Abstract
Cardiovascular development is initiated soon after gastrulation as bilateral precardiac mesoderm is progressively symmetrically determined at both sides of the developing embryo. The precardiac mesoderm subsequently fused at the embryonic midline constituting an embryonic linear heart tube. As development progress, the embryonic heart displays the first sign of left-right asymmetric morphology by the invariably rightward looping of the initial heart tube and prospective embryonic ventricular and atrial chambers emerged. As cardiac development progresses, the atrial and ventricular chambers enlarged and distinct left and right compartments emerge as consequence of the formation of the interatrial and interventricular septa, respectively. The last steps of cardiac morphogenesis are represented by the completion of atrial and ventricular septation, resulting in the configuration of a double circuitry with distinct systemic and pulmonary chambers, each of them with distinct inlets and outlets connections. Over the last decade, our understanding of the contribution of multiple growth factor signaling cascades such as Tgf-beta, Bmp and Wnt signaling as well as of transcriptional regulators to cardiac morphogenesis have greatly enlarged. Recently, a novel layer of complexity has emerged with the discovery of non-coding RNAs, particularly microRNAs and lncRNAs. Herein, we provide a state-of-the-art review of the contribution of non-coding RNAs during cardiac development. microRNAs and lncRNAs have been reported to functional modulate all stages of cardiac morphogenesis, spanning from lateral plate mesoderm formation to outflow tract septation, by modulating major growth factor signaling pathways as well as those transcriptional regulators involved in cardiac development.
1. Growth Factor Signalling in Cardiac Morphogenesis
Specification, differentiation and development of cardiac progenitors are subjected to intense paracrine regulation mediated by different growth factors, particularly bone morphogenetic proteins (Bmps), fibroblastic growth factors (Fgfs), transforming growth factors (Tfgs) and Wnt signaling [1,2,3]. Importantly, these growth factor signals also contribute to additional cardiac morphogenetic processes, such as proepicardial/epicardial development [4,5,6,7,8,9], endocardial cushion formation [10,11,12,13,14,15,16] and outflow tract remodeling [14,17,18,19]. In addition, several other signaling pathways are also required for discrete cardiovascular morphogenetic processes, such as Hippo pathway that is fundamental for cardiomyocyte proliferation and organ size determination [20,21,22], and Notch and neuregulin signaling pathways [23,24,25,26,27,28,29] that are required for ventricular morphogenesis and maturation. Evidence of the functional role of non-coding RNAs impacting on these signaling pathways has been recently reported as detailed in the following subheadings.
2. Transcriptional Control of Cardiac Morphogenesis
Cardiac morphogenesis is orchestrated by a large number of transcriptional regulators. Precardiac mesoderm is configured by Mesp1 and Mesp2 expression [30,31,32]. Soon thereafter, the cardiogenic cells are characterized by the expression of cardiac enriched transcription factors such as Gata4, Nkx2.5, Mef2c, Srf that start conferring the initial cardiomyocyte properties and providing positional clues to develop the initial cardiac straight tube [33,34,35,36,37,38,39,40,41,42]. Rightward bending is governed by a differential outgrowth regulated by Prrx1 [43] while Pitx2 imprints left-sided positional cues to the linear heart tube [44,45,46,47,48]. Subsequently, as cardiac chambers emerge, distinct members of the T-box family display regional expression, providing transcriptional cues for the formation of the arterial pole by Tbx1 expression [49,50], the prospective atrioventricular canal and thus part of the slow components of the cardiac conduction system by expression of Tbx2 and Tbx3 [51,52], the left ventricle and atrial chambers by Tbx5 [53,54,55,56] and the external epicardial lining by Tbx18 [57,58]. In addition, systemic and pulmonary ventricles are characterized by Hand1 and Hand2 transcriptional inputs [59,60,61,62], while atrial and ventricular chambers are characterized by Coup-TFII [63], Hey1 and Hey2 expression [64,65], respectively. Thus, overall, an intricate transcriptional regulation governs cardiac morphogenesis, providing evidence of a complex and exquisite regulatory network that involves many transcription factors.
3. The Emergence of a Novel Layer of Gene Regulation: Post-Transcriptional Regulation by Non-Coding RNAs
Whereas transcriptional regulation constitutes a major step governing and defining the molecular mechanisms that direct cardiac morphogenesis and cardiovascular cell differentiation, a novel layer of gene regulation is emerging with the discovery of non-coding RNAs. Non-coding RNAs are broadly classified according to their length in two distinct categories, small non-coding RNA if smaller than 200 nucleotides and long non-coding RNAs if larger than 200 nucleotides [66,67]. Small non-coding RNAs are represented by distinct RNA types such as snoRNAs, siRNAs, piRNAs and the most extensively studied and abundantly expressed microRNAs [68,69]. In all cases, these small non-coding RNAs exerts their function as post-transcriptional regulators, as detailed below. On the other hand, long non-coding RNAs represent a wide and large array of non-coding RNAs that can be classified according to the genomic location (promoter associated, sense, antisense, bidirectional, intronic, intergenic, 3′UTR associated, 5’UTR associated lncRNAs) and/or their functional properties [70,71,72]. Importantly, lncRNAs can be located in distinct subcellular compartments [73,74], such as the nucleus and the cytoplasm, exerting distinct functional roles, i.e., at transcriptional and post-transcriptional, respectively.
4. Biogenesis and Function of microRNAs and lncRNAs
MicroRNAs represent the most studied subtype of small non-coding RNAs [69,70]. microRNAs display temporal and spatial differential expression in both embryonic and adult tissues, contributing thus to both embryonic development and tissue homeostasis [75]. Impaired expression and/or function of microRNAs also lead to pathological conditions [76,77]. Importantly, microRNAs are highly conserved during evolution, ranging from C. elegans to humans. MicroRNAs are encoded in the nucleus, by transcription of precursors microRNA molecules that are normally transcribed by RNA polymerase II. In certain genomic localization, microRNAs are clustered in such a way that the primary transcript contains multiple microRNA precursors, such as in the case of miR-23/miR-27/miR-24 cluster, a transcript thus named pri-miRNA. Pri-miRNA is then processed by RNAses such as Drosha and Dgcr8 to generate distinct pre-miRNA molecules, i.e., pre-miR-23, pre-miR-27 and pre-miR-24, that are subsequently exported to the cytoplasm by exportin-5/Ran protein complex. Within the cytoplasm, the pre-miRNA is processed into a mature microRNA duplex by Dicer RNAse and loaded into the RISC complex in which one of the double-stranded microRNA molecule is degraded. The mature single-stranded microRNA molecule within the RISC complex is able now to scan other RNA molecules for sequence homology of its seed sequence provoking RNA target cleavage, translation repression and/or RNA deadenylation [78,79] In most cases, the final output consequence is a decrease on the miRNA/protein target abundance.
Long non-coding RNAs also display tissue-specific expression during embryogenesis and tissue homeostasis, being their role in pathology also emerging [73,80,81]. However, their tissue expression levels are, on average, 10-fold lower that mRNAs and microRNAs and they are poorly conserved during evolution. LncRNAs are transcribed in the nucleus, in most cases by RNA polymerase II, and display genomic organization similar to coding RNAs, i.e., introns and exons which are processed by alternative splicing. LcnRNAs are, in most cases, 5′ capped and 3′ polyadenylated and are distinctly distributed in the nucleus or the cytoplasm or both subcellular compartments simultaneously [73,82]. Importantly, although their sequence length can, in some cases, excess 10 Kb, the lncRNAs do not contain open reading frames or if they do, they only provide small oligopeptides or polypeptides that, in most cases, it is unclear if they are indeed translated and/or functional. At the functional level, lncRNAs can exert multiple tasks, such as modulation of transcription factor function, alternative splicing and histone modification (acetylation and/or methylation) at the transcriptional level. On other hand at the post-transcriptional level, they can serve as scaffold for protein post-transcriptional modifications (phosphorylation, protein degradation), multimeric protein complex formation or as competing endogenous RNAs for microRNA sponging [73,80,81]. Furthermore, lncRNAs can also be processed to be loaded into extracellular vesicles, such as exosomes, and thus participate in intercellular communication processes [83,84].
5. Post-Transcriptional Control of Precardiac Mesoderm Formation by ncRNAs
Bmp signaling plays key determinant roles during cardiogenesis, particularly on the early stages of precardiac mesoderm specification [85,86,87]. Different murine mutant models for Bmp2, Bmp4 and Bmp7 have revealed the importance of these growth factors in cardiogenesis. For example, Bmp2 knock-out mice are embryonic lethal at E7.5 displaying an exocoelomic location of the heart and as well as cardiac differentiation defects [88,89,90]. Bmp4 deficient mice showed even an earlier embryonic lethality than Bmp2 null mutants [91]. Bmp4 induces the expression of Brachyury and Nanog, which in turn induces differentiation of the cardiogenic mesoderm while Bmp2 is necessary for the correct expression of two key factors for early cardiogenesis, Nkx2.5 and Gata4 [92,93]. In this context, it is important to highlight that miR-130 has been recently identified as a key molecular regulator of Bmp2-Fgf8 signaling during early cardiac specification. Bmp2 induces while Fgf8 represses, Nkx2.5 and Gata4 in early precardiac mesoderm. Bmp2 also induces miR-130 expression which directly targets Erk1/2 and thus influences Fgf8 expression [94] (Figure 1A,D).
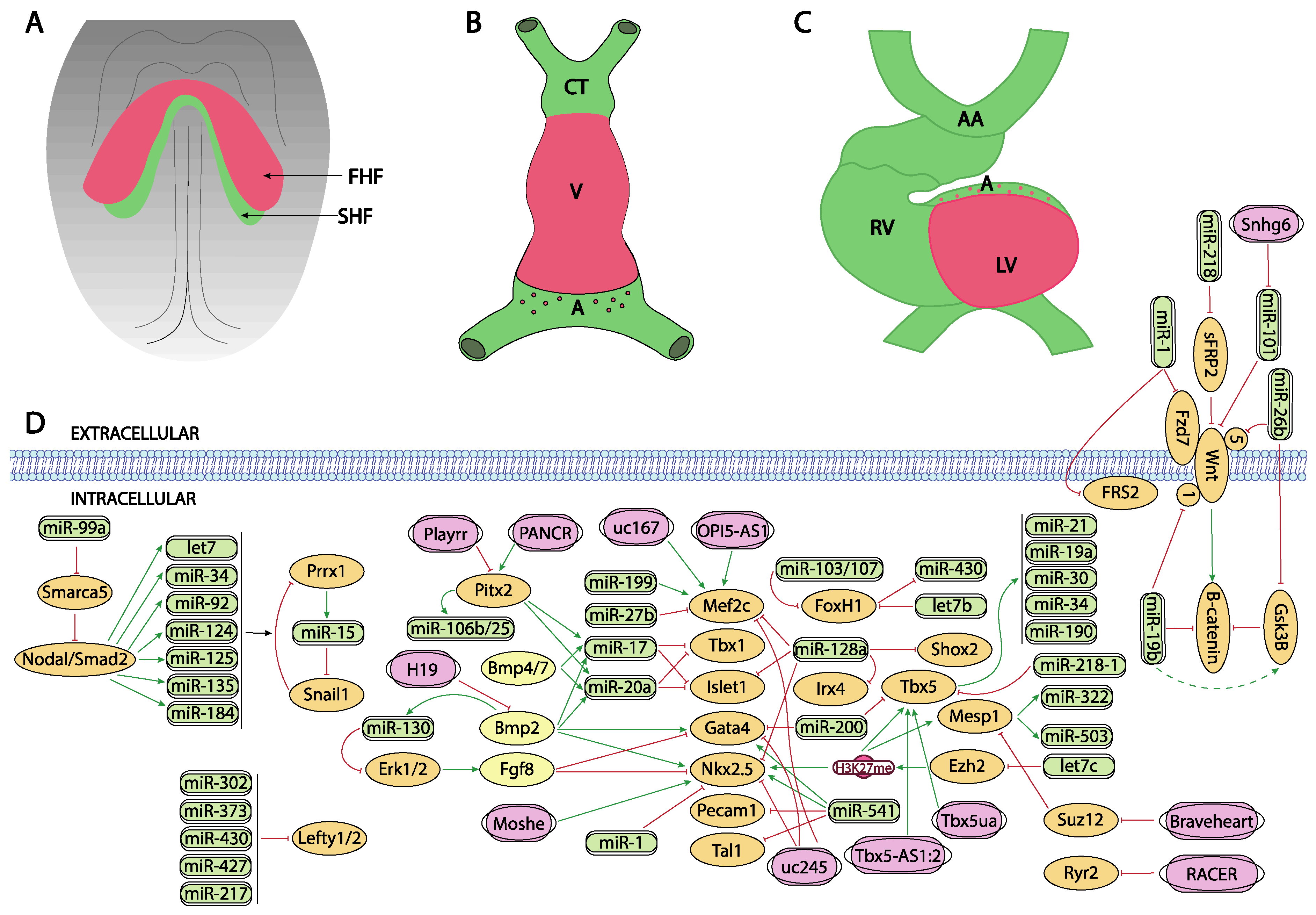
Figure 1.
Schematic representation of the main developmental events during the early stages of cardiovascular development, ranging from the configuration of the cardiac crescent (A), the early cardiac straight tube (B) and the rightward looping (C). (D) represent the current molecular interaction between microRNAs and lncRNAs with distinct growth factors and transcription factors involved in these developmental processes. FHF, first heart field; SHF, second heart field, CT conotruncus, V, ventricle, A, atrium, RV, right ventricle, LV, left ventricle, AA, aortic arches.
