. Geographical location specific composition of cultured microbiota and Lactobacillus occurrence in human breast milk in China. Food Funct. 2019, 10, 554–564. [Google Scholar] [CrossRef]
- Biagi, E.; Quercia, S.; Aceti, A.; Beghetti, I.; Rampelli, S.; Turroni, S.; Faldella, G.; Candela, M.; Brigidi, P.; Corvaglia, L. The bacterial ecosystem of mother’s milk and infant’s mouth and gut. Front. Microbiol. 2017, 8, 1214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costello, E.K.; Carlisle, E.M.; Bik, E.M.; Morowitz, M.J.; Relman, D.A. Microbiome assembly across multiple body sites in low-birthweight infants. mBio 2013, 4, e00782-13. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.E.; Carrothers, J.M.; Lackey, K.A.; Beatty, N.F.; Brooker, S.L.; Peterson, H.K.; Steinkamp, K.M.; York, M.A.; Shafii, B.; Price, W.J.; et al. Strong multivariate relations exist among milk, oral, and fecal microbiomes in mother-infant dyads during the first six months postpartum. J. Nutr. 2019, 149, 902–914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ling, Z.; Kong, J.; Jia, P.; Wei, C.; Wang, Y.; Pan, Z.; Huang, W.; Li, L.; Chen, H.; Xiang, C. Analysis of oral microbiota in children with dental caries by PCR-DGGE and barcoded pyrosequencing. Microb. Ecol. 2010, 60, 677–690. [Google Scholar] [CrossRef]
- Chen, C.; Hemme, C.; Beleno, J.; Shi, Z.J.; Ning, D.; Qin, Y.; Tu, Q.; Jorgensen, M.; He, Z.; Wu, L.; et al. Oral microbiota of periodontal health and disease and their changes after nonsurgical periodontal therapy. ISME J. 2018, 12, 1210–1224. [Google Scholar] [CrossRef]
- Sulyanto, R.M.; Thompson, Z.A.; Beall, C.J.; Leys, E.J.; Griffen, A.L. The predominant oral microbiota is acquired early in an organized pattern. Sci. Rep. 2019, 9, 10550. [Google Scholar] [CrossRef] [Green Version]
- Oba, P.M.; Holscher, H.D.; Mathai, R.A.; Kim, J.; Swanson, K.S. Diet influences the oral microbiota of infants during the first six months of life. Nutrients 2020, 12, 3400. [Google Scholar] [CrossRef]
- Timby, N.; Domellof, M.; Holgerson, P.L.; West, C.E.; Lonnerdal, B.; Hernell, O.; Johansson, I. Oral microbiota in infants fed a formula supplemented with bovine milk fat globule membranes—A randomized controlled trial. PLoS ONE 2017, 12, e0169831. [Google Scholar] [CrossRef]
- Mason, M.R.; Chambers, S.; Dabdoub, S.M.; Thikkurissy, S.; Kumar, P.S. Characterizing oral microbial communities across dentition states and colonization niches. Microbiome 2018, 6, 67. [Google Scholar] [CrossRef]
- Dzidic, M.; Collado, M.C.; Abrahamsson, T.; Artacho, A.; Stensson, M.; Jenmalm, M.C.; Mira, A. Oral microbiome development during childhood: An ecological succession influenced by postnatal factors and associated with tooth decay. ISME J. 2018, 12, 2292–2306. [Google Scholar] [CrossRef] [PubMed]
- Matsuyama, M.; Gomez-Arango, L.F.; Fukuma, N.M.; Morrison, M.; Davies, P.S.W.; Hill, R.J. Breastfeeding: A key modulator of gut microbiota characteristics in late infancy. J. Dev. Orig. Health Dis. 2019, 10, 206–213. [Google Scholar] [CrossRef] [PubMed]
- Walsh, C.; Lane, J.A.; van Sinderen, D.; Hickey, R.M. Human milk oligosaccharides: Shaping the infant gut microbiota and supporting health. J. Funct. Foods 2020, 72, 104074. [Google Scholar] [CrossRef]
- Asnicar, F.; Manara, S.; Zolfo, M.; Truong, D.T.; Scholz, M.; Armanini, F.; Ferretti, P.; Gorfer, V.; Pedrotti, A.; Tett, A.; et al. Studying vertical microbiome transmission from mothers to infants by strain-level metagenomic profiling. mSystems 2017, 2, e00164-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milani, C.; Mancabelli, L.; Lugli, G.A.; Duranti, S.; Turroni, F.; Ferrario, C.; Mangifesta, M.; Viappiani, A.; Ferretti, P.; Gorfer, V.; et al. Exploring vertical transmission of Bifidobacteria from mother to child. Appl. Environ. Microbiol. 2015, 81, 7078–7087. [Google Scholar] [CrossRef] [Green Version]
- Jost, T.; Lacroix, C.; Braegger, C.P.; Rochat, F.; Chassard, C. Vertical mother-neonate transfer of maternal gut bacteria via breastfeeding. Environ. Microbiol. 2014, 16, 2891–2904. [Google Scholar] [CrossRef]
- Duranti, S.; Lugli, G.A.; Mancabelli, L.; Armanini, F.; Turroni, F.; James, K.; Ferretti, P.; Gorfer, V.; Ferrario, C.; Milani, C.; et al. Maternal inheritance of bifidobacterial communities and bifidophages in infants through vertical transmission. Microbiome 2017, 5, 66. [Google Scholar] [CrossRef]
- Koren, O.; Goodrich, J.K.; Cullender, T.C.; Spor, A.; Laitinen, K.; Backhed, H.K.; Gonzalez, A.; Werner, J.J.; Angenent, L.T.; Knight, R.; et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell 2012, 150, 470–480. [Google Scholar] [CrossRef] [Green Version]
- Carrothers, J.M.; York, M.A.; Brooker, S.L.; Lackey, K.A.; Williams, J.E.; Shafii, B.; Price, W.J.; Settles, M.L.; McGuire, M.A.; McGuire, M.K. Fecal microbial community structure is stable over time and related to variation in macronutrient and micronutrient intakes in lactating women. J. Nutr. 2015, 145, 2379–2388. [Google Scholar] [CrossRef] [Green Version]
- Canul-Medina, G.; Fernandez-Mejia, C. Morphological, hormonal, and molecular changes in different maternal tissues during lactation and post-lactation. J. Physiol. Sci. 2019, 69, 825–835. [Google Scholar] [CrossRef]