The role of Wnt signaling in early cardiogenesis is still a matter of debate. Several reports pointed out that Wnt signaling is required for cardiac tissue formation emanating from the early mesoderm [95,96,97] while others reported that cardiac specification can occur independent of Wnt/beta-catenin signaling [98]. Overall, these data support the notion of a tight temporal and tissue specific role for Wnt signaling during early cardiogenesis [99,100]. Importantly, robust evidence on the role of Wnt signaling in cardiogenesis have been found during in vitro embryonic stem cell-derived cardiogenesis [101,102] and the implication of microRNA regulation in this setting has been documented.
Wang et al. [103] reported that miR-218 modulates Wnt signaling in mouse cardiac stem cells, by directly targeting sFRP2 that act as negative regulator of Wnt, thus promoting proliferation while inhibiting differentiation (Figure 1D). More recently, Wang et al. [104] reported the functional role of miR-26b regulating cardiomyocyte differentiation by direct interaction with Gsk3beta and Wnt5a while Liu et al. [105] and Qin et al. [106] analyzed the role of miR-19b in P19 mouse carcinoma cells, demonstrating that miR-19b directly targets Wnt1 and indirectly enhanced Gsk3beta and beta-catenin expression (Figure 1D). Lu et al. [107] investigated the consequences of miR-1 in cardiomyocyte commitment from human cardiovascular progenitors and reported that Fzd7 and Frs2 are direct targets of miR-1 (Figure 1D). Overall, these data illustrate the functional role of microRNAs modulating Wnt signalling in cardiogenesis, yet most of these evidence is reported using in vitro experimental models.
Several lines of evidence reported the role of Tgf-beta signaling in cardiomyogenic commitment [108,109]. While the functional role of Tgf-beta signaling is thus crucial for multiple steps of heart development, our current understanding of the impact of microRNAs regulating this signaling pathway in cardiogenesis is still incipient. No evidence has been reported on the modulation of Tgf-beta signaling in early cardiomyogenic commitment nor during cardiomyocyte proliferation.
While at present there is no experimental evidence that microRNAs can directly target Mesp1 and/or Mesp2, there is indirect evidence of the functional role regulating the activity of these key cardiogenic transcriptional factors. Coppola et al. [110] demonstrated that miR-99a/let-7c are involved in the control of cardiomyogenesis, in part, by altering epigenetic factors. By over-expressing and inhibiting let7c expression in mouse embryonic stem cells, these authors demonstrate that cardiomyogenesis was promoted by mesodermal specification genes such as T/Bra and nodal as well as cardiac differentiation genes such as Mesp1, Nkx2.5 and Tbx5 were upregulated. Importantly, the functional role of let-7c is restricted to the early phase of mesoderm formation. Let7c induced upregulation of these transcription factors by directly targeting the Polycomb complex group protein Ezh2 and thus modifying H3K27me3 marks from the promoters of crucial cardiac transcription factors. On the other hand, miR-99a represses cardiac differentiation via the nucleosome-remodeling factor Smarca5, attenuating the Nodal/Smad2 signaling (Figure 1A,D).
Additional evidence of the functional role of microRNAs in the early stages of precardiac mesoderm specification was reported by Chen et al. [111] since deletion of Dgcr8 microprocessor, a key protein involved in microRNA biogenesis, in Mesp1 cardiovascular progenitor cells lead to dilated cardiomyopathy, due to defective cardiomyocyte differentiation. These authors identified miR-541 as a highly expressed microRNA in early mouse hearts (E9.5) that partly rescued the Dgcr8 deletion Mesp1 cardiovascular progenitor defects by downregulating angiogenic genes. Shen et al. [112] identified miR-322/miR-503 as the most enriched microRNAs in Mesp1 expressing cells. Ectopic expression of miR-322/miR-503 mimicked the endogenous temporal expression of genes driving cardiomyocyte specification while inhibiting neural lineage development (Figure 1A,D).
The precardiac mesoderm commitment into the cardiogenic lineage is primarily directed by the combinatorial action of several transcription factors such as Gata4, Mef2c and Nkx2.5. At present limited information is available about the regulation of Gata4 by microRNAs during early cardiogenesis. Yao et al. [113] described that miR-200b targets GATA4, modulating thus myosin heavy chain expression (Figure 1A,D). On the other hand, additional evidence is reported during cardiac hypertrophy as Gata4 is modulated by miR-26 [114] and during hypoxia by miR-200 [115]. While there is scarce indirect evidence of microRNAs targeting Nkx2.5 during early stages of cardiogenesis in mice [116], modulation of miR-1 by Nkx2.5 is essential for early cardiogenesis in Drosophila [117] (Figure 1A,D). Additional indirect evidence is also reported in mice, including therein miR-1, miR133 and miR-128b, and demonstrating a key role for miR-128b in cardiogenesis [118]. However, it needs to be considered that some controversies are emanating in this context, as the cardiac miRnome of precardiac mesoderm and early committed cardiomyocytes is not significantly altered in absence of Nkx2.5 [119]. As previously mentioned, Coppola et al. [110] reported that let-7c upregulated genes involved in mesoderm specification as well as cardiac differentiation markers such Mesp1, Nkx2.5 and Tbx5. Evidence of Nkx2.5 regulation by these microRNAs, i.e., miR-1 and miR-133 is also reported in other biological contexts [120].
Finally, similarly scarce reports have demonstrated the regulation of Mef2c by microRNAs in the cardiovascular context. Chinchilla et al. [121] firstly reported that miR-27b directly targets Mef2c, while more recently Chen et al. [122] demonstrated that miR-199 inhibition leads to cardiomyocyte differentiation by up-regulating Mef2c expression in embryonic stem cells (Figure 1A,D). On the other hand, additional evidence on Mef2c regulation by microRNAs has been reported during skeletal muscle development, such as miR-449 [123], miR-194 [124], miR-204 [125], miR-1 [126], miR-206 [126] and miR-214 [127], while miR-488 regulates Mef2c in vascular smooth muscle cells [128].
LncRNAs are versatile RNA molecules that can modulate gene expression either at transcriptional or post-transcriptional level depending on their subcellular localization. To date, our understanding of the functional role of lncRNAs during early stages of heart development is still incipient. A fundamental role for Braveheart lncRNA has been reported during early precardiac mesoderm commitment. Braveheart is required for the activation of a core cardiovascular gene network by acting upstream of Mesp1. Mechanistically, Braveheart interacts with SUZ12, a component of polycomb-repressive complex 2 (PRC2), during cardiomyocyte differentiation [129] (Figure 1A,D). Importantly, Braveheart administration is sufficient for mesenchymal stem cells to differentiate into cardiogenic lineage in vitro [130]. Curiously, besides its early developmental role, Braveheart is also expressed in the adult heart [131,132] and it is regulated by distinct core cardiac transcription factors [131]. Additional evidence on functional role of lncRNAs during early cardiomyogenic lineage specification has been obtained only using in vitro systems. For example, uc.245 lncRNA over-expression downregulates the expression of several cardiomyogenic-specific molecular markers such as Nkx2.5, Gata4, Mef2c in P19 cells [132,133]. Similarly, uc.167 overexpression inhibits proliferation while promoting apoptosis in P19 cells and regulates Mef2c expression [134]. However, the molecular mechanisms underlying such effects remains unexplored. Interestingly OIP5-AS1 lncRNA interacts with Mef2c mRNA promoting myogenic gene expression in vitro using a skeletal muscle cell line [135] (Figure 1A,D).
Besides Braveheart, Moshe lncRNA, an antisense transcript located upstream of Gata6 locus, has also been reported to fine-tune early heart development. Moshe knock-down during cardiogenesis leads to significant repression of Nkx2.5 in cardiac progenitor stages. RNA immunoprecipitation assays demonstrate that Moshe activates Nkx2.5 gene expression via direct binding to its promoter region [136] (Figure 1A,D).
Fendrr, a lncRNA located in the vicinity of Foxf1 locus and with enhanced expression at the caudal end of the lateral plate mesoderm is essential for heart and body wall development [137]. Fendrr knock out mice are embryonic lethal at E12.5 and displayed hypoplasia of the ventricular chamber and interventricular septum. Mechanistically, Mef2c, Gata6 and Nkx2.5 are upregulated in embryonic hearts whereas Gata6, Foxf1, Pitx2 and Irx3 were also upregulated in the caudal end of the lateral plate mesoderm in this mutant (Figure 2D). Regulation of the expression of these transcription factors is mediated by histone modification at their promoters [138].
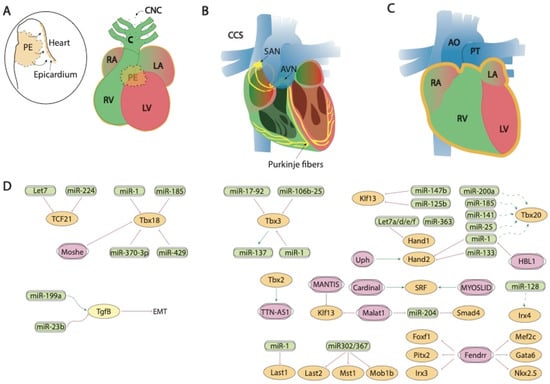
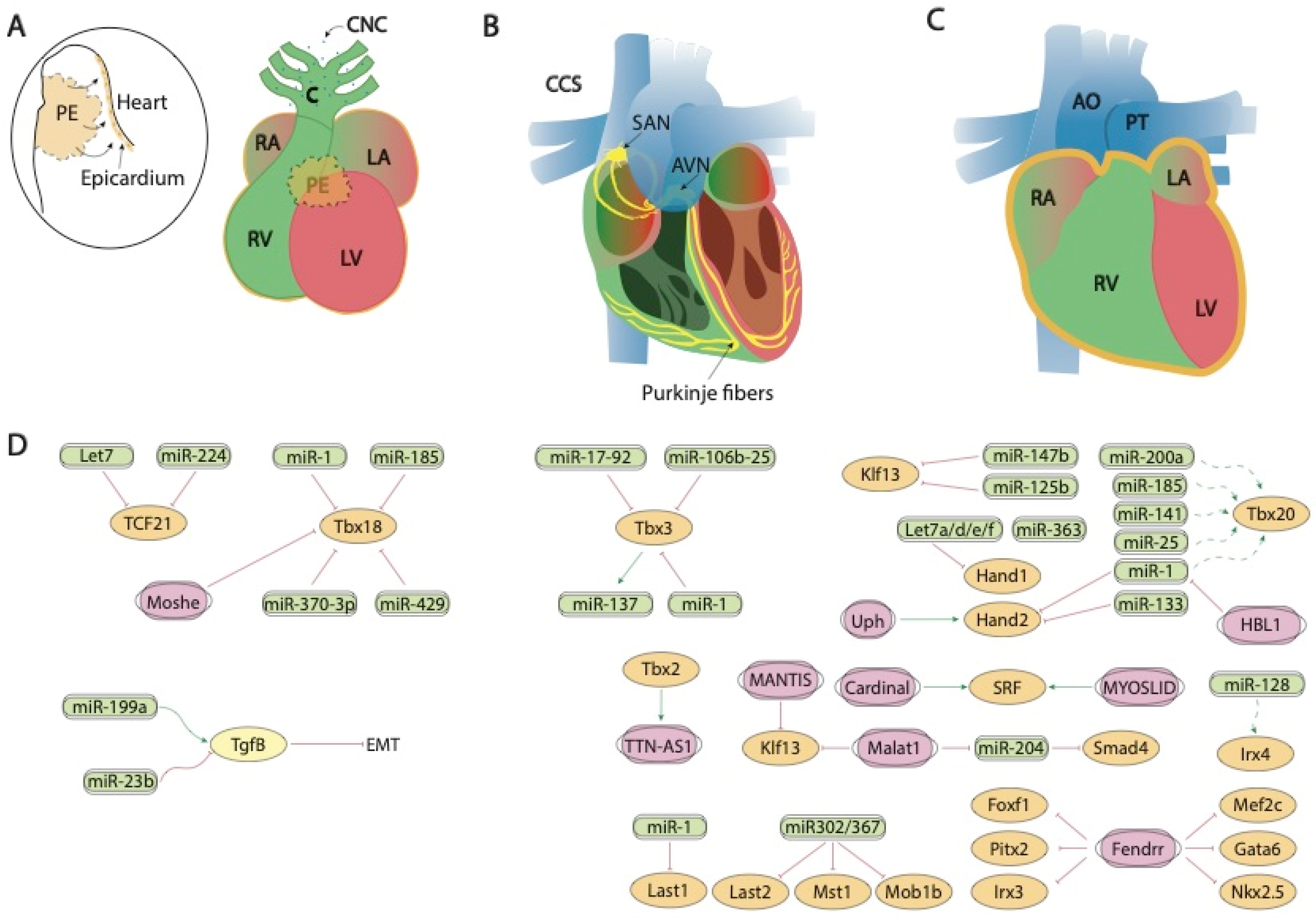
Figure 2.
Schematic representation of the mid-developmental events during cardiovascular development, ranging from the epicardial lining of the developing heart and the arrival of the cardiac neural crest cells (A), the early configuration of the cardiac conduction system (B) and chamber septation process (C). (D) represent the current molecular interaction between microRNAs and lncRNAs with distinct growth factors and transcription factors involved in these developmental processes. PE, proepicardium, CNC, cardiac neural crest, CCS, cardiac conduction system, RA, right atrium, LA, left atrium, RV, right ventricle, LV, left ventricle, SAN, sinoatrial node, AVN, atrioventricular node, AO, aorta, PT, pulmonary trunk.
6. Post-Transcriptional Control of Heart Fields Deployment by ncRNAs
Several transcription factors have been identified as key regulators of second heart field deployment, among which the most relevant are Foxh1, Isl1 and Tbx5 as detailed below. Forkhead box protein family are important components of the signaling pathways that instruct cardiogenesis and embryonic heart development [138]. Particularly, Foxh1 acts as a mesodermal specification inductor during gastrulation and, as the development proceeds its expression is restricted to the developing heart [139,140,141]. In this way, Foxh1 modulates Mef2c and Pitx2 transcription factors during the anterior heart field (AHF) formation which is required for right ventricle (RV) and outflow tract (OFT) development during the heart looping stage [141]. Foxh1 systemic mutants are arrested at the cardiac linear heart tube stage [141]. In zebrafish model, it has been demonstrated that Foxh1 is a direct downstream target of let-7b and miR-103/107 microRNA family [142,143] (Figure 1A,D). In the first steps of development, miR-103/107 modulate Foxh1 expression which together with Smad3 are able to modulate Nodal expression in the lateral plate mesoderm. Impaired expression of this gene can disturb left-right signaling genes generating cardiac edema and kidney failure [143]. More recently, it has been demonstrated that Foxh1 represses miR-430, via non-canonical regulation during early development in zebrafish, balancing Nodal signaling which controls early zygotic gene expression [144] (Figure 1A,D). In other contexts, Foxh1 promotes breast cancer cell proliferation through Wnt/β-catenin signaling pathway activation. In line with that, some authors conclude that let-7b may control cell cycle progression at least in part through the downregulation of Foxh1 [142,145].
Isl1 is a LIM-homeodomain transcription factor important for the development of multiple organs during embryogenesis. In particular, it has a crucial role during the secondary heart field (SHF) formation by directing the contribution of progenitor cells to the deployment of the right ventricle (RV), outflow tract (OFT) and the atria [146,147,148] during cardiovascular development.
Several microRNAs have been reported to modulate Isl1 expression in this context. At the first steps of cardiac development, miR-128a modulates cardiac progenitor cells differentiation into different cardiomyocytes subpopulations through the modulation of Isl1 as well as such as Nkx2.5, Mef2c, Irx4 and Shox2 [118] (Figure 1A,D). However, whether such regulatory effects are direct or indirect remains to be established. It has also been demonstrated that BMP-signaling regulates the miR-17-92 cluster that modulate the molecular pathway which promotes SHF myocardial differentiation. Moreover, bioinformatics analyses have shown miR-17 and miR-20a seed sequences in the Isl1 3′UTR and those interactions were validated by luciferase assays (Figure 1A,D). These data reveal the mechanism underlying Bmp-regulated OFT myocardial differentiation and indicate that Bmp-regulated miRNA activity is critical for fine-tuning cardiac progenitor genes [149].
Regulation of Isl1 by microRNAs has also been reported in distinct cardiac pathological conditions. In vitro experiments of cardiac hypertrophy stimulation with angiotensin II has evidenced that Isl-1+, Sca-1+, c-kit+ porcine cardiac progenitor cells exhibited high levels of Mef2c, Gata4, miR-29a and miR-21 [150]. Moreover, in another pathological context, such as type 2 diabetes mellitus, miR-128-3p targets Isl1 promoting cardiovascular calcification and insulin resistance modulating Wnt pathway [151].
Tbx5 is a T-box transcription factor family member which has a key role during forelimb and cardiac development. The broad spatiotemporal expression of Tbx5 during development is generally conserved in vertebrates, for example, in human hearts. Tbx5 is expressed in the epicardium, myocardium, and endocardium of embryonic and adult hearts [152]. In zebrafish differential microRNAs and mRNAs expression were analyzed in Tbx5 deficient embryos, identifying miR-19a, miR-30, miR-34, miR-190 and miR-21 as differentially expressed microRNAs [153]. Importantly, some of these microRNAs are already known to be involved in cardiac development [154,155,156,157]. For example, miR-19a is essential for correct heart development [154], and, of particular importance is miR-34a, which has been previously described playing a protecting role against cardiac remodeling in a stress situation [155]. Furthermore, miR-30 modulates Robo1, which is implicated in heart tube formation [156] and miR-21 could modulate Ndrg4 causing several cardiac defects [157].
Moreover, Tbx5 and miR-218-1 have a correlated expression in mouse cardiomyocytes. In this line, it has been shown that, in zebrafish, Tbx5 and miR-218-1 dysregulation affect early heart development, concretely, miR-218-1 modulation can rescue cardiac defects generated by Tbx5 over expression, acting as a mediator of Tbx5 during cardiogenesis [158]. In addition, Tbx5 has been reported to be directly targeted by miR-200 [159] during cardiogenesis (Figure 1A,D).
Evidence on the regulatory role of lncRNAs modulating SHF transcription factors has been also reported in vitro by Hou et al. [130] demonstrating that Braveheart enhances Isl1 expression as well as other cardiac enriched transcription factors such as Nkx2.5, Gata4, Gata6 during the process of transdifferentiation of mesenchymal stem cells into cells with the cardiogenic phenotype.
Importantly, Moshe knock-down also increases expression of SHF lineage genes such as Isl1, as well as several transcription factors relevant for subsequent morphogenetic processes such as Hand2, Tbx2, Shox2 and Tbx18. Therefore, these data suggest that Moshe is a heart-enriched lncRNA that controls a sophisticated network of cardiogenesis by repressing genes in SHF via Nkx2.5 during cardiac development [136] (Figure 1A,D).
Modulation of Tbx5, another SHF transcription factor has also been reported to be modulated by lncRNAs. TBX5-AS1:2 lncRNA regulates Tbx5 expression via modulation of its promoter methylation status. Curiously, TBX5-AS1:2 lncRNA is dysregulated in Tetralogy of Fallot patients, suggesting a plausible role in the onset of this cardiac congenital defect [160]. Additionally, Tbx5ua, a lncRNA close to Tbx5 locus is required for proper ventricular wall development, and thus being necessary for embryonic development, since Tbx5ua mouse knock-out embryonic lethal [161] (Figure 1A,D). Besides lncRNAs regulating Tbx5, evidence on the transcriptional regulation of Tbx5 on non-coding RNAs has been recently elucidated by Yang et al. [162]. Over 2600 novel lncRNAs were identified as Tbx5-dependent lncRNAs and these authors further identified RACER as a novel lncRNAs that modulate the expression of the calcium-handling gene Ryr2 (Figure 1A,D).
7. Post-Transcriptional Control of Sidedness and Cardiac Looping by ncRNAs
The establishment of sidedness and thus the configuration of left-right asymmetry is a complex developmental process in which multiple growth factors are involved, such as nodal, lefty-1 and lefty-2, among others [163,164], that ultimately converge into the activation of Pitx2 [165,166,167,168]. Importantly, Pitx2 is dispensable for correct cardiac looping in mice [169,170] and recently Prrx1 transcription factor has emerged as a key player in this developmental process [43].
Our current understanding of the functional role of microRNAs modulating left-right signaling is progressively emerging, yet most of the evidence has been obtained using in vitro models. Several lines of evidence demonstrated that lefties (lefty-1 and lefty-2) are modulated by miR-302, miR-373 miR-430 and miR-427 in human embryonic stem cells (hESCs) [171,172,173] provoking alterations in early hESCs differentiation. Similarly, the role of miR-430 has been confirmed in zebrafish early development, by using target protectors [174], while miR-217 regulates lefty-1 in mouse embryonic stem cells development, modulating thus mesendoderm formation [175]. In the Japanese flounder, squint is modulated by miR-430 [176] (Figure 1B–D).
Similarly, the role of microRNAs in Wnt signaling during cardiogenesis is limited [177] while increasing evidence has been reported in vitro [103,104,105,106], as previously stated. Importantly, in vivo evidence demonstrated that miR-19b overexpression in zebrafish embryos impaired left-right cardiac signaling leading to impaired cardiac development. Beta-catenin was identified as a direct miR-19b, supporting the notion of impaired Wnt signaling in this experimental model [177] (Figure 1B–D).
Pitx2 is a homeobox transcription factor which is expressed quite early in the embryo [167,178,179,180,181]. Pitx2 expression is restricted to the left lateral plate mesoderm of different species and several studies have demonstrated that Pitx2 is important for transmitting positional information from LPM to organ primordia such as heart, lung and gut [182,183,184]. Moreover, during development and adulthood Pitx2 expression remains in the developing heart, predisposing to atrial arrhythmias [167,185,186,187].
Our current understanding of the impact of microRNAs during embryonic Pitx2 expression is still incipient. Wang et al. [185] demonstrated that Pitx2 positively regulates miR-17-92 and miR-106b-25 repressing the sinoatrial node genetic program (Figure 1B–D). On the other hand, several studies have evidenced that Pitx2 modulates the expression of several microRNAs that contribute to Atrial Fibrillation (AF), such as miR-106-25, miR-17-92, miR-29a, miR-200, miR-203, miR-21, miR-208ab, miR-1, miR-26b and miR-106ab [185,188,189,190,191,192]. The lack of Pitx2 expression in the left atrial appendage impairs the expression of a microRNA battery (miR-21, miR-106a, miR-203 and miR-208ab down-regulated, and miR-1, miR-26b, miR-29a, miR-106b and miR-200 up-regulated) through a Wnt8a and Wnt11 modulation, leading to a calcium, sodium and potassium channel remodeling during AF [185,188,189].
Prrx1 is a paired-like homeobox transcription factor that modulates EMT during embryonic development as well as in pathological conditions such as cancer [193]. During embryonic development, EMT is also regulated by other transcription factors such as Snail, Zeb and Twist. Fazilaty et al. [194] evidenced a feedback mechanism among these EMT inductive factors, regulated by microRNAs. Particularly, they reported that Snail1 represses Prrx1 whereas Prrx1 represses Snail1 via miR-15 regulation. Additionally, Nodal signaling in the left LPM downregulates Prrx1 and Snail1 via the upregulation of some microRNAs, such as let7, miR-34, miR-92, miR-124, miR-125, miR-135 and miR-184 [195]. It has been demonstrated that miR-34a and miR-92a modulate Prrx1 expression in a narrow time window (Figure 1B–D). If these miRNAs are deregulated Prrx1 expression is impaired, and consequently, a mesocardia is observed in embryos due to a loss of L/R asymmetry [195].
Regulation of Pitx2 left-right transcription factor by lncRNAs has been reported in two distinct biological contexts. Within early developmental stages, it has been evidenced that the lncRNA Pitx2 locus asymmetric regulated RNA (Playrr) and Pitx2 displayed contralateral expression, in other words, while Pitx2 has a left side pattern expression, Playrr is expressed in the right side of the embryo. Furthermore, there is a mutual antagonism between them that is regulated by chromosome structure. As Pitx2 expression needs to be dynamic during development, Pitx2 and Playrr interactions are tightly regulated to provide a proper Pitx2 dose for gut development [196] (Figure 1B–D). However, no evidence is reported that such interaction also applies to cardiovascular development. In humans, lncRNA Pitx2 adjacent noncoding RNA (PANCR) and Pitx2 are co-expressed during cardiomyocyte differentiation and within human left atrium. However, functional role remains unclear, particularly in atrial fibrillation, a Pitx2-dependent arrhythmogenic defect [197] (Figure 1B–D).
8. Post-Transcriptional Control of Proepicardium/Epicardium Formation by ncRNAs
The proepicardium is a transitory structure that contributes to the developing heart by extruding cells into the external surface of the embryonic myocardium, lining it and subsequently experimenting an epithelial-to-mesenchymal transition that promotes these cells to contribute to distinct cardiovascular cell types, particular to the coronary vasculature and the cardiac fibroskeleton [198,199,200,201].
Tgf-beta signaling plays multiple roles during cardiogenesis, including epicardial development [202,203,204,205,206,207,208,209,210]. The role of microRNAs regulating epicardial development has been reported by Brønnum et al. [211], demonstrating enhanced miR-21 expression. In addition, Pontemezzo et al. [212] have shown selective miR-200c downregulation after Tfg-beta administration, influencing therefore the epithelial-to-mesenchymal transition of epicardial cells.