- Hunt, K.M.; Foster, J.A.; Forney, L.J.; Schutte, U.M.; Beck, D.L.; Abdo, Z.; Fox, L.K.; Williams, J.E.; McGuire, M.K.; McGuire, M.A. Characterization of the diversity and temporal stability of bacterial communities in human milk. PLoS ONE 2011, 6, e21313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moossavi, S.; Sepehri, S.; Robertson, B.; Bode, L.; Goruk, S.; Field, C.J.; Lix, L.M.; de Souza, R.J.; Becker, A.B.; Mandhane, P.J.; et al. Composition and variation of the human milk microbiota are influenced by maternal and early-life factors. Cell Host Microbe 2019, 25, 324–335.e324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hermansson, H.; Kumar, H.; Collado, M.C.; Salminen, S.; Isolauri, E.; Rautava, S. Breast milk microbiota Is shaped by mode of delivery and intrapartum antibiotic exposure. Front. Nutr. 2019, 6, 4. [Google Scholar] [CrossRef] [Green Version]
- Wan, Y.; Jiang, J.; Lu, M.; Tong, W.; Zhou, R.; Li, J.; Yuan, J.; Wang, F.; Li, D. Human milk microbiota development during lactation and its relation to maternal geographic location and gestational hypertensive status. Gut Microbes 2020, 11, 1438–1449. [Google Scholar] [CrossRef]
- Khodayar-Pardo, P.; Mira-Pascual, L.; Collado, M.C.; Martinez-Costa, C. Impact of lactation stage, gestational age and mode of delivery on breast milk microbiota. J. Perinatol. 2014, 34, 599–605. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.E.; Carrothers, J.M.; Lackey, K.A.; Beatty, N.F.; York, M.A.; Brooker, S.L.; Shafii, B.; Price, W.J.; Settles, M.L.; McGuire, M.A.; et al. Human milk mcrobial community structure is relatively stable and related to variations in macronutrient and micronutrient intakes in healthy lactating women. J. Nutr. 2017, 147, 1739–1748. [Google Scholar] [CrossRef] [PubMed]
- Cabrera-Rubio, R.; Collado, M.C.; Laitinen, K.; Salminen, S.; Isolauri, E.; Mira, A. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am. J. Clin. Nutr. 2012, 96, 544–551. [Google Scholar] [CrossRef] [Green Version]
- Li, S.W.; Watanabe, K.; Hsu, C.C.; Chao, S.H.; Yang, Z.H.; Lin, Y.J.; Chen, C.C.; Cao, Y.M.; Huang, H.C.; Chang, C.H.; et al. Bacterial composition and diversity in breast milk samples from mothers living in Taiwan and Mainland China. Front. Microbiol. 2017, 8, 965. [Google Scholar] [CrossRef]
- Moossavi, S.; Miliku, K.; Sepehri, S.; Khafipour, E.; Azad, M.B. The prebiotic and probiotic properties of human milk: Implications for infant immune development and pediatric asthma. Front. Pediatr. 2018, 6, 197. [Google Scholar] [CrossRef] [Green Version]
- Sela, D.A.; Mills, D.A. Nursing our microbiota: Molecular linkages between bifidobacteria and milk oligosaccharides. Trends Microbiol. 2010, 18, 298–307. [Google Scholar] [CrossRef] [Green Version]
- Sela, D.A.; Chapman, J.; Adeuya, A.; Kim, J.H.; Chen, F.; Whitehead, T.R.; Lapidus, A.; Rokhsar, D.S.; Lebrilla, C.B.; German, J.B.; et al. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc. Natl. Acad. Sci. USA 2008, 105, 18964–18969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marcobal, A.; Barboza, M.; Froehlich, J.W.; Block, D.E.; German, J.B.; Lebrilla, C.B.; Mills, D.A. Consumption of human milk oligosaccharides by gut-related microbes. J. Agric. Food Chem. 2010, 58, 5334–5340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pace, R.M.; Williams, J.E.; Robertson, B.; Lackey, K.A.; Meehan, C.L.; Price, W.J.; Foster, J.A.; Sellen, D.W.; Kamau-Mbuthia, E.W.; Kamundia, E.W.; et al. Variation in human milk compostion is related to differeces in milk and infant fecal microbial communities. Microorganisms 2021, 9, 1153. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Li, M.; Wu, S.; Lebrilla, C.B.; Chapkin, R.S.; Ivanov, I.; Donovan, S.M. Fecal microbiota composition of breast-fed infants is correlated with human milk oligosaccharides consumed. J. Pediatr. Gastroenterol. Nutr. 2015, 60, 825–833. [Google Scholar] [CrossRef] [Green Version]
- Laursen, M.F.; Pekmez, C.T.; Larsson, M.W.; Lind, M.V.; Yonemitsu, C.; Larnkjær, A.; Mølgaard, C.; Bode, L.; Dragsted, L.O.; Michaelsen, K.F.; et al. Maternal milk microbiota and oligosaccharides contribute to the infant gut microbiota assembly. ISME Commun. 2021, 1, 21. [Google Scholar] [CrossRef]
- Ramani, S.; Stewart, C.J.; Laucirica, D.R.; Ajami, N.J.; Robertson, B.; Autran, C.A.; Shinge, D.; Rani, S.; Anandan, S.; Hu, L.; et al. Human milk oligosaccharides, milk microbiome and infant gut microbiome modulate neonatal rotavirus infection. Nat. Commun. 2018, 9, 5010. [Google Scholar] [CrossRef] [Green Version]
- De Leoz, M.L.; Kalanetra, K.M.; Bokulich, N.A.; Strum, J.S.; Underwood, M.A.; German, J.B.; Mills, D.A.; Lebrilla, C.B. Human milk glycomics and gut microbial genomics in infant feces show a correlation between human milk oligosaccharides and gut microbiota: A proof-of-concept study. J. Proteome Res. 2015, 14, 491–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crusell, M.K.W.; Hansen, T.H.; Nielsen, T.; Allin, K.H.; Ruhlemann, M.C.; Damm, P.; Vestergaard, H.; Rorbye, C.; Jorgensen, N.R.; Christiansen, O.B.; et al. Gestational diabetes is associated with change in the gut microbiota composition in third trimester of pregnancy and postpartum. Microbiome 2018, 6, 89. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zheng, J.; Shi, W.; Du, N.; Xu, X.; Zhang, Y.; Ji, P.; Zhang, F.; Jia, Z.; Wang, Y.; et al. Dysbiosis of maternal and neonatal microbiota associated with gestational diabetes mellitus. Gut 2018, 67, 1614–1625. [Google Scholar] [CrossRef] [PubMed]