At transcriptional level, three distinct transcription factors have been reported to play essential roles in proepicardium and epicardium development, namely, Tcf21, Tbx18 and Wt1. Tcf21 is a transcription factor of the “helix-loop-helix” (HLH) family that is expressed in different tissues derived from the mesoderm, including therein the epicardium [213,214]. In the epicardium, one of the main functions of Tcf1 is to regulate the EMT and the subsequent differentiation and specification of cells derived from the epicardium (EPDC) towards different cell types [214]. The expression of Tcf21 in this tissue is regulated by various miRNAs, such as let-7 [215,216] or miR-224, which binds directly to the Tcf21 3′UTR [217] (Figure 2A,D). While the impact of Tcf21 regulation by microRNAs has not been reported to date during embryonic development, it is important to highlight that several examples of Tcf21-microRNA modulation has been reported in cardiac physiopathological conditions. In Caucasian and East Asian populations, an SNP (rs12190287) in the 3′UTR region of Tcf21 prevents miR-224 binding, resulting in impaired Tcf21 expression, a condition related to the development of alterations in the ventricular septum of the heart [218]. A deregulation of Tcf21 produced at this SNP has also been associated with the risk of coronary heart disease [219,220,221]. The lack of miR-224 binding generates inadequate cell differentiation because of alterations in activated proteins 1 (AP-1) and platelet-derived growth factor (PDGF) [222]. Furthermore, Tcf21 deregulation causes greater fibroblast development, since this factor acts as an antagonist in the MYOCD-SRF signaling pathway [223]. Another miRNA that regulates Tcf21 is miR-146a. In an Iranian population study, another SNP (rs2910164) is produced in the sequence of the precursor of miR-146a, altering the structure and expression of mature miRNA [224].
Tbx18 is involved in the development of the venous pole, the epicardial development and the development of cardiomyocytes of the interventricular septum [51]. Several miRNAs that regulate the expression of this transcription factor have been described, such as miR-1 and miR-185 in chicken [159] or miR-429 in humans [191]. miR-429 [225] and miR-370-3p [226] regulate the expression of Tbx18 during the development of the sinus node and thus the cardiac pacemaker action potential (Figure 2A,D).
Although it has been widely described that Wt1 is a key transcription factor for epicardial development ([227,228,229,230], non-coding RNAs that regulate the expression of Wt1 during cardiogenesis have not yet been identified.
At present, evidence of the regulation proepicardium/epicardium formation transcription factors by lncRNAs has only been reported for Tbx18, as previously mentioned by Moshe [136], while evidence for Tcf21 is lacking and for Wt1 has only been reported in other biological contexts [231,232,233] (Figure 2A,D).
9. Post-Transcriptional Control of Conduction System Development by ncRNAs
The cardiac conduction system constitutes the electrical wiring of the heart and it is originated from myocardial precursors sleeves located at the junction of the cardiac chambers [234]. Formation of the cardiac conduction system is highly dynamic during development and several transcription factors such as Tbx2, Tbx3, Irx3 and Irx5 have been reported to play fundamental roles during its configuration as detailed below.
Tbx3 is a cardiac transcription factor involved in the development of the sinoatrial node (SAN) and atrioventricular canal (AVC) [235,236]. Several miRNAs that regulate the expression of Tbx3 have been described, such as miR-17-92 and miR-106b-25, which are also regulated by Pitx2c on the left side. Pitx2c acts by positively regulating miR-17-92 and miR-106b-25 on the left side thus suppressing the development of the sinoatrial node, as both miRNAs negatively regulate Tbx3 expression. However, on the right side, there is no expression of Pitx2c, allowing the activation of the Shox2 signaling pathway and subsequently of Tbx3 [237]. Also important is the role played by miR-1 on Tbx3, controlling the differentiation of sinoatrial precursors (Figure 2B,D). In addition to confirming this by computer analysis [238], it was also experimentally demonstrated that the overexpression of miR-1 in mESC-derived cells produces a decrease in the expression of Tbx3, thus reducing the development of SAN [239]. Tbx3 is also involved in the pluripotency of embryonic stem cells [240]. miR-137 acts as a direct target of Tbx3 repressing its expression, since an overexpression of this miRNA produces a significant decrease in cell proliferation [241] (Figure 2B,D). Tbx3 is able to modulate other factors that also intervene in the regulation of differentiation, such as Oct4, Sox2 or Nanog [242,243]. Such regulation is mediated by phosphatidylinositol-3-OH kinase-Akt and mitogen-activated protein kinase pathways, preferentially stimulating Nanog [244]. Therefore, although miR-137 does not have a direct target on Nanog, its expression can be regulated by the action of Tbx3 [241]. For Tbx2 and Irx3, non-coding RNAs that control the expression of Tbx2 and/or Irx3 during cardiogenesis have not been described at the moment, while scarce evidence is reported for Irx5 [245] in cardiac pathological conditions.
In line with the evidence reported during proepicardium formation, scarce evidence is currently available for the functional role of lncRNA regulating transcription factors with key relevant roles during conduction system formation. TTN-AS1 is enriched during cardiac development and its expression is regulated by TBX2, suggesting a plausible of TTN-AS1 during cardiac conduction development [246] (Figure 2B,D).
10. Post-Transcriptional Control of Chamber Morphogenesis and Valve Development by ncRNAs
Soon after looping, distinct atrial and ventricular cardiac chambers are configured. Within the atrial chambers pectinated muscles are formed while within the ventricular chamber, the embryonic myocardium is initially configured into a trabecular meshwork that subsequently, as development proceeds is replaced by the progressive formation of a compact myocardial layer.
Several growth factors are implicated in different phases of cardiac chamber morphogenesis, in particular, Notch and Neuregulin play a fundamental role in establishing the trabeculated myocardium while Hippo signaling is essential for organ size.
Notch signaling is widely implicated in cardiogenesis, particularly establishing a cross-talk communication between myocardium and endocardium during chamber formation [247,248,249]. Despite the prominent role of Notch signaling during cardiogenesis, no evidence of microRNAs regulation has been reported directly targeting Notch1 in this context.
Neuregulin signal is essential for the correct ventricular chamber trabeculation [250,251,252,253]. To date, there is no evidence of microRNAs regulating neuregulin in the cardiovascular context, yet Sun et al. [254] identified several microRNAs that are modulated by neuregulin administration. Neuregulin administration in murine embryonic stem cells increased miR-296-3p and miR-200c while decreased miR-465b-5p expression. Importantly, manipulation of these microRNAs, i.e., inhibition of miR-296-3p or miR-200c decreased and inhibition of miR-465-5p, respectively, promoted differentiation of embryonic stem cells into the cardiac lineage (Figure 3A,C).
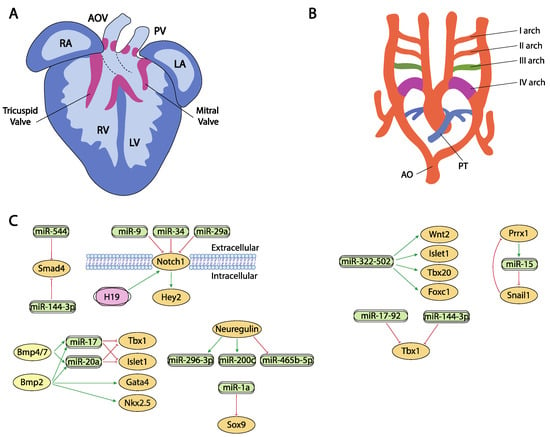
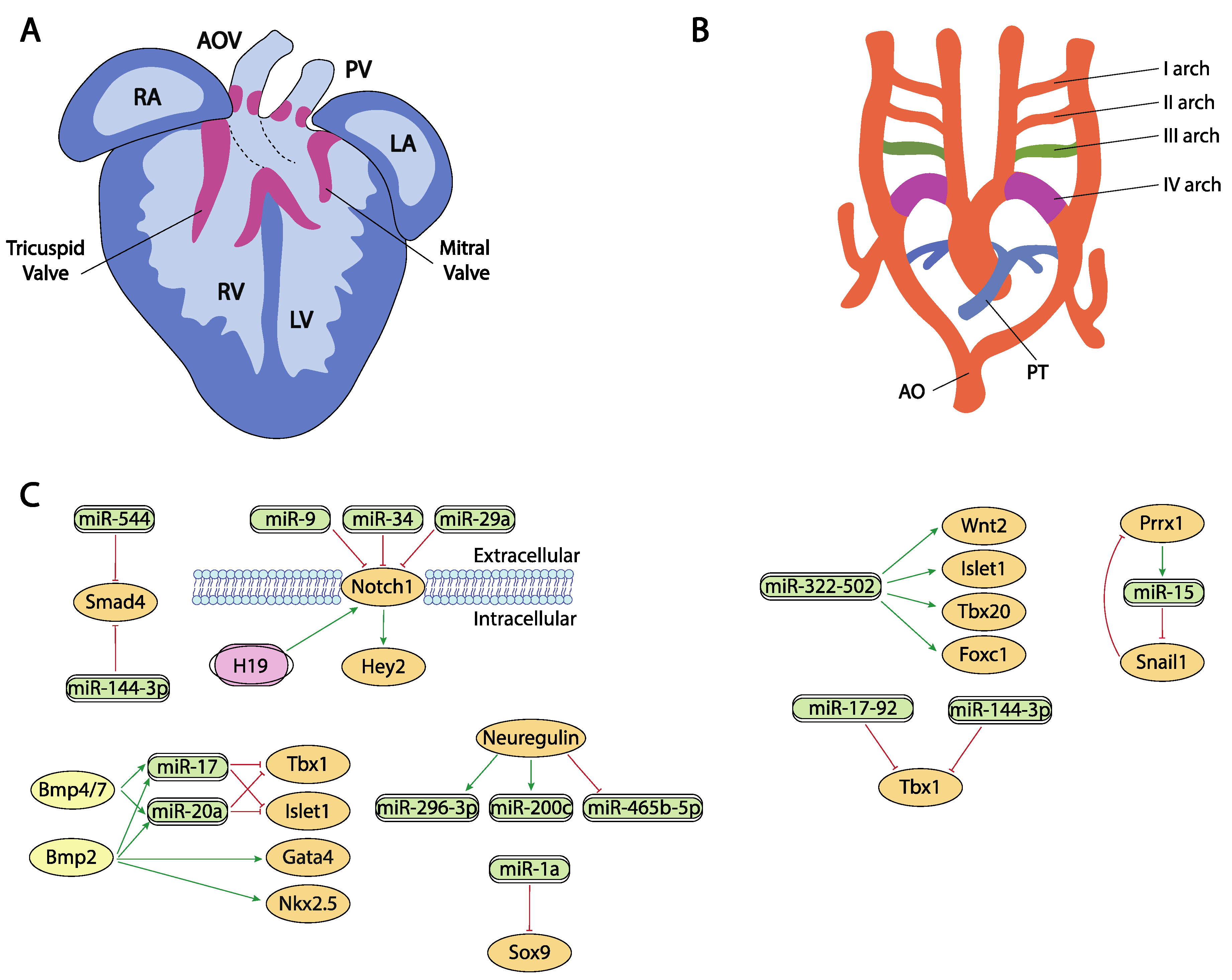
Figure 3.
Schematic representation of the cardiac chamber and valvular morphogenesis (A) and the configuration and remodeling of the aortic arches (B). (C) represent the current molecular interaction between microRNAs and lncRNAs with distinct growth factors and transcription factors involved in these developmental processes. RA, right atrium, LA, left atrium, RV, right ventricle, LV, left ventricle, AO, aorta, PT, pulmonary trunk.
Hippo signaling is primarily involved during cardiac development by controlling cardiomyocyte proliferation and thus heart size [255,256] and also plays pivotal roles in cardiac regeneration [20,257] and diseases [258,259]. It has been reported that the miRNA cluster miR302–367 negatively regulates the Hippo pathway in vivo, by directly targeting three key components of this pathway, i.e., Last2, Mst1 and Mob1b thus promoting cardiomyocyte proliferation and dedifferentiation. More recently Xie et al. [259] demonstrated that miR-10b also regulated human embryonic stem cell-derived cardiomyocyte proliferation via directly interacting with Last1, which is a major component of the Hippo pathway (Figure 2C,D).
Several transcription factors have been implicated in different phases of cardiac chamber morphogenesis, being particularly important for atrial vs. ventricular chamber development the transcription factors Hey1, Hey2, Irx4, Coup-tfII and Tbx20, while Hand1 and Hand2 are of importance to systemic vs. pulmonary development of the ventricular chambers and Foxm1, Hop, Kfl13 and Srf for the development of compact vs. trabecular myocardium.
Indirect evidence has been reported on the plausible role of miR-34 regulating distinct components of the Notch signaling pathway, including therein Hey2 [260], as well as for miR-128 regulating Irx4 [118], however no direct interactions between microRNAs and Hey1, Hey2, Irx4 or Coup-tfII in the cardiovascular context have been reported to date (Figure 2C,D). Importantly, several reports in other biological contexts have been widely reported [261,262,263,264,265,266,267].
Another key factor for cardiovascular development is Tbx20, which is involved in processes such as the differentiation of cardiac chambers and the proliferation and differentiation of cardiomyocytes [268]. Despite the importance of this factor during cardiogenesis, only indirect evidence for miR-1, miR-25, miR-141, miR-185 and miR-200a has been described distinctly modulating Tbx20 expression during cardiogenesis [159] (Figure 2C,D).
On the other hand, several reports are available about the functional role of distinct microRNAs modulating the expression of the Hand1 and Hand2 transcription factors. It has been reported that miR-363 downregulates Hand1, suggesting that miR-363 contributes to the generation of a functional left ventricle [269,270], and also Hand1 is modulated by let7 microRNA family members (mmu-let-7a/7d/7e/7f) at different stages in mouse heart development [271]. On the other hand, miR-1 and miR-133a directly target Hand2, balancing proliferation and differentiation during cardiogenesis [272,273,274] (Figure 2C,D).
Klf13 is a transcriptional factor important during cardiogenesis that physically interact with Gata4 [275]. At present two distinct microRNAs have been shown to directly interact with Klf13, miR-147b [276] and miR-125b [277] (Figure 2C,D).
Serum response factor (SRF) is a member of the superfamily MADS-box (MCM1, agamous, deficient, and SRF) of transcriptional factors and is highly expressed through embryonic, fetal and postnatal development of the heart. Srf regulates multiple target genes by the presence in their promoter of serum response elements, such as genes involved in metabolism, extracellular matrix deposition, gene transcription and protein translation [278]. Several microRNAs regulate Srf expression in distinct cardiovascular contexts, such as miR-483-3p in endothelial progenitor cells [279], miR-22 in human umbilical vein endothelial cells [280], miR-23, miR-93, miR-486 in cardiomyocytes [281] and miR-181 in vascular smooth muscle cells [282]. On the other hand, no evidence is reported on the ncRNA regulation of Foxm1 and Hop, despite their crucial role during cardiogenesis [283,284].
In contrast to our understanding of the impact of lncRNA during proepicardium and conduction system development, evidence of the role of lncRNA during chamber formation and valve development is widely emerging. Evidence on the modulation of Bmp2 has been reported in cardiac valves, mediated by H19 lncRNA. H19 administration silenced Notch1 expression by modulating the recruitment of p53 to its promoter and consequently, Bmp2 and Runx2 are downregulated, as they are downstream targets of Notch1 signalling [285] (Figure 3A,C). In addition, Hand2 transcription is modulated by Upperhand (Uph) [286], and this lncRNA is essential for proper cardiac development, as systemic deletion caused hypoplasia of the right ventricle and septal defects [287] (Figure 2C,D).
Furthermore, Cardinal, a myocardin adjacent lncRNA, regulates SRF-dependent mitogenic gene transcription as reported by Anderson et al. [288], while MYOSLID, an SRF-dependent lncRNA, is a direct transcriptional target of SRF and play essential roles regulating role differentiation while blocking proliferation in vascular smooth muscle cells. Curiously decreased MYOSLID expression does not affect SRF expression [289] (Figure 2C,D).
Liu et al. [290] described a regulatory network where HBL1 lncRNA acts as a functional repressor of miR-1, thus inhibiting Tbx20 expression and cardiomyocyte differentiation. Smad4 is also regulated by lncRNAs. Xiao et al. [291] demonstrated that this transcriptional factor is downregulated by miR-204 which can be regulated by Malat1 lncRNA acting thus as a miR-204 sponge (Figure 2C,D). Additionally, Klf2 is also regulated by lncRNAs but in other biological contexts, such as modulating monocyte adhesion to endothelial cells by MANTIS lncRNA [292] or acute myocardial infarction by MALAT1 [293] (Figure 2C,D).
11. Post-Transcriptional Control of Outflow Tract and Atrioventricular Septation by ncRNAs
Septation of the heart involves the formation of distinct muscular septa, such as the primary and secondary atrial septa in the atrial chambers and the interventricular septum in the ventricular chambers, as well as remodeling of the sinoatrial, atrioventricular and conotruncal endocardial cushions eventually leading to the formation of a four-chambered heart. Within these developmental processes, Bmp, Wnt, Tgf-beta and Notch signaling play essential roles.
Besides their role in early cardiac specification, Bmp2 and Bmp4 also play an essential role in the differentiation of the outflow tract myocardium derived from the secondary cardiac field [18,19,294,295]. In this context, it has been reported that Bmp2 and Bmp4 induced the expression of two microRNAs encoded by the miR17-92 cluster, miR-17 and miR-20a [296]. Seed sequences of both microRNAs recognized the 3′UTRs from two cardiac progenitor genes, Islet1 and Tbx1 [297,298,299]. The absence of Bmp2/Bmp4 expression results in a down-regulation of miR-17 and miR-20a and thus an increase in Islet1 and Tbx1 expression (Figure 3A,C). Gain and/or loss of function assay and dual-luciferase assays have shown that both microRNAs repress the expression of Islet1 and Tbx1. The down-regulation of miR-17 and miR-20a leads to sustained expression of both transcription factors in cardiac progenitors that prevent myocardial OFT differentiation. As a consequence of failure to silence Islet1 and Tbx1, the Bmp2/4 CKO mutants show a reduced and thickened OFT myocardium accompanied by a down-regulation of sarcomeric proteins such as Mhy6 or Mhy7 [296].
Bai et al. [294] demonstrated a pivotal role of Bmp4/7-miR 17-92 cluster in the development of OFT cushions. Bmp4 mutant mice have shown that it is necessary for OFT septation and cushion development [300] (Figure 3A,C). Interestingly, Bmp4 deficiency increases Bmp7 expression, suggesting a possible compensation mechanism and therefore the involvement in the same morphogenetic process. However, Bmp7 deletion does not result in a visible phenotype, demonstrating that its possible role is residual compared to Bmp4 [300]. Functional assays demonstrated that both induce miR-17-92 expression, which is necessary to correct outflow tract cushion development by inducing epithelial to mesenchymal transition (EMT) [294]. The miR-17-92 cluster triggers Vgfa2 mRNA, which represses EMT of the atrioventricular canal (AVC) [301]. Interestingly, SHF Bmp4/Bmp7 deficiency and the miR-17-92 cluster show similar phenotypes, resulting in a defect of EMT and a reduction in cardiac neural crest entry, with the consequent patent trunk arteriosus showing a function linked in the development of the outflow tract [300].
Functional role of Wnt signaling governing valve formation is extensively documented [302,303,304,305,306,307,308]. However, no evidence has been reported on the role of microRNAs regulating Wnt signaling during cardiac valve development yet. There are compiling evidence in cardiac valve pathological conditions, such as valvular calcification by miR-29b [309] and inflammation by miR-27a [310], as well as in other cardiac pathological conditions such as atrial fibrillation [311] regulating Wnt3 by miR-27b and ischemia/reperfusion regulating Wnt/β-catenin signaling pathway by miR-148b [10].
Tgf-beta signaling also plays a pivotal role in endocardial cushion formation [312,313,314,315,316,317,318,319], where indirect evidence for miR-23 and miR-199a was reported by Lagendijk et al. [320] and Bonet et al. [321] during cardiac valve formation, respectively (Figure 2A,D).
Notch signaling is widely implicated during valve formation [322,323,324,325,326,327]. Despite the prominent role of Notch signaling during cardiogenesis, only miR-34 has been reported as a direct modulator of Notch1 in embryonic endocardial cells [261] while additional evidence on the role of microRNAs regulating Notch signaling in aortic valve calcification has been reported for miR-34 and miR-29a [328,329,330] (Figure 3A,C).
Our current understanding of the transcriptional regulation of the cardiac muscular septa is still incipient, whereas several transcription factors such as Klf2, Sox9, Smad4 and Odd1 have been involved in endocardial cushion septation. Kuppel-like factor 2 (Klf2) is a very important molecule involved in angiogenesis and endothelial vascular formation and proliferation [331,332]. To date most of the evidence provided on the regulation of Klf2 by microRNAs are reported in pathological conditions, miR-92 and miR-32 regulate Klf2 expression in endothelial cells in acute myocardial infarction [331,333]. Similar evidence on the transcriptional control of Klf2 regulating microRNA expression has been reported in distinct biological settings, such as shear stress-inducing expression of miR-30 family members [334], miR-23b, miR-145 and miR-143 [332,335] while reducing the expression of miR-126-5p [332]. More recently, Sindi et al. [335] have shown that Klf2 has the ability to induce the presence of miR-181-5p and miR-324-5p in exosomes, reducing the vascular remodeling in pulmonary hypertension.
Sex-determining region Y box (Sox9) is a very important regulator of chondrocyte differentiation [336,337]. In cardiovascular development, it plays a crucial role regulating endocardial cushion formation and septation. In this context, Li et al. [337] demonstrated the existence of a regulation network between Sox9 and miR-1a as this miRNA downregulates Sox9 expression, correlating those findings with the onset of ventricular septal defects (Figure 3A,C). Additionally, in the context of cardiac fibrosis, Cui et al. [336] established a correlation between miR-145 and Sox9 and verified those finding by dual luciferase assay while Cheng et al. [338] reported a direct regulation of Sox9 by miR-30e in an experimental model of myocardial ischemia-reperfusion injury.
Smad4 plays multiple roles in distinct developmental and adult tissues and thus is widely expressed. As part of the Tgf-beta signaling, its role during endocardial cushion formation has been widely documented [339,340]. Regulation of Smad4 by microRNAs have been documented in distinct cardiovascular settings [339,340], such as regulation by miR-34 and miR-122 in cardiac fibrosis [340], by miR-146a-5p in exosomes of human cardiac-resident mesenchymal progenitor cells (CPC) [341], by miR-26a in hypertension-induced myocardial fibrosis [342], yet the only evidence on the regulatory role of microRNAs in cardiovascular development has been established by Dong et al. [343]. These authors observed Smad4 downregulation and miR-144-3p and miR-544 up-regulation correlating with the presence of several cardiovascular defects (Figure 3A,C).
12. Post-Transcriptional Control of Aortic Arch Development by ncRNAs
Finally, a critical step in cardiac development requires the exquisite remodeling of the arterial and venous connections to provide appropriate systemic and venous connections and thus the establishment of a closed and double circulatory system. In this context, several transcription factors such as Tbx1, Foxc1, Foxc2, Prrx1, Prrx2 have been involved.
The Tbx1 gene is a member of the T-box transcription factor family which participates in the arrangement of pharyngeal arch endoderm, differentiation and migration of neural crest cells migration and cardiac outflow tract development [344]. During myocardial differentiation of cardiac progenitors in the secondary heart field, Tbx1 is directly downregulated by miR-17-92 cluster under the control of Bmp signaling [149]. Although Tbx1 is currently described as one of the core genes implicated in congenital heart disease (CHD), the molecular mechanisms underlying its role in the pathogenesis of this abnormality is not fully understood [345]. Recent studies, described by Cao et al. [346] showed that miR-144 inhibits cardiomyocyte proliferation and promotes cardiomyocyte apoptosis by targeting the 3′UTR of Tbx1 via regulating Jak2/Stat1 signaling pathway (Figure 3B,C).
The transcription factor Forkhead box C1 (Foxc1) is essential for cell proliferation, migration, invasion, as well as vascular formation and maturation [347,348]. Foxc1 is also implicated in the regulation of early cardiomyogenesis and the functional properties of ESC-derived cardiomyocytes [349]. Although its specific regulation remains largely unknown, some studies described non-coding RNAs as promising posttranscriptional regulators of this factor. In heart development, miR-322/-502 cluster up-regulated cardiac genes, such as Wnt2, Isl1, Tbx20 or Foxc1, involved in vascular endothelial cell differentiation [112] (Figure 3B,C). A negative correlation between Foxc1 and miR-511-3p expression levels has been discovered by Henn et al. [350] in a model for in vivo induction of neoangiogenesis.
Forkhead box C2 (Foxc2) participates in blood and lymphatic vessel development as well as regulating adipose cell metabolism [351]. This transcription factor is a predicted target of miR-199a-5p, a microRNA which has been implicated in cardiac remodeling regulation and ventricular hypertrophy [352]. In varicose vein tissues, the downregulation of this microRNA may promote vascular smooth muscle cells (VSMC) proliferation by upregulating Foxc2 [353].
The paired-related homeobox genes Prrx1 and Prrx2 are highly expressed in mesenchymal tissues throughout development, with the cardiovascular system being one of those with the highest transcript levels of Prrx genes reported. These transcriptional factors are implicated in matrix modulation and in the inhibition of adipogenesis activating TGF-β signaling pathway [354]. Prrx1, and other transcription factors such as Zeb or Snail, also plays an important role in the epithelial to mesenchymal transition (EMT) that occurs during embryonic heart development. In this process, Prrx1 attenuates Snail1 expression through direct activation of the miR-15 family promoting a decrease in stem cell properties [194] (Figure 3B,C).
At present, none of the main transcription factors currently involved in aortic arch development, such as Tbx1, Prrx1, Prrx2, Foxc1 and Foxc2 have been reported to be regulated by lncRNAs. However, there is evidence that the long-non-coding RNA (lncRNA) LINC00242 competitively binds to miR-141 and promotes Foxc1 expression accelerating the angiogenesis in gastric cancer [355] while in hepatocellular cancer, Foxc1 binds to the upstream region of HOTAIR, a lncRNA highly expressed in cardiac tissues that may epigenetically regulate embryonic heart development by recruiting PRC2 [356].
13. Conclusions and Future Perspectives
Cardiovascular development is a complex developmental process in which multiple cell types are involved [357]. Over the last decades, our understanding of the cellular contribution to the developing heart has been enormous, particularly dissecting the deployment of distinct heart fields to the nascent cardiac tube [358,359] as well as the extracardiac contributions emanating from the epicardium and the cardiac neural crest [227,360]. Similarly, our understanding of molecular cascades governing these morphogenetic events has been equally impressive, by dissecting the functional role of multiple growth factors and transcription factors during cardiogenesis [357]. Recently, a novel layer of complexity has emerged with the discovery of non-coding RNAs and their impact on the gene regulatory networks that governed cardiac development. In this review, we have summarized our current understanding of the functional role of both microRNAs and lncRNAs at different stages of heart development. It is important to highlight that although our insights on the regulatory roles of these non-coding RNAs is still incipient, there is convincing evidence of their impact on early aspects of heart formation, ranging from early precardiac mesoderm modulation [94] to cardiac chamber formation and aortic arch remodeling [345]. It is important also to highlight in this context that although we are currently lacking evidence of the functional role of microRNAs and lncRNAs in a large number of molecular determinants that play essential roles during cardiogenesis, there is ample evidence in other biological contexts. For example, regulation of microRNAs by Bmp4 has been reported in other biological settings, such as angiogenesis [361] as well as during cardiac valve calcification, i.e., miR-141, miR-486, miR-204, miR-30 [362,363,364,365]. Similarly, a large body of evidence have been reported on the microRNA modulation of Tgf-beta signaling in cardiac physiopathology, particularly on fibrosis [366,367,368,369,370,371,372,373] involving distinct microRNAs, such as miR-34a/miR-93 [366], miR-21 [367], miR-155 [368], miR-27 [369], miR-22 [370], miR-23 [371] and miR-30c [372].