- Maher, S.E.; O’Brien, E.C.; Moore, R.L.; Byrne, D.F.; Geraghty, A.A.; Saldova, R.; Murphy, E.F.; Van Sinderen, D.; Cotter, P.D.; McAuliffe, F.M. The association between the maternal diet and the maternal and infant gut microbiome: A systematic review. Br. J. Nutr. 2020, 1–29. [Google Scholar] [CrossRef] [Green Version]
- Keohane, D.M.; Ghosh, T.S.; Jeffery, I.B.; Molloy, M.G.; O’Toole, P.W.; Shanahan, F. Microbiome and health implications for ethnic minorities after enforced lifestyle changes. Nat. Med. 2020, 26, 1089–1095. [Google Scholar] [CrossRef] [PubMed]
- Durazzi, F.; Sala, C.; Castellani, G.; Manfreda, G.; Remondini, D.; De Cesare, A. Comparison between 16S rRNA and shotgun sequencing data for the taxonomic characterization of the gut microbiota. Sci. Rep. 2021, 11, 3030. [Google Scholar] [CrossRef] [PubMed]
- Barrett, H.L.; Gomez-Arango, L.F.; Wilkinson, S.A.; McIntyre, H.D.; Callaway, L.K.; Morrison, M.; Dekker Nitert, M. A vegetarian diet is a major determinant of gut microbiota composition in early pregnancy. Nutrients 2018, 10, 890. [Google Scholar] [CrossRef] [Green Version]
- Mandal, S.; Godfrey, K.M.; McDonald, D.; Treuren, W.V.; Bjørnholt, J.V.; Midtvedt, T.; Moen, B.; Rudi, K.; Knight, R.; Brantsæter, A.L.; et al. Fat and vitamin intakes during pregnancy have stronger relations with a pro-inflammatory maternal microbiota than does carbohydrate intake. Microbiome 2016, 4, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roytio, H.; Mokkala, K.; Vahlberg, T.; Laitinen, K. Dietary intake of fat and fibre according to reference values relates to higher gut microbiota richness in overweight pregnant women. Br. J. Nutr. 2017, 118, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Kota, S.K.; Gayatri, K.; Jammula, S.; Kota, S.K.; Krishna, S.V.; Meher, L.K.; Modi, K.D. Endocrinology of parturition. Indian J. Endocrinol. Metab. 2013, 17, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Edwards, S.M.; Cunningham, S.A.; Dunlop, A.L.; Corwin, E.J. The maternal gut microbiome during pregnancy. MCN Am. J. Matern. Child Nurs. 2017, 42, 310–317. [Google Scholar] [CrossRef]
- Grajeda, R.; Perez-Escamilla, R. Stress during labor and delivery is associated with delayed onset of lactation among urban Guatemalan women. J. Nutr. 2002, 132, 3055–3060. [Google Scholar] [CrossRef]
- Ilchmann-Diounou, H.; Menard, S. Psychological stress, intestinal barrier dysfunctions, and autoimmune disorders: An overview. Front. Immunol. 2020, 11, 1823. [Google Scholar] [CrossRef]
- Ward, T.L.; Hosid, S.; Ioshikhes, I.; Altosaar, I. Human milk metagenome: A functional capacity analysis. BMC Microbiol. 2013, 13, 116. [Google Scholar] [CrossRef] [Green Version]
- Jimenez, E.; de Andres, J.; Manrique, M.; Pareja-Tobes, P.; Tobes, R.; Martinez-Blanch, J.F.; Codoner, F.M.; Ramon, D.; Fernandez, L.; Rodriguez, J.M. Metagenomic analysis of milk of healthy and mastitis-suffering women. J. Hum. Lact. 2015, 31, 406–415. [Google Scholar] [CrossRef]
- Grice, E.A.; Kong, H.H.; Conlan, S.; Deming, C.B.; Davis, J.; Young, A.C.; Program, N.C.S.; Bouffard, G.G.; Blakesley, R.W.; Murray, P.R.; et al. Topographical and temporal diversity of the human skin microbiome. Science 2009, 324, 1190–1192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otto, M. Staphylococcus epidermidis—The ‘accidental’ pathogen. Nat. Rev. Microbiol. 2009, 7, 555–567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moles, L.; Gomez, M.; Moroder, E.; Bustos, G.; Melgar, A.; Del Campo, R.; Rodriguez, J.M. Staphylococcus epidermidis in feedings and feces of preterm neonates. PLoS ONE 2020, 15, e0227823. [Google Scholar] [CrossRef] [Green Version]
- Dong, Y.; Speer, C.P. The role of Staphylococcus epidermidis in neonatal sepsis: Guarding angel or pathogenic devil? Int. J. Med. Microbiol. 2014, 304, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Soeorg, H.; Metsvaht, T.; Eelmae, I.; Merila, M.; Treumuth, S.; Huik, K.; Jurna-Ellam, M.; Ilmoja, M.L.; Lutsar, I. The role of breast milk in the colonization of neonatal gut and skin with coagulase-negative staphylococci. Pediatr. Res. 2017, 82, 759–767. [Google Scholar] [CrossRef]
- Boix-Amoros, A.; Collado, M.C.; Mira, A. Relationship between milk microbiota, bacterial load, macronutrients, and human cells during lactation. Front. Microbiol. 2016, 7, 492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Craig, S.J.; Blankenberg, D.; Parodi, A.C.L.; Paul, I.M.; Birch, L.L.; Savage, J.S.; Marini, M.E.; Stokes, J.L.; Nekrutenko, A.; Reimherr, M. Child weight gain trajectories linked to oral microbiota composition. Sci. Rep. 2018, 8, 14030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, J.; Fiscella, K.A.; Gill, S.R. Oral microbiome: Possible harbinger for children’s health. Int. J. Oral Sci. 2020, 12, 12. [Google Scholar] [CrossRef]
- Reddy, R.M.; Weir, W.B.; Barnett, S.; Heiden, B.T.; Orringer, M.B.; Lin, J.; Chang, A.C.; Carrott, P.W.; Lynch, W.R.; Beer, D.G.; et al. Increased variance in oral and gastric microbiome correlates with esophagectomy anastomotic leak. Ann. Thorac. Surg. 2018, 105, 865–870. [Google Scholar] [CrossRef] [Green Version]
- Blod, C.; Schlichting, N.; Schulin, S.; Suttkus, A.; Peukert, N.; Stingu, C.S.; Hirsch, C.; Elger, W.; Lacher, M.; Buhligen, U.; et al. The oral microbiome-the relevant reservoir for acute pediatric appendicitis? Int. J. Colorectal Dis. 2018, 33, 209–218. [Google Scholar] [CrossRef] [Green Version]
- Premaraj, T.S.; Vella, R.; Chung, J.; Lin, Q.; Panier, H.; Underwood, K.; Premaraj, S.; Zhou, Y. Ethnic variation of oral microbiota in children. Sci. Rep. 2020, 10, 14788. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Quinque, D.; Horz, H.P.; Li, M.; Rzhetskaya, M.; Raff, J.A.; Hayes, M.G.; Stoneking, M. Comparative analysis of the human saliva microbiome from different climate zones: Alaska, Germany, and Africa. BMC Microbiol. 2014, 14, 316. [Google Scholar] [CrossRef] [Green Version]
- Blekhman, R.; Goodrich, J.K.; Huang, K.; Sun, Q.; Bukowski, R.; Bell, J.T.; Spector, T.D.; Keinan, A.; Ley, R.E.; Gevers, D.; et al. Host genetic variation impacts microbiome composition across human body sites. Genome Biol. 2015, 16, 191. [Google Scholar] [CrossRef] [Green Version]
- Herrero, E.R.; Slomka, V.; Bernaerts, K.; Boon, N.; Hernandez-Sanabria, E.; Passoni, B.B.; Quirynen, M.; Teughels, W. Antimicrobial effects of commensal oral species are regulated by environmental factors. J. Dent. 2016, 47, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Uehara, Y.; Kikuchi, K.; Nakamura, T.; Nakama, H.; Agematsu, K.; Kawakami, Y.; Maruchi, N.; Totsuka, K. H2O2 produced by viridans group streptococci may contribute to inhibition of methicillin-resistant Staphylococcus aureus colonization of oral cavities in newborns. Clin. Infect. Dis. 2001, 32, 1408–1413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Shehri, S.; Henman, M.; Charles, B.G.; Cowley, D.; Shaw, P.N.; Liley, H.; Tomarchio, A.; Punyadeera, C.; Duley, J.A. Collection and determination of nucleotide metabolites in neonatal and adult saliva by high performance liquid chromatography with tandem mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2013, 931, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Al-Shehri, S.S.; Knox, C.L.; Liley, H.G.; Cowley, D.M.; Wright, J.R.; Henman, M.G.; Hewavitharana, A.K.; Charles, B.G.; Shaw, P.N.; Sweeney, E.L.; et al. Breastmilk-saliva interactions boost innate immunity by regulating the oral microbiome in early infancy. PLoS ONE 2015, 10, e0135047. [Google Scholar] [CrossRef] [Green Version]
- Sweeney, E.L.; Al-Shehri, S.S.; Cowley, D.M.; Liley, H.G.; Bansal, N.; Charles, B.G.; Shaw, P.N.; Duley, J.A.; Knox, C.L. The effect of breastmilk and saliva combinations on the in vitro growth of oral pathogenic and commensal microorganisms. Sci. Rep. 2018, 8, 15112. [Google Scholar] [CrossRef] [Green Version]
- Hurley, E.; Mullins, D.; Barrett, M.P.; O’Shea, C.A.; Kinirons, M.; Ryan, C.A.; Stanton, C.; Whelton, H.; Harris, H.M.B.; O’Toole, P.W. The microbiota of the mother at birth and its influence on the emerging infant oral microbiota from birth to 1 year of age: A cohort study. J. Oral Microbiol. 2019, 11, 1599652. [Google Scholar] [CrossRef] [Green Version]
- Lif Holgerson, P.; Esberg, A.; Sjodin, A.; West, C.E.; Johansson, I. A longitudinal study of the development of the saliva microbiome in infants 2 days to 5 years compared to the microbiome in adolescents. Sci. Rep. 2020, 10, 9629. [Google Scholar] [CrossRef] [PubMed]
- Kahharova, D.; Brandt, B.W.; Buijs, M.J.; Peters, M.; Jackson, R.; Eckert, G.; Katz, B.; Keels, M.A.; Levy, S.M.; Fontana, M.; et al. Maturation of the oral microbiome in caries-free toddlers: A longitudinal study. J. Dent. Res. 2020, 99, 159–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kennedy, B.; Peura, S.; Hammar, U.; Vicenzi, S.; Hedman, A.; Almqvist, C.; Andolf, E.; Pershagen, G.; Dicksved, J.; Bertilsson, S.; et al. Oral microbiota development in early childhood. Sci. Rep. 2019, 9, 19025. [Google Scholar] [CrossRef]
- Kaan, A.M.M.; Kahharova, D.; Zaura, E. Acquisition and establishment of the oral microbiota. Periodontol. 2000 2021, 86, 123–141. [Google Scholar] [CrossRef] [PubMed]
- Al-Shehri, S.S.; Sweeney, E.L.; Cowley, D.M.; Liley, H.G.; Ranasinghe, P.D.; Charles, B.G.; Shaw, P.N.; Vagenas, D.; Duley, J.A.; Knox, C.L. Deep sequencing of the 16S ribosomal RNA of the neonatal oral microbiome: A comparison of breast-fed and formula-fed infants. Sci. Rep. 2016, 6, 38309. [Google Scholar] [CrossRef] [Green Version]
- Holgerson, P.L.; Vestman, N.R.; Claesson, R.; Ohman, C.; Domellof, M.; Tanner, A.C.; Hernell, O.; Johansson, I. Oral microbial profile discriminates breast-fed from formula-fed infants. J. Pediatr. Gastroenterol. Nutr. 2013, 56, 127–136. [Google Scholar] [CrossRef] [Green Version]
- Salli, K.; Soderling, E.; Hirvonen, J.; Gursoy, U.K.; Ouwehand, A.C. Influence of 2′-fucosyllactose and galacto-oligosaccharides on the growth and adhesion of Streptococcus mutans. Br. J. Nutr. 2020, 124, 824–831. [Google Scholar] [CrossRef]
- Abranches, J.; Zeng, L.; Kajfasz, J.K.; Palmer, S.R.; Chakraborty, B.; Wen, Z.T.; Richards, V.P.; Brady, L.J.; Lemos, J.A. Biology of oral Streptococci. Microbiol. Spectr. 2018, 6, 6.5.11. [Google Scholar] [CrossRef]