Similar evidence is also applicable for lncRNAs. A recent study reported increased expression of SNHG6 lncRNA in ventricular septal defects patients while functionally SNHG6 lncRNA overexpression modulates miR-101 and Wnt/beta signaling, supporting a plausible role in congenital heart disease [373]. Zhao et al. [374] identified that NORAD directly binds to miR-590-3p in human umbilical vein endothelial cells and miR-590-3p directly target proangiogenic agents, such as Vegf, Fgf1 and Fgf2. On the other hand, Sun et al., [375], identified that TUG1 served as a sponge for miR-590 and Fgf1 is a direct target of miR-590. Importantly, TUG1 expression is increased in AMI tissues and cardiac fibroblasts treated with Tgf-β1. More recently, evidence of Wnt regulated lncRNA Walras was reported, demonstrating a role for this lncRNA in cardiomyocyte cytoarchitecture in the context of atrial fibrillation [376]. Overall, these data support the notion that in next coming years we will greatly enlarge our understanding of the impact of microRNAs and lncRNAs, their interactive networks on each and every aspect of cardiac development. Moreover, it is also expected that a novel class of non-coding RNA, i.e., circRNAs, that at present have been scarcely implicated in cardiac development [377], will also pop up.
Funding
This research was funded by Junta de Andalucia Research Council, grant-in aid number CTS-446 to D.F. and AA, FEDER-UJA grant (1259108) to D.F. and A.A. and by a grant from the Ministry of Science, Innovation and Universities of the Spanish Government (PID2019-107492GB-I00) to D.F. and A.A.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Van Wijk, B.; Moorman, A.F.M.; van den Hoff, M.J.B. Role of bone morphogenetic proteins in cardiac differentiation. Cardiovasc. Res. 2007, 74, 244–255. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.N.; Green, J.; Wang, Z.; Deng, Y.; Qiao, M.; Peabody, M.; Zhang, Q.; Ye, J.; Yan, Z.; Denduluri, S.; et al. Bone Morphogenetic Protein (BMP) signaling in development and human diseases. Genes Dis. 2014, 1, 87–105. [Google Scholar] [CrossRef] [PubMed]
- Khosravi, F.; Ahmadvand, N.; Bellusci, S.; Sauer, H. The Multifunctional Contribution of FGF Signaling to Cardiac Development, Homeostasis, Disease and Repair. Front. Cell Dev. Biol. 2021, 9, 672935. [Google Scholar] [CrossRef] [PubMed]
- Kruithof, B.P.; van Wijk, B.; Somi, S.; Julio, M.K.-D.; Pomares, J.M.P.; Weesie, F.; Wessels, A.; Moorman, A.F.; Hoff, M.J.V.D. BMP and FGF regulate the differentiation of multipotential pericardial mesoderm into the myocardial or epicardial lineage. Dev. Biol. 2006, 295, 507–522. [Google Scholar] [CrossRef] [PubMed]
- Van Wijk, B.; van den Berg, G.; Abu-Issa, R.; Barnett, P.; van der Velden, S.; Schmidt, M.; Ruijter, J.M.; Kirby, M.L.; Moorman, A.F.; Hoff, M.J.V.D. Epicardium and Myocardium Separate From a Common Precursor Pool by Crosstalk Between Bone Morphogenetic Protein– and Fibroblast Growth Factor–Signaling Pathways. Circ. Res. 2009, 105, 431–441. [Google Scholar] [CrossRef]
- Torlopp, A.; Schlueter, J.; Brand, T. Role of fibroblast growth factor signaling during proepicardium formation in the chick embryo. Dev. Dyn. 2010, 239, 2393–2403. [Google Scholar] [CrossRef]
- Ishii, Y.; Garriock, R.J.; Navetta, A.M.; Coughlin, L.E.; Mikawa, T. BMP Signals Promote Proepicardial Protrusion Necessary for Recruitment of Coronary Vessel and Epicardial Progenitors to the Heart. Dev. Cell 2010, 19, 307–316. [Google Scholar] [CrossRef]
- Andrés-Delgado, L.; Ernst, A.; Galardi-Castilla, M.; Bazaga, D.; Peralta, M.; Münch, J.; González-Rosa, J.M.; Marques, I.; Tessadori, F.; de la Pompa, J.L.; et al. Actin dynamics and the Bmp pathway drive apical extrusion of proepicardial cells. Development 2019, 146, dev174961. [Google Scholar] [CrossRef]
- Andrés-Delgado, L.; Galardi-Castilla, M.; Münch, J.; Peralta, M.; Ernst, A.; González-Rosa, J.M.; Tessadori, F.; Santamaría, L.; Bakkers, J.; Vermot, J.; et al. Notch and Bmp signaling pathways act coordinately during the formation of the proepicardium. Dev. Dyn. 2020, 249, 1455–1469. [Google Scholar] [CrossRef]
- Yamagishi, T.; Ando, K.; Nakamura, H. Roles of TGFβ and BMP during valvulo–septal endocardial cushion formation. Anat. Sci. Int. 2009, 84, 77–87. [Google Scholar] [CrossRef]
- Kruithof, B.P.; Duim, S.N.; Moerkamp, A.T.; Goumans, M.-J. TGFβ and BMP signaling in cardiac cushion formation: Lessons from mice and chicken. Differentiation 2012, 84, 89–102. [Google Scholar] [CrossRef]
- Saxon, J.G.; Baer, D.R.; Barton, J.A.; Hawkins, T.; Wu, B.; Trusk, T.C.; Harris, S.E.; Zhou, B.; Mishina, Y.; Sugi, Y. BMP2 expression in the endocardial lineage is required for AV endocardial cushion maturation and remodeling. Dev. Biol. 2017, 430, 113–128. [Google Scholar] [CrossRef]
- Inai, K.; Burnside, J.L.; Hoffman, S.; Toole, B.P.; Sugi, Y. BMP-2 Induces Versican and Hyaluronan That Contribute to Post-EMT AV Cushion Cell Migration. PLoS ONE 2013, 8, e77593. [Google Scholar] [CrossRef]
- Zhang, J.; Chang, J.Y.-F.; Huang, Y.; Lin, X.; Luo, Y.; Schwartz, R.J.; Martin, J.F.; Wang, F. The FGF-BMP Signaling Axis Regulates Outflow Tract Valve Primordium Formation by Promoting Cushion Neural Crest Cell Differentiation. Circ. Res. 2010, 107, 1209–1219. [Google Scholar] [CrossRef]
- Sugi, Y.; Ito, N.; Szebenyi, G.; Myers, K.; Fallon, J.F.; Mikawa, T.; Markwald, R.R. Fibroblast growth factor (FGF)-4 can induce proliferation of cardiac cushion mesenchymal cells during early valve leaflet formation. Dev. Biol. 2003, 258, 252–263. [Google Scholar] [CrossRef]
- Nakajima, Y.; Yamagishi, T.; Hokari, S.; Nakamura, H. Mechanisms involved in valvuloseptal endocardial cushion formation in early cardiogenesis: Roles of transforming growth factor (TGF)-beta and bone morphogenetic protein (BMP). Anat. Rec. 2000, 258, 119–127. [Google Scholar] [CrossRef]
- Todorovic, V.; Frendewey, D.; Gutstein, D.E.; Chen, Y.; Freyer, L.; Finnegan, E.; Liu, F.; Murphy, A.; Valenzuela, D.; Yancopoulos, G.; et al. Long form of latent TGF-β binding protein 1 (Ltbp1L) is essential for cardiac outflow tract septation and remodeling. Development 2007, 134, 3723–3732. [Google Scholar] [CrossRef]
- Zhang, R.; Cao, P.; Yang, Z.; Wang, Z.; Wu, J.-L.; Chen, Y.; Pan, Y. Heparan Sulfate Biosynthesis Enzyme, Ext1, Contributes to Outflow Tract Development of Mouse Heart via Modulation of FGF Signaling. PLoS ONE 2015, 10, e0136518. [Google Scholar] [CrossRef]
- Beppu, H.; Malhotra, R.; Beppu, Y.; Lepore, J.J.; Parmacek, M.S.; Bloch, K.D. BMP type II receptor regulates positioning of outflow tract and remodeling of atrioventricular cushion during cardiogenesis. Dev. Biol. 2009, 331, 167–175. [Google Scholar] [CrossRef]
- Wang, J.; Liu, S.; Heallen, T.; Martin, J.F. The Hippo pathway in the heart: Pivotal roles in development, disease, and regeneration. Nat. Rev. Cardiol. 2018, 15, 672–684. [Google Scholar] [CrossRef]
- Heallen, T.; Zhang, M.; Wang, J.; Bonilla-Claudio, M.; Klysik, E.; Johnson, R.L.; Martin, J.F. Hippo Pathway Inhibits Wnt Signaling to Restrain Cardiomyocyte Proliferation and Heart Size. Science 2011, 332, 458–461. [Google Scholar] [CrossRef]
- Zhou, Q.; Li, L.; Zhao, B.; Guan, K.-L. The Hippo Pathway in Heart Development, Regeneration, and Diseases. Circ. Res. 2015, 116, 1431–1447. [Google Scholar] [CrossRef]
- D’Amato, G.; Luxán, G.; Mínguez, J.L.D.L.P. Notch signalling in ventricular chamber development & cardiomyopathy. FEBS J. 2016, 283, 4223–4237. [Google Scholar] [CrossRef]
- D’Amato, G.; Luxán, G.; Nieto, G.D.M.; Poveda, B.M.; Torroja, C.; Walter, W.; Bochter, M.S.; Benedito, R.; Cole, S.E.; Martinez, F.; et al. Sequential Notch activation regulates ventricular chamber development. Nat. Cell Biol. 2015, 18, 7–20. [Google Scholar] [CrossRef]
- Samsa, L.; Givens, C.; Tzima, E.; Stainier, D.; Qian, L.; Liu, J. Cardiac contraction activates endocardial Notch signaling to modulate chamber maturation in zebrafish. Development 2015, 142, 4080–4091. [Google Scholar] [CrossRef]
- Yang, Y.; Li, B.; Zhang, X.; Zhao, Q.; Lou, X. The zinc finger protein Zfpm1 modulates ventricular trabeculation through Neuregulin-ErbB signalling. Dev. Biol. 2019, 446, 142–150. [Google Scholar] [CrossRef]
- Zhao, Y.-Y.; Sawyer, D.R.; Baliga, R.R.; Opel, D.J.; Han, X.; Marchionni, M.A.; Kelly, R.A. Neuregulins Promote Survival and Growth of Cardiac Myocytes. J. Biol. Chem. 1998, 273, 10261–10269. [Google Scholar] [CrossRef]
- Hertig, C.M.; Kubalak, S.W.; Wang, Y.; Chien, K.R. Synergistic Roles of Neuregulin-1 and Insulin-like Growth Factor-I in Activation of the Phosphatidylinositol 3-Kinase Pathway and Cardiac Chamber Morphogenesis. J. Biol. Chem. 1999, 274, 37362–37369. [Google Scholar] [CrossRef] [PubMed]
- Lai, D.; Liu, X.; Forrai, A.; Wolstein, O.; Michalicek, J.; Ahmed, I.; Garratt, A.N.; Birchmeier, C.; Zhou, M.; Hartley, L.; et al. Neuregulin 1 Sustains the Gene Regulatory Network in Both Trabecular and Nontrabecular Myocardium. Circ. Res. 2010, 107, 715–727. [Google Scholar] [CrossRef] [PubMed]
- Saga, Y.; Miyagawa-Tomita, S.; Takagi, A.; Kitajima, S.; Miyazaki, J.-I.; Inoue, T. MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development 1999, 126, 3437–3447. [Google Scholar] [CrossRef]
- Saga, Y. Mesp1 Expression Is the Earliest Sign of Cardiovascular Development. Trends Cardiovasc. Med. 2000, 10, 345–352. [Google Scholar] [CrossRef]
- Kitajima, S.; Takagi, A.; Inoue, T.; Saga, Y. MesP1 and MesP2 are essential for the development of cardiac mesoderm. Development 2000, 127, 3215–3226. [Google Scholar] [CrossRef] [PubMed]
- Laverriere, A.C.; MacNeill, C.; Mueller, C.; Poelmann, R.E.; Burch, J.B.; Evans, T. GATA-4/5/6, a subfamily of three transcription factors transcribed in developing heart and gut. J. Biol. Chem. 1994, 269, 23177–23184. [Google Scholar] [CrossRef]
- Charron, F.; Nemer, M. GATA transcription factors and cardiac development. Semin. Cell Dev. Biol. 1999, 10, 85–91. [Google Scholar] [CrossRef]