- Djais, A.A.; Theodorea, C.F.; Mashima, I.; Otomo, M.; Saitoh, M.; Nakazawa, F. Identification and phylogenetic analysis of oral Veillonella species isolated from the saliva of Japanese children. F1000Research 2019, 8, 616. [Google Scholar] [CrossRef]
- Mashima, I.; Theodorea, C.F.; Djais, A.A.; Kunihiro, T.; Kawamura, Y.; Otomo, M.; Saitoh, M.; Tamai, R.; Kiyoura, Y. Veillonella nakazawae sp. nov., an anaerobic Gram-negative coccus isolated from the oral cavity of Japanese children. Int. J. Syst. Evol. Microbiol. 2021, 71, 004583. [Google Scholar] [CrossRef]
- Brown, M.M.; Horswill, A.R. Staphylococcus epidermidis-skin friend or foe? PLoS Pathog. 2020, 16, e1009026. [Google Scholar] [CrossRef] [PubMed]
- Devang Divakar, D.; Muzaheed; Aldeyab, S.S.; Alfawaz, S.A.; AlKheraif, A.A.; Ahmed Khan, A. High proportions of Staphylococcus epidermidis in dental caries harbor multiple classes of antibiotics resistance, significantly increase inflammatory interleukins in dental pulps. Microb. Pathog. 2017, 109, 29–34. [Google Scholar] [CrossRef]
- Jian, C.; Carpen, N.; Helve, O.; de Vos, W.M.; Korpela, K.; Salonen, A. Early-life gut microbiota and its connection to metabolic health in children: Perspective on ecological drivers and need for quantitative approach. EBioMedicine 2021, 69, 103475. [Google Scholar] [CrossRef] [PubMed]
- Vatanen, T.; Franzosa, E.A.; Schwager, R.; Tripathi, S.; Arthur, T.D.; Vehik, K.; Lernmark, A.; Hagopian, W.A.; Rewers, M.J.; She, J.X.; et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature 2018, 562, 589–594. [Google Scholar] [CrossRef] [PubMed]
- Backhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H.; et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 2015, 17, 690–703. [Google Scholar] [CrossRef] [Green Version]
- Garrido, D.; Barile, D.; Mills, D.A. A molecular basis for bifidobacterial enrichment in the infant gastrointestinal tract. Adv. Nutr. 2012, 3, 415S–421S. [Google Scholar] [CrossRef]
- Sakanaka, M.; Gotoh, A.; Yoshida, K.; Odamaki, T.; Koguchi, H.; Xiao, J.Z.; Kitaoka, M.; Katayama, T. Varied pathways of infant gut-associated Bifidobacterium to assimilate human milk oligosaccharides: Prevalence of the gene set and its correlation with Bifidobacteria-rich microbiota formation. Nutrients 2019, 12, 71. [Google Scholar] [CrossRef] [Green Version]
- Lawson, M.A.E.; O’Neill, I.J.; Kujawska, M.; Gowrinadh Javvadi, S.; Wijeyesekera, A.; Flegg, Z.; Chalklen, L.; Hall, L.J. Breast milk-derived human milk oligosaccharides promote Bifidobacterium interactions within a single ecosystem. ISME J. 2020, 14, 635–648. [Google Scholar] [CrossRef] [Green Version]
- LoCascio, R.G.; Ninonuevo, M.R.; Freeman, S.L.; Sela, D.A.; Grimm, R.; Lebrilla, C.B.; Mills, D.A.; German, J.B. Glycoprofiling of bifidobacterial consumption of human milk oligosaccharides demonstrates strain specific, preferential consumption of small chain glycans secreted in early human lactation. J. Agric. Food Chem. 2007, 55, 8914–8919. [Google Scholar] [CrossRef]
- Laursen, M.F.; Sakanaka, M.; von Burg, N.; Morbe, U.; Andersen, D.; Moll, J.M.; Pekmez, C.T.; Rivollier, A.; Michaelsen, K.F.; Molgaard, C.; et al. Bifidobacterium species associated with breastfeeding produce aromatic lactic acids in the infant gut. Nat. Microbiol. 2021, 6, 1367–1382. [Google Scholar] [CrossRef]
- Ehrlich, A.M.; Pacheco, A.R.; Henrick, B.M.; Taft, D.; Xu, G.; Huda, M.N.; Mishchuk, D.; Goodson, M.L.; Slupsky, C.; Barile, D.; et al. Indole-3-lactic acid associated with Bifidobacterium-dominated microbiota significantly decreases inflammation in intestinal epithelial cells. BMC Microbiol. 2020, 20, 357. [Google Scholar] [CrossRef] [PubMed]
- Pannaraj, P.S.; Li, F.; Cerini, C.; Bender, J.M.; Yang, S.; Rollie, A.; Adisetiyo, H.; Zabih, S.; Lincez, P.J.; Bittinger, K.; et al. Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr. 2017, 171, 647–654. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.J.; Lynch, D.B.; Murphy, K.; Ulaszewska, M.; Jeffery, I.B.; O’Shea, C.A.; Watkins, C.; Dempsey, E.; Mattivi, F.; Tuohy, K.; et al. Evolution of gut microbiota composition from birth to 24 weeks in the INFANTMET Cohort. Microbiome 2017, 5, 4. [Google Scholar] [CrossRef] [Green Version]
- Fallani, M.; Amarri, S.; Uusijarvi, A.; Adam, R.; Khanna, S.; Aguilera, M.; Gil, A.; Vieites, J.M.; Norin, E.; Young, D.; et al. Determinants of the human infant intestinal microbiota after the introduction of first complementary foods in infant samples from five European centres. Microbiology 2011, 157, 1385–1392. [Google Scholar] [CrossRef] [Green Version]
- Ray, C.; Kerketta, J.A.; Rao, S.; Patel, S.; Dutt, S.; Arora, K.; Pournami, F.; Bhushan, P. Human milk oligosaccharides: The journey ahead. Int. J. Pediatr. 2019, 2019, 2390240. [Google Scholar] [CrossRef] [Green Version]
- Wicinski, M.; Sawicka, E.; Gebalski, J.; Kubiak, K.; Malinowski, B. Human milk oligosaccharides: Health benefits, potential applications in infant formulas, and pharmacology. Nutrients 2020, 12, 266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bode, L. Human milk oligosaccharides: Every baby needs a sugar mama. Glycobiology 2012, 22, 1147–1162. [Google Scholar] [CrossRef] [Green Version]