- Belaguli, N.S.; Sepulveda, J.L.; Nigam, V.; Charron, F.; Nemer, M.; Schwartz, R.J. Cardiac Tissue Enriched Factors Serum Response Factor and GATA-4 Are Mutual Coregulators. Mol. Cell. Biol. 2000, 20, 7550–7558. [Google Scholar] [CrossRef]
- Durocher, D.; Charron, F.; Warren, R.; Schwartz, R.J.; Nemer, M. The cardiac transcription factors Nkx2-5 and GATA-4 are mutual cofactors. EMBO J. 1997, 16, 5687–5696. [Google Scholar] [CrossRef]
- Jiang, Y.; Evans, T. TheXenopusGATA-4/5/6 Genes Are Associated with Cardiac Specification and Can Regulate Cardiac-Specific Transcription during Embryogenesis. Dev. Biol. 1996, 174, 258–270. [Google Scholar] [CrossRef][Green Version]
- Harvey, R. NK-2Homeobox Genes and Heart Development. Dev. Biol. 1996, 178, 203–216. [Google Scholar] [CrossRef]
- Biben, C.; Harvey, R.P. Homeodomain factor Nkx2-5 controls left/right asymmetric expression of bHLH gene eHand during murine heart development. Genes Dev. 1997, 11, 1357–1369. [Google Scholar] [CrossRef]
- Lin, Q.; Schwarz, J.; Bucana, C.; Olson, E.N. Control of Mouse Cardiac Morphogenesis and Myogenesis by Transcription Factor MEF2C. Science 1997, 276, 1404–1407. [Google Scholar] [CrossRef]
- Croissant, J.D.; Kim, J.-H.; Eichele, G.; Goering, L.; Lough, J.; Prywes, R.; Schwartz, R.J. Avian Serum Response Factor Expression Restricted Primarily to Muscle Cell Lineages Is Required for α-Actin Gene Transcription. Dev. Biol. 1996, 177, 250–264. [Google Scholar] [CrossRef]
- Kim, S.; Ip, H.S.; Lu, M.M.; Clendenin, C.; Parmacek, M.S. A serum response factor-dependent transcriptional regulatory program identifies distinct smooth muscle cell sublineages. Mol. Cell. Biol. 1997, 17, 2266–2278. [Google Scholar] [CrossRef]
- Ocaña, O.H.; Coskun, H.; Minguillón, C.; Murawala, P.; Tanaka, E.M.; Galceran, J.; Muñoz-Chápuli, R.; Nieto, M.A. A right-handed signalling pathway drives heart looping in vertebrates. Nature 2017, 549, 86–90. [Google Scholar] [CrossRef]
- Ammirabile, G.; Tessari, A.; Pignataro, V.; Szumska, D.; Sardo, F.S.; Benes, J.; Balistreri, M.; Bhattacharya, S.; Sedmera, D.; Campione, M. Pitx2 confers left morphological, molecular, and functional identity to the sinus venosus myocardium. Cardiovasc. Res. 2011, 93, 291–301. [Google Scholar] [CrossRef]
- Campione, M.; Ros, M.; Icardo, J.M.; Piedra, E.; Christoffels, V.M.; Schweickert, A.; Blum, M.; Franco, D.; Moorman, A.F. Pitx2 Expression Defines a Left Cardiac Lineage of Cells: Evidence for Atrial and Ventricular Molecular Isomerism in the iv/iv Mice. Dev. Biol. 2001, 231, 252–264. [Google Scholar] [CrossRef]
- Campione, M.; Acosta, L.; Martinez, S.; Icardo, J.; Aranega, A.; Franco, D. Pitx2 and Cardiac Development: A Molecular Link between Left/Right Signaling and Congenital Heart Disease. Cold Spring Harb. Symp. Quant. Biol. 2002, 67, 89–96. [Google Scholar] [CrossRef]
- Franco, D.; Campione, M.; Kelly, R.; Zammit, P.S.; Buckingham, M.; Lamers, W.H.; Moorman, A.F.M. Multiple transcriptional domains, with distinct left and right components, in the atrial chambers of the developing heart. Circ. Res. 2000, 87, 984–991. [Google Scholar] [CrossRef]
- Campione, M.; Franco, D. Current Perspectives in Cardiac Laterality. J. Cardiovasc. Dev. Dis. 2016, 3, 34. [Google Scholar] [CrossRef]
- Xu, H.; Morishima, M.; Wylie, J.N.; Schwartz, R.J.; Bruneau, B.G.; Lindsay, E.A.; Baldini, A. Tbx1has a dual role in the morphogenesis of the cardiac outflow tract. Development 2004, 131, 3217–3227. [Google Scholar] [CrossRef]
- Zhang, Z.; Huynh, T.; Baldini, A. Mesodermal expression ofTbx1is necessary and sufficient for pharyngeal arch and cardiac outflow tract development. Development 2006, 133, 3587–3595. [Google Scholar] [CrossRef]
- Greulich, F.; Rudat, C.; Kispert, A. Mechanisms of T-box gene function in the developing heart. Cardiovasc. Res. 2011, 91, 212–222. [Google Scholar] [CrossRef] [PubMed]
- Aanhaanen, W.T.; Brons, J.F.; Domínguez, J.N.; Rana, M.S.; Norden, J.; Airik, R.; Wakker, V.; Vries, C.D.G.-D.; Brown, N.A.; Kispert, A.; et al. The Tbx2 + Primary Myocardium of the Atrioventricular Canal Forms the Atrioventricular Node and the Base of the Left Ventricle. Circ. Res. 2009, 104, 1267–1274. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, J.K.; Ohgi, M.; Koshiba-Takeuchi, K.; Shiratori, H.; Sakaki, I.; Ogura, K.; Saijoh, Y.; Ogura, T. Tbx5specifies the left/right ventricles and ventricular septum position during cardiogenesis. Development 2003, 130, 5953–5964. [Google Scholar] [CrossRef] [PubMed]
- Moskowitz, I.P.G.; Pizard, A.; Patel, V.V.; Bruneau, B.; Kim, J.B.; Kupershmidt, S.; Roden, D.; Berul, C.I.; Seidman, C.E.; Seidman, J.G. The T-Box transcription factor Tbx5 is required for the patterning and maturation of the murine cardiac conduction system. Development 2004, 131, 4107–4116. [Google Scholar] [CrossRef]
- Bruneau, B.G.; Logan, M.; Davis, N.; Levi, T.; Tabin, C.J.; Seidman, J.G.; Seidman, C.E. Chamber-Specific Cardiac Expression of Tbx5 and Heart Defects in Holt–Oram Syndrome. Dev. Biol. 1999, 211, 100–108. [Google Scholar] [CrossRef]
- Bruneau, B.; Nemer, G.; Schmitt, J.P.; Charron, F.; Robitaille, L.; Caron, S.; Conner, D.A.; Gessler, M.; Nemer, M.; Seidman, C.E.; et al. A Murine Model of Holt-Oram Syndrome Defines Roles of the T-Box Transcription Factor Tbx5 in Cardiogenesis and Disease. Cell 2001, 106, 709–721. [Google Scholar] [CrossRef]
- Greulich, F.; Farin, H.F.; Schuster-Gossler, K.; Kispert, A. Tbx18 function in epicardial development. Cardiovasc. Res. 2012, 96, 476–483. [Google Scholar] [CrossRef]
- Wu, S.-P.; Dong, X.-R.; Regan, J.N.; Su, C.; Majesky, M.W. Tbx18 regulates development of the epicardium and coronary vessels. Dev. Biol. 2013, 383, 307–320. [Google Scholar] [CrossRef]
- Firulli, A.B.; McFadden, D.G.; Lin, Q.; Srivastava, D.; Olson, E.N. Heart and extra-embryonic mesodermal defects in mouse embryos lacking the bHLH transcription factor Hand1. Nat. Genet. 1998, 18, 266–270. [Google Scholar] [CrossRef]
- Riley, P.; Anaon-Cartwight, L.; Cross, J.C. The Hand1 bHLH transcription factor is essential for placentation and cardiac morphogenesis. Nat. Genet. 1998, 18, 271–275. [Google Scholar] [CrossRef]
- Srivastava, D.; Cserjesi, P.; Olson, E.N. A Subclass of bHLH Proteins Required for Cardiac Morphogenesis. Science 1995, 270, 1995–1999. [Google Scholar] [CrossRef]
- Thomas, T.; Yamagishi, H.; Overbeek, P.A.; Olson, E.N.; Srivastava, D. The bHLH Factors, dHAND and eHAND, Specify Pulmonary and Systemic Cardiac Ventricles Independent of Left–Right Sidedness. Dev. Biol. 1998, 196, 228–236. [Google Scholar] [CrossRef]
- Pereira, F.; Qiu, Y.; Zhou, G.; Tsai, M.-J.; Tsai, S.Y. The orphan nuclear receptor COUP-TFII is required for angiogenesis and heart development. Genes Dev. 1999, 13, 1037–1049. [Google Scholar] [CrossRef]
- Fischer, A.; Schumacher, N.; Maier, M.; Sendtner, M.; Gessler, M. The Notch target genes Hey1 and Hey2 are required for embryonic vascular development. Genes Dev. 2004, 18, 901–911. [Google Scholar] [CrossRef]
- Nakagawaab, O.; Nakagawaab, M.; Richardson, J.A.; Olson, E.N.; Srivastava, D. HRT1, HRT2, and HRT3: A New Subclass of bHLH Transcription Factors Marking Specific Cardiac, Somitic, and Pharyngeal Arch Segments. Dev. Biol. 1999, 216, 72–84. [Google Scholar] [CrossRef]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef]
- Hombach, S.; Kretz, M. Non-coding RNAs: Classification, Biology and Functioning. Adv. Exp. Med. Biol. 2016, 937, 3–17. [Google Scholar] [CrossRef]
- Ounzain, S.; Crippa, S.; Pedrazzini, T. Small and long non-coding RNAs in cardiac homeostasis and regeneration. Biochim. et Biophys. Acta 2013, 1833, 923–933. [Google Scholar] [CrossRef]
- Chen, J.; Wang, D.-Z. microRNAs in cardiovascular development. J. Mol. Cell. Cardiol. 2012, 52, 949–957. [Google Scholar] [CrossRef]
- Beermann, J.; Piccoli, M.-T.; Viereck, J.; Thum, T. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol. Rev. 2016, 96, 1297–1325. [Google Scholar] [CrossRef]
- Poller, W.; Dimmeler, S.; Heymans, S.; Zeller, T.; Haas, J.; Karakas, M.; Leistner, D.M.; Jakob, P.; Nakagawa, S.; Blankenberg, S.; et al. Non-coding RNAs in cardiovascular diseases: Diagnostic and therapeutic perspectives. Eur. Heart J. 2018, 39, 2704–2716. [Google Scholar] [CrossRef] [PubMed]
- Dykes, I.M.; Emanueli, C. Transcriptional and Post-transcriptional Gene Regulation by Long Non-coding RNA. Genom. Proteom. Bioinform. 2017, 15, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Shi, Z.-M.; Chang, Y.-N.; Hu, Z.-M.; Qi, H.-X.; Hong, W. The ways of action of long non-coding RNAs in cytoplasm and nucleus. Gene 2014, 547, 1–9. [Google Scholar] [CrossRef]
- Wojciechowska, A.; Braniewska, A.; Kozar-Kamińska, K. MicroRNA in cardiovascular biology and disease. Adv. Clin. Exp. Med. 2017, 26, 868–874. [Google Scholar] [CrossRef]
- Barwari, T.; Joshi, A.; Mayr, M. MicroRNAs in Cardiovascular Disease. J. Am. Coll. Cardiol. 2016, 68, 2577–2584. [Google Scholar] [CrossRef]
- Wong, L.L.; Wang, J.; Liew, O.W.; Richards, A.M.; Chen, Y.-T. MicroRNA and Heart Failure. Int. J. Mol. Sci. 2016, 17, 502. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Lin, S.; Gregory, R.I. MicroRNA biogenesis pathways in cancer. Nat. Cancer 2015, 15, 321–333. [Google Scholar] [CrossRef]
- Ransohoff, J.D.; Wei, Y.; Khavari, P.A. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 2018, 19, 143–157. [Google Scholar] [CrossRef]
- Bhan, A.; Soleimani, M.; Mandal, S.S. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res. 2017, 77, 3965–3981. [Google Scholar] [CrossRef]
- Dahariya, S.; Paddibhatla, I.; Kumar, S.; Raghuwanshi, S.; Pallepati, A.; Gutti, R.K. Long non-coding RNA: Classification, biogenesis and functions in blood cells. Mol. Immunol. 2019, 112, 82–92. [Google Scholar] [CrossRef]
- Yuan, Z.; Huang, W. New Developments in Exosomal lncRNAs in Cardiovascular Diseases. Front. Cardiovasc. Med. 2021, 8, 709169. [Google Scholar] [CrossRef]
- Zheng, D.; Huo, M.; Li, B.; Wang, W.; Piao, H.; Wang, Y.; Zhu, Z.; Li, D.; Wang, T.; Liu, K. The Role of Exosomes and Exosomal MicroRNA in Cardiovascular Disease. Front. Cell Dev. Biol. 2021, 8, 616161. [Google Scholar] [CrossRef]