- Le Doare, K.; Holder, B.; Bassett, A.; Pannaraj, P.S. Mother’s milk: A purposeful contribution to the development of the infant microbiota and immunity. Front. Immunol. 2018, 9, 361. [Google Scholar] [CrossRef] [Green Version]
- Davis, E.C.; Wang, M.; Donovan, S.M. The role of early life nutrition in the establishment of gastrointestinal microbial composition and function. Gut Microbes 2017, 8, 143–171. [Google Scholar] [CrossRef]
- Lyons, K.E.; Ryan, C.A.; Dempsey, E.M.; Ross, R.P.; Stanton, C. Breast milk, a source of beneficial microbes and associated benefits for infant health. Nutrients 2020, 12, 1039. [Google Scholar] [CrossRef]
- Zivkovic, A.M.; Lewis, Z.T.; German, J.B.; Mills, D.A. Establishment of a milk-oriented microbiota (MOM) in early life: How babies meet their MOMs. Funct. Food Rev. 2013, 5, 3–12. [Google Scholar]
- Donovan, S.M.; Wang, M.; Li, M.; Friedberg, I.; Schwartz, S.L.; Chapkin, R.S. Host-microbe interactions in the neonatal intestine: Role of human milk oligosaccharides. Adv. Nutr. 2012, 3, 450S–455S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borewicz, K.; Gu, F.; Saccenti, E.; Arts, I.C.W.; Penders, J.; Thijs, C.; van Leeuwen, S.S.; Lindner, C.; Nauta, A.; van Leusen, E.; et al. Correlating infant faecal microbiota composition and human milk oligosaccharide consumption by microbiota of one-month old breastfed infants. Mol. Nutr. Food Res. 2019, 63, e1801214. [Google Scholar] [CrossRef] [PubMed]
- Borewicz, K.; Gu, F.; Saccenti, E.; Hechler, C.; Beijers, R.; de Weerth, C.; van Leeuwen, S.S.; Schols, H.A.; Smidt, H. The association between breastmilk oligosaccharides and faecal microbiota in healthy breastfed infants at two, six, and twelve weeks of age. Sci. Rep. 2020, 10, 4270. [Google Scholar] [CrossRef] [PubMed]
- Plows, J.F.; Berger, P.K.; Jones, R.B.; Alderete, T.L.; Yonemitsu, C.; Najera, J.A.; Khwajazada, S.; Bode, L.; Goran, M.I. Longitudinal changes in human milk oligosaccharides (HMOs) over the course of 24 months of lactation. J. Nutr. 2021, 151, 876–882. [Google Scholar] [CrossRef] [PubMed]
- Minami, J.; Odamaki, T.; Hashikura, N.; Abe, F.; Xiao, J.Z. Lysozyme in breast milk is a selection factor for bifidobacterial colonisation in the infant intestine. Benef. Microbes 2016, 7, 53–60. [Google Scholar] [CrossRef]
- Gopalakrishna, K.P.; Hand, T.W. Influence of maternal milk on the neonatal intestinal microbiome. Nutrients 2020, 12, 823. [Google Scholar] [CrossRef] [Green Version]
- Gotoh, A.; Katoh, T.; Sakanaka, M.; Ling, Y.; Yamada, C.; Asakuma, S.; Urashima, T.; Tomabechi, Y.; Katayama-Ikegami, A.; Kurihara, S.; et al. Sharing of human milk oligosaccharides degradants within bifidobacterial communities in faecal cultures supplemented with Bifidobacterium bifidum. Sci. Rep. 2018, 8, 13958. [Google Scholar] [CrossRef]
- Yu, Z.T.; Chen, C.; Newburg, D.S. Utilization of major fucosylated and sialylated human milk oligosaccharides by isolated human gut microbes. Glycobiology 2013, 23, 1281–1292. [Google Scholar] [CrossRef] [Green Version]
- Chia, L.W.; Mank, M.; Blijenberg, B.; Bongers, R.S.; van Limpt, K.; Wopereis, H.; Tims, S.; Stahl, B.; Belzer, C.; Knol, J. Cross-feeding between Bifidobacterium infantis and Anaerostipes caccae on lactose and human milk oligosaccharides. Benef. Microbes 2021, 12, 69–83. [Google Scholar] [CrossRef]
- Morrow, A.L.; Ruiz-Palacios, G.M.; Jiang, X.; Newburg, D.S. Human-milk glycans that inhibit pathogen binding protect breast-feeding infants against infectious diarrhea. J. Nutr. 2005, 135, 1304–1307. [Google Scholar] [CrossRef] [Green Version]
- Hegar, B.; Wibowo, Y.; Basrowi, R.W.; Ranuh, R.G.; Sudarmo, S.M.; Munasir, Z.; Atthiyah, A.F.; Widodo, A.D.; Supriatmo; Kadim, M.; et al. The role of two human milk oligosaccharides, 2′-fucosyllactose and lacto-n-neotetraose, in infant nutrition. Pediatr. Gastroenterol. Hepatol. Nutr. 2019, 22, 330–340. [Google Scholar] [CrossRef]
- Andreas, N.J.; Kampmann, B.; Mehring Le-Doare, K. Human breast milk: A review on its composition and bioactivity. Early Hum. Dev. 2015, 91, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Coppa, G.V.; Pierani, P.; Zampini, L.; Carloni, I.; Carlucci, A.; Gabrielli, O. Oligosaccharides in human milk during different phases of lactation. Acta Paediatr. Suppl. 1999, 88, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Saben, J.L.; Sims, C.R.; Abraham, A.; Bode, L.; Andres, A. Human milk oligosaccharide concentrations and infant intakes are associated with maternal overweight and obesity and predict infant growth. Nutrients 2021, 13, 446. [Google Scholar] [CrossRef]
- Cheema, A.S.; Stinson, L.F.; Rea, A.; Lai, C.T.; Payne, M.S.; Murray, K.; Geddes, D.T.; Gridneva, Z. Human milk lactose, insulin, and glucose relative to infant body composition during exclusive breastfeeding. Nutrients 2021, 13, 3724. [Google Scholar] [CrossRef] [PubMed]
- Cheema, A.S.; Lai, C.T.; Dymock, M.; Rae, A.; Geddes, D.T.; Payne, M.S.; Stinson, L.F. Impact of expression mode and timing of sample collection, relative to milk ejection, on human milk bacterial DNA profiles. J. Appl. Microbiol. 2021, 131, 988–995. [Google Scholar] [CrossRef]
- Seferovic, M.D.; Mohammad, M.; Pace, R.M.; Engevik, M.; Versalovic, J.; Bode, L.; Haymond, M.; Aagaard, K.M. Maternal diet alters human milk oligosaccharide composition with implications for the milk metagenome. Sci. Rep. 2020, 10, 22092. [Google Scholar] [CrossRef]
- Kent, J.C.; Mitoulas, L.R.; Cregan, M.D.; Ramsay, D.T.; Doherty, D.A.; Hartmann, P.E. Volume and frequency of breastfeedings and fat content of breast milk throughout the day. Pediatrics 2006, 117, e387–e395. [Google Scholar] [CrossRef] [Green Version]
- Kent, J.C.; Hepworth, A.R.; Sherriff, J.L.; Cox, D.B.; Mitoulas, L.R.; Hartmann, P.E. Longitudinal changes in breastfeeding patterns from 1 to 6 months of lactation. Breastfeed. Med. 2013, 8, 401–407. [Google Scholar] [CrossRef] [Green Version]
- Stinson, L.; Hallingstrom, M.; Barman, M.; Viklund, F.; Keelan, J.; Kacerovsky, M.; Payne, M.; Jacobsson, B. Comparison of bacterial DNA profiles in mid-trimester amniotic fluid samples from preterm and term deliveries. Front. Microbiol. 2020, 11, 415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheema, A.S.; Stinson, L.F.; Lai, C.T.; Geddes, D.T.; Payne, M.S. DNA extraction method influences human milk bacterial profiles. J. Appl. Microbiol. 2020, 130, 142–156. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lenth, R.V. Least-Squares Means: TheRPackagelsmeans. J. Stat. Softw. 2016, 69, 1–33. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Pinheiro, J.; Bates, D.; DebRoy, S.; Sarkar, D.; R Core Team. Nlme: Linear and Nonlinear Mixed Effects Models; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]

| Characteristics (n = 10) | Mean ± SD (Min–Max) or n (%) |
|---|---|
| Maternal | |
| Age at infant birth (years) | 31.60 ± 2.42 (28–35) |
| Parity | 2.50 ± 0.50 (2–3) |
| Infant | |
| Male (n, (%)) | 4 (40.0%) |
| Gestational age (weeks) | 39.26 ± 1.11 (36.6–40.2) |
| Birth weight (grams) | 3622.50 ± 234.11 (3320–4020) |
| Birth length (cm) | 51.55 ± 1.39 (49–54) |
| 24-h milk intake (grams) | 837.80 ± 131.17 (580–1040) |
| Time Points | Maternal Faecal | Human Milk | Infant Oral | Infant Faecal | ||||
|---|---|---|---|---|---|---|---|---|
| MD (95% CI) | p-Value | MD (95% CI) | p-Value | MD (95% CI) | p-Value | MD (95% CI) | p-Value | |
| Richness (Observed OTUs) | ||||||||
| 2–5 days vs. 30 days | −72.27 (−118.35, −26.19) | 0.0023 | −4.18 (−49.06, 40.7) | 0.8543 | 33.3 (−10.35, 76.95) | 0.1339 | −3.9 (−47.55, 39.75) | 0.8602 |
| 2–5 days vs. 60 days | −44.82 (−90.9, 1.26) | 0.0565 | −3.58 (−48.46, 41.3) | 0.8750 | 36.3 (−7.35, 79.95) | 0.1025 | −14.7 (−58.35, 28.95) | 0.5070 |
| 2–5 days vs. 90 days | −71.68 (−116.55, −26.8) | 0.0019 | −7.28 (−52.16, 37.6) | 0.7491 | 32.4 (−11.25, 76.05) | 0.1447 | 3.2 (−40.45, 46.85) | 0.8851 |
| 2–5 days vs. 120 days | −60.98 (−105.85, −16.1) | 0.0080 | −32.28 (−77.16, 12.6) | 0.1574 | 33.6 (−10.05, 77.25) | 0.1305 | −17.5 (−61.15, 26.15) | 0.4297 |
| 30 days vs. 60 days | 27.44 (−18.56, 73.45) | 0.2406 | 0.6 (−43.05, 44.25) | 0.9784 | 3 (−40.65, 46.65) | 0.8922 | −10.8 (−54.45, 32.85) | 0.6258 |
| 30 days vs. 90 days | 0.59 (−44.29, 45.47) | 0.9793 | −3.1 (−46.75, 40.55) | 0.8887 | −0.9 (−44.55, 42.75) | 0.9676 | 7.1 (−36.55, 50.75) | 0.7485 |
| 30 days vs. 120 days | 11.29 (−33.59, 56.17) | 0.6200 | −28.1 (−71.75, 15.55) | 0.2055 | 0.3 (−43.35, 43.95) | 0.9892 | −13.6 (−57.25, 30.05) | 0.5393 |
| 60 days vs. 90 days | −26.85 (−71.73, 18.02) | 0.2392 | −3.7 (−47.35, 39.95) | 0.8673 | −3.9 (−47.55, 39.75) | 0.8602 | 17.9 (−25.75, 61.55) | 0.4193 |
| 60 days vs. 120 days | −16.15 (−61.03, 28.72) | 0.4783 | −28.7 (−72.35, 14.95) | 0.1960 | −2.7 (−46.35, 40.95) | 0.9029 | −2.8 (−46.45, 40.85) | 0.8994 |
| 90 days vs. 120 days | 10.7 (−32.95, 54.35) | 0.6290 | −25 (−68.65, 18.65) | 0.2598 | 1.2 (−42.45, 44.85) | 0.9568 | −20.7 (−64.35, 22.95) | 0.3505 |
| Shannon diversity | ||||||||
| 2–5 days vs. 30 days | −0.82 (−1.61, −0.02) | 0.0435 | −0.11 (−0.88, 0.66) | 0.7775 | 0.5 (−0.25, 1.25) | 0.1909 | 0.06 (−0.69, 0.81) | 0.8763 |
| 2–5 days vs. 60 days | −0.54 (−1.33, 0.25) | 0.1824 | −0.19 (−0.96, 0.58) | 0.6237 | 0.6 (−0.15, 1.35) | 0.1168 | −0.32 (−1.08, 0.43) | 0.3936 |
| 2–5 days vs. 90 days | −0.92 (−1.69, −0.15) | 0.0199 | −0.27 (−1.04, 0.5) | 0.4857 | 0.61 (−0.14, 1.36) | 0.1093 | 0.15 (−0.6, 0.9) | 0.7030 |
| 2–5 days vs. 120 days | −0.71 (−1.49, 0.06) | 0.0691 | −0.69 (−1.46, 0.09) | 0.0809 | 0.55 (−0.2, 1.3) | 0.1516 | −0.2 (−0.95, 0.55) | 0.5983 |
| 30 days vs. 60 days | 0.28 (−0.51, 1.07) | 0.4876 | −0.08 (−0.83, 0.67) | 0.8305 | 0.1 (−0.65, 0.85) | 0.7927 | −0.38 (−1.13, 0.37) | 0.3134 |
| 30 days vs. 90 days | −0.1 (−0.87, 0.67) | 0.7937 | −0.16 (−0.91, 0.59) | 0.6697 | 0.11 (−0.64, 0.86) | 0.7671 | 0.09 (−0.66, 0.84) | 0.8215 |
| 30 days vs. 120 days | 0.1 (−0.67, 0.87) | 0.7962 | −0.58 (−1.33, 0.17) | 0.1319 | 0.05 (−0.7, 0.8) | 0.8989 | −0.26 (−1.01, 0.49) | 0.4950 |
| 60 days vs. 90 days | −0.38 (−1.15, 0.39) | 0.3309 | −0.08 (−0.83, 0.67) | 0.8316 | 0.01 (−0.74, 0.76) | 0.9734 | 0.47 (−0.28, 1.22) | 0.2178 |
| 60 days vs. 120 days | −0.18 (−0.95, 0.59) | 0.6499 | −0.49 (−1.24, 0.26) | 0.1954 | −0.05 (−0.8, 0.7) | 0.8920 | 0.12 (−0.63, 0.87) | 0.7438 |
| 90 days vs. 120 days | 0.2 (−0.55, 0.95) | 0.5933 | −0.41 (−1.16, 0.34) | 0.2786 | −0.06 (−0.81, 0.69) | 0.8657 | −0.35 (−1.1, 0.4) | 0.3642 |
| Time Points | Maternal Faecal | Human Milk | Infant Oral | Infant Faecal |
|---|---|---|---|---|
| Bray-Curtis Dissimilarity | ||||
| 2–5 days vs 30 days | 0.0365 | 0.4227 | 0.0029 | 0.3971 |
| 2–5 days vs. 60 days | 0.1037 | 0.1354 | 0.0211 | 0.4323 |
| 2–5 days vs. 90 days | 0.3725 | 0.3969 | <0.0001 | 0.1590 |
| 2–5 days vs. 120 days | 0.8534 | 0.2948 | 0.0043 | 0.4046 |
| 30 days vs. 60 days | 0.6497 | 0.0289 | 0.6791 | 0.4193 |
| 30 days vs. 90 days | 0.3358 | 0.0741 | 0.0081 | 0.2825 |
| 30 days vs. 120 days | 0.3102 | 0.0513 | 0.0415 | 0.5278 |
| 60 days vs. 90 days | 0.4660 | 0.6015 | 0.1343 | 0.2471 |
| 60 days vs. 120 days | 0.5362 | 0.2185 | 0.6746 | 0.5739 |
| 90 days vs. 120 days | 0.6205 | 0.5657 | 0.2726 | 0.3194 |
| Sample Types | Day 2–5 | Day 30 | Day 60 | Day 90 | Day 120 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| MD (95% CI) | p-Value | MD (95% CI) | p-Value | MD (95% CI) | p-Value | MD (95% CI) | p-Value | MD (95% CI) | p-Value | |
| Richness (Observed OTUs) | ||||||||||
| HM vs. IF | 7.92 (−36.96, 52.8) | 0.7280 | 8.2 (−35.45, 51.85) | 0.7112 | −3.2 (−46.85, 40.45) | 0.8851 | 18.4 (−25.25, 62.05) | 0.4064 | 22.7 (−20.95, 66.35) | 0.3060 |
| HM vs. IO | −64.08 (−108.96, −19.2) | 0.0054 | −26.6 (−70.25, 17.05) | 0.2306 | −24.2 (−67.85, 19.45) | 0.2753 | −24.4 (−68.05, 19.25) | 0.2713 | 1.8 (−41.85, 45.45) | 0.9352 |
| HM vs. MF | −127.81 (−173.89, −81.72) | <0.0001 | −195.89 (−240.77, −151.01) | <0.0001 | −169.05 (−213.92, −124.17) | <0.0001 | −192.2 (−235.85, −148.55) | <0.0001 | −156.5 (−200.15, −112.85) | <0.0001 |
| IF vs. IO | −72 (−115.65, −28.35) | 0.0014 | −34.8 (−78.45, 8.85) | 0.1174 | −21 (−64.65, 22.65) | 0.3436 | −42.8 (−86.45, 0.85) | 0.0546 | −20.9 (−64.55, 22.75) | 0.3458 |
| IF vs. MF | −135.72 (−180.6, −90.85) | <0.0001 | −204.09 (−248.97, −159.21) | <0.0001 | −165.85 (−210.72, −120.97) | <0.0001 | −210.6 (−254.25, −166.95) | <0.0001 | −179.2 (−222.85, −135.55) | <0.0001 |
| IO vs. MF | −63.72 (−108.6, −18.85) | 0.0057 | −169.29 (−214.17, −124.41) | <0.0001 | −144.85 (−189.72, −99.97) | <0.0001 | −167.8 (−211.45, −124.15) | <0.0001 | −158.3 (−201.95, −114.65) | <0.0001 |
| Shannon diversity | ||||||||||
| HM vs. IF | −0.06 (−0.83, 0.71) | 0.8746 | 0.11 (−0.64, 0.86) | 0.7765 | −0.19 (−0.94, 0.56) | 0.6090 | 0.36 (−0.39, 1.11) | 0.3498 | 0.42 (−0.33, 1.17) | 0.2666 |
| HM vs. IO | −1.21 (−1.98, −0.44) | 0.0023 | −0.6 (−1.35, 0.15) | 0.1160 | −0.42 (−1.17, 0.33) | 0.2718 | −0.33 (−1.08, 0.42) | 0.3931 | 0.02 (−0.73, 0.77) | 0.9512 |
| HM vs. MF | −1.95 (−2.74, −1.16) | <0.0001 | −2.65 (−3.42, −1.88) | <0.0001 | −2.29 (−3.06, −1.52) | <0.0001 | −2.59 (−3.34, −1.84) | <0.0001 | −1.98 (−2.73, −1.23) | <0.0001 |
| IF vs. IO | −1.15 (−1.9, −0.4) | 0.0029 | −0.71 (−1.46, 0.04) | 0.0640 | −0.22 (−0.97, 0.53) | 0.5559 | −0.68 (−1.43, 0.07) | 0.0747 | −0.4 (−1.15, 0.35) | 0.2937 |
| IF vs. MF | −1.89 (−2.66, −1.12) | <0.0001 | −2.76 (−3.53, −1.99) | <0.0001 | −2.1 (−2.87, −1.33) | <0.0001 | −2.95 (−3.7, −2.2) | <0.0001 | −2.4 (−3.15, −1.65) | <0.0001 |
| IO vs. MF | −0.74 (−1.51, 0.03) | 0.0603 | −2.05 (−2.82, −1.28) | <0.0001 | −1.87 (−2.65, −1.1) | <0.0001 | −2.27 (−3.02, −1.52) | <0.0001 | −2 (−2.75, −1.25) | <0.0001 |
| Sample Types | Day 2–5 | Day 30 | Day 60 | Day 90 | Day 120 |
|---|---|---|---|---|---|
| Bray-Curtis Dissimilarity | |||||
| Human milk vs. Infant faecal | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 |
| Human milk vs. Infant oral | 0.0218 | 0.0001 | 0.0563 | 0.0004 | <0.0001 |
| Human milk vs. Maternal faecal | 0.0004 | <0.0001 | 0.0002 | <0.0001 | <0.0001 |
| Infant faecal vs. Infant oral | <0.0001 | 0.0006 | <0.0001 | <0.0001 | <0.0001 |
| Infant faecal vs. Maternal faecal | 0.0002 | <0.0001 | <0.0001 | <0.0001 | <0.0001 |
| Infant oral vs. Maternal faecal | <0.0001 | <0.0001 | 0.0069 | <0.0001 | <0.0001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheema, A.S.; Trevenen, M.L.; Turlach, B.A.; Furst, A.J.; Roman, A.S.; Bode, L.; Gridneva, Z.; Lai, C.T.; Stinson, L.F.; Payne, M.S.; et al. Exclusively Breastfed Infant Microbiota Develops over Time and Is Associated with Human Milk Oligosaccharide Intakes. Int. J. Mol. Sci. 2022, 23, 2804. https://doi.org/10.3390/ijms23052804
Cheema AS, Trevenen ML, Turlach BA, Furst AJ, Roman AS, Bode L, Gridneva Z, Lai CT, Stinson LF, Payne MS, et al. Exclusively Breastfed Infant Microbiota Develops over Time and Is Associated with Human Milk Oligosaccharide Intakes. International Journal of Molecular Sciences. 2022; 23(5):2804. https://doi.org/10.3390/ijms23052804
Chicago/Turabian StyleCheema, Ali Sadiq, Michelle Louise Trevenen, Berwin Ashoka Turlach, Annalee June Furst, Ana Sophia Roman, Lars Bode, Zoya Gridneva, Ching Tat Lai, Lisa Faye Stinson, Matthew Scott Payne, and et al. 2022. "Exclusively Breastfed Infant Microbiota Develops over Time and Is Associated with Human Milk Oligosaccharide Intakes" International Journal of Molecular Sciences 23, no. 5: 2804. https://doi.org/10.3390/ijms23052804
APA StyleCheema, A. S., Trevenen, M. L., Turlach, B. A., Furst, A. J., Roman, A. S., Bode, L., Gridneva, Z., Lai, C. T., Stinson, L. F., Payne, M. S., & Geddes, D. T. (2022). Exclusively Breastfed Infant Microbiota Develops over Time and Is Associated with Human Milk Oligosaccharide Intakes. International Journal of Molecular Sciences, 23(5), 2804. https://doi.org/10.3390/ijms23052804