- Barron, M.; Gao, M.; Lough, J. Requirement for BMP and FGF signaling during cardiogenic induction in non-precardiac mesoderm is specific, transient, and cooperative. Dev. Dyn. 2000, 218, 383–393. [Google Scholar] [CrossRef]
- Lough, J.; Barron, M.; Brogley, M.; Sugi, Y.; Bolender, D.L.; Zhu, X. Combined BMP-2 and FGF-4, but Neither Factor Alone, Induces Cardiogenesis in Non-Precardiac Embryonic Mesoderm. Dev. Biol. 1996, 178, 198–202. [Google Scholar] [CrossRef][Green Version]
- Schultheiss, T.M.; Burch, J.B.; Lassar, A.B. A role for bone morphogenetic proteins in the induction of cardiac myogenesis. Genes Dev. 1997, 11, 451–462. [Google Scholar] [CrossRef]
- Zhang, H.; Bradley, A. Mice deficient for BMP2 are nonviable and have defects in amnion/chorion and cardiac development. Development 1996, 122, 2977–2986. [Google Scholar] [CrossRef]
- Sugi, Y.; Yamamura, H.; Okagawa, H.; Markwald, R.R. Bone morphogenetic protein-2 can mediate myocardial regulation of atrioventricular cushion mesenchymal cell formation in mice. Dev. Biol. 2004, 269, 505–518. [Google Scholar] [CrossRef]
- Luna-Zurita, L.; Prados, B.; Grego-Bessa, J.; Luxán, G.; del Monte, G.; Benguría, A.; Adams, R.H.; Pérez-Pomares, J.M.; de la Pompa, J.L. Integration of a Notch-dependent mesenchymal gene program and Bmp2-driven cell invasiveness regulates murine cardiac valve formation. J. Clin. Investig. 2010, 120, 3493–3507. [Google Scholar] [CrossRef]
- Winnier, G.; Blessing, M.; Labosky, P.A.; Hogan, B.L. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 1995, 9, 2105–2116. [Google Scholar] [CrossRef] [PubMed]
- Waldo, K.L.; Kumiski, D.H.; Wallis, K.T.; Stadt, H.A.; Hutson, M.R.; Platt, D.H.; Kirby, M.L. Conotruncal myocardium arises from a secondary heart field. Development 2001, 128, 3179–3188. [Google Scholar] [CrossRef]
- Bernardo, A.S.; Faial, T.; Gardner, L.; Niakan, K.K.; Ortmann, D.; Senner, C.E.; Callery, E.M.; Trotter, M.W.; Hemberger, M.; Smith, J.C.; et al. BRACHYURY and CDX2 Mediate BMP-Induced Differentiation of Human and Mouse Pluripotent Stem Cells into Embryonic and Extraembryonic Lineages. Cell Stem Cell 2011, 9, 144–155. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Sanchez, C.; Franco, D.; Bonet, F.; Garcia-Lopez, V.; Aranega, A.; Martinez, F.B. Negative Fgf8-Bmp2 feed-back is regulated by miR-130 during early cardiac specification. Dev. Biol. 2015, 406, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, C.A.; Eisenberg, L.M. WNT11 promotes cardiac tissue formation of early mesoderm. Dev. Dyn. 1999, 216, 45–58. [Google Scholar] [CrossRef]
- Ueno, S.; Weidinger, G.; Osugi, T.; Kohn, A.D.; Golob, J.L.; Pabon, L.; Reinecke, H.; Moon, R.T.; Murry, C.E. Biphasic role for Wnt/beta-catenin signaling in cardiac specification in zebrafish and embryonic stem cells. Proc. Natl. Acad. Sci. USA 2007, 104, 9685–9690. [Google Scholar] [CrossRef]
- Afouda, B.A.; Hoppler, S. Different requirements for GATA factors in cardiogenesis are mediated by non-canonical Wnt signaling. Dev. Dyn. 2011, 240, 649–662. [Google Scholar] [CrossRef]
- Samuel, L.J.; Latinkić, B.V. Early Activation of FGF and Nodal Pathways Mediates Cardiac Specification Independently of Wnt/β-Catenin Signaling. PLoS ONE 2009, 4, e7650. [Google Scholar] [CrossRef]
- Klaus-Bergmann, A.; Saga, Y.; Taketo, M.M.; Tzahor, E.; Birchmeier, W. Distinct roles of Wnt/beta-catenin and Bmp signaling during early cardiogenesis. Proc. Natl. Acad. Sci. USA 2007, 104, 18531–18536. [Google Scholar] [CrossRef]
- Gessert, S.; Kühl, M. The Multiple Phases and Faces of Wnt Signaling During Cardiac Differentiation and Development. Circ. Res. 2010, 107, 186–199. [Google Scholar] [CrossRef]
- Onizuka, T.; Yuasa, S.; Kusumoto, D.; Shimoji, K.; Egashira, T.; Ohno, Y.; Kageyama, T.; Tanaka, T.; Hattori, F.; Fujita, J.; et al. Wnt2 accelerates cardiac myocyte differentiation from ES-cell derived mesodermal cells via non-canonical pathway. J. Mol. Cell. Cardiol. 2011, 52, 650–659. [Google Scholar] [CrossRef]
- Medley, T.L.; Furtado, M.; Lam, N.T.; Idrizi, R.; Williams, D.; Verma, P.J.; Costa, M.; Kaye, D.M. Effect of Oxygen on Cardiac Differentiation in Mouse iPS Cells: Role of Hypoxia Inducible Factor-1 and Wnt/Beta-Catenin Signaling. PLoS ONE 2013, 8, e80280. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, J.; Cui, J.; Sun, M.; Du, W.; Chen, T.; Ming, X.; Zhang, L.; Tian, J.; Li, J.; et al. MiR218 Modulates Wnt Signaling in Mouse Cardiac Stem Cells by Promoting Proliferation and Inhibiting Differentiation through a Positive Feedback Loop. Sci. Rep. 2016, 6, 20968. [Google Scholar] [CrossRef]
- Wang, D.; Liu, C.; Wang, Y.; Wang, W.; Wang, K.; Wu, X.; Li, Z.; Zhao, C.; Li, L.; Peng, L. Impact of miR-26b on cardiomyocyte differentiation in P19 cells through regulating canonical/non-canonical Wnt signalling. Cell Prolif. 2017, 50, e12371. [Google Scholar] [CrossRef]
- Liu, X.; Yang, L.; Wang, H.; Xu, G.; Zhu, S.; Li, M.; Hu, X.; Zhu, J.; Zhu, C.; Xu, J.; et al. Effects of miR-19b knockdown on the cardiac differentiation of P19 mouse embryonic carcinoma cells. Mol. Med. Rep. 2014, 11, 2504–2512. [Google Scholar] [CrossRef]
- Qin, D.-N.; Qian, L.; Hu, D.-L.; Yu, Z.-B.; Han, S.-P.; Zhu, C.; Wang, X.; Hu, X. Effects of miR-19b Overexpression on Proliferation, Differentiation, Apoptosis and Wnt/β-Catenin Signaling Pathway in P19 Cell Model of Cardiac Differentiation In Vitro. Cell Biophys. 2013, 66, 709–722. [Google Scholar] [CrossRef]
- Lu, T.-Y.; Lin, B.; Li, Y.; Arora, A.; Han, L.; Cui, C.; Coronnello, C.; Sheng, Y.; Benos, P.V.; Yang, L. Overexpression of microRNA-1 promotes cardiomyocyte commitment from human cardiovascular progenitors via suppressing WNT and FGF signaling pathways. J. Mol. Cell. Cardiol. 2013, 63, 146–154. [Google Scholar] [CrossRef]
- Xu, J.; Gruber, P.J.; Chien, K.R. SMAD4 Is Essential for Human Cardiac Mesodermal Precursor Cell Formation. Stem Cells 2018, 37, 216–225. [Google Scholar] [CrossRef]
- López-Sánchez, C.; Climent, V.; Schoenwolf, G.; Álvarez, I.; García-Martínez, V. Induction of cardiogenesis by Hensen’s node and fibroblast growth factors. Cell Tissue Res. 2002, 309, 237–249. [Google Scholar] [CrossRef]
- Coppola, A.; Romito, A.; Borel, C.; Gehrig, C.; Gagnebin, M.; Falconnet, E.; Izzo, A.; Altucci, L.; Banfi, S.; Antonarakis, S.; et al. Cardiomyogenesis is controlled by the miR-99a/let-7c cluster and epigenetic modifications. Stem Cell Res. 2014, 12, 323–337. [Google Scholar] [CrossRef]
- Chen, X.; Wang, L.; Huang, R.; Qiu, H.; Wang, P.; Wu, D.; Zhu, Y.; Ming, J.; Wang, Y.; Wang, J.; et al. Dgcr8 deletion in the primitive heart uncovered novel microRNA regulating the balance of cardiac-vascular gene program. Protein Cell 2019, 10, 327–346. [Google Scholar] [CrossRef]
- Shen, X.; Soibam, B.; Benham, A.; Xu, X.; Chopra, M.; Peng, X.; Yu, W.; Bao, W.; Liang, R.; Azares, A.; et al. miR-322/-503 cluster is expressed in the earliest cardiac progenitor cells and drives cardiomyocyte specification. Proc. Natl. Acad. Sci. USA 2016, 113, 9551–9556. [Google Scholar] [CrossRef]
- Yao, C.-X.; Wei, Q.-X.; Zhang, Y.-Y.; Wang, W.-P.; Xue, L.-X.; Yang, F.; Zhang, S.-F.; Xiong, C.-J.; Li, W.-Y.; Wei, Z.-R.; et al. miR-200b targets GATA-4 during cell growth and differentiation. RNA Biol. 2013, 10, 465–480. [Google Scholar] [CrossRef]
- Han, M.; Yang, Z.; Sayed, D.; He, M.; Gao, S.; Lin, L.; Yoon, S.; Abdellatif, M. GATA4 expression is primarily regulated via a miR-26b-dependent post-transcriptional mechanism during cardiac hypertrophy. Cardiovasc. Res. 2012, 93, 645–654. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, S.; Guo, C.; Li, J.; Sang, W. Downregulation of miR-200c protects cardiomyocytes from hypoxia-induced apoptosis by targeting GATA-4. Int. J. Mol. Med. 2017, 39, 1589–1596. [Google Scholar] [CrossRef]
- Kay, M.; Soltani, B.M.; Aghdaei, F.H.; Ansari, H.; Baharvand, H. Hsa-miR-335 regulates cardiac mesoderm and progenitor cell differentiation. Stem Cell Res. Ther. 2019, 10, 191. [Google Scholar] [CrossRef]
- Qian, L.; Wythe, J.D.; Liu, J.; Cartry, J.; Vogler, G.; Mohapatra, B.; Otway, R.T.; Huang, Y.; King, I.N.; Maillet, M.; et al. Tinman/Nkx2-5 acts via miR-1 and upstream of Cdc42 to regulate heart function across species. J. Cell Biol. 2011, 193, 1181–1196. [Google Scholar] [CrossRef] [PubMed]
- Hoelscher, S.C.; Stich, T.; Diehm, A.; Lahm, H.; Dreßen, M.; Zhang, Z.; Neb, I.; Aherrahrou, Z.; Erdmann, J.; Schunkert, H.; et al. miR-128a Acts as a Regulator in Cardiac Development by Modulating Differentiation of Cardiac Progenitor Cell Populations. Int. J. Mol. Sci. 2020, 21, 1158. [Google Scholar] [CrossRef] [PubMed]
- Arasaratnam, D.; Bell, K.M.; Sim, C.B.; Koutsis, K.; Anderson, D.J.; Qian, E.L.; Stanley, E.G.; Elefanty, A.G.; Cheung, M.M.; Oshlack, A.; et al. The role of cardiac transcription factor NKX2-5 in regulating the human cardiac miRNAome. Sci. Rep. 2019, 9, 15928. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Wu, D.; Wang, C.; Guo, Z.; Li, B.; Zuo, Z. Hexabromocyclododecane exposure induces cardiac hypertrophy and arrhythmia by inhibiting miR-1 expression via up-regulation of the homeobox gene Nkx2.5. J. Hazard. Mater. 2015, 302, 304–313. [Google Scholar] [CrossRef]
- Chinchilla, A.; Lozano, E.; Daimi, H.; Esteban, F.J.; Crist, C.; Aranega, A.E.; Franco, D. MicroRNA profiling during mouse ventricular maturation: A role for miR-27 modulating Mef2c expression. Cardiovasc. Res. 2010, 89, 98–108. [Google Scholar] [CrossRef]
- Chen, H.-P.; Wen, J.; Tan, S.-R.; Kang, L.-M.; Zhu, G.-C. MiR-199a-3p inhibition facilitates cardiomyocyte differentiation of embryonic stem cell through promotion of MEF2C. J. Cell. Physiol. 2019, 234, 23315–23325. [Google Scholar] [CrossRef]
- Xu, M.; Chen, X.; Chen, D.; Yu, B.; Li, M.; He, J.; Huang, Z. MicroRNA-499-5p regulates skeletal myofiber specification via NFATc1/MEF2C pathway and Thrap1/MEF2C axis. Life Sci. 2018, 215, 236–245. [Google Scholar] [CrossRef]
- Shi, Y.; Mao, X.; Cai, M.; Hu, S.; Lai, X.; Chen, S.; Jia, X.; Wang, J.; Lai, S. miR-194-5p negatively regulates the proliferation and differentiation of rabbit skeletal muscle satellite cells. Mol. Cell. Biochem. 2020, 476, 425–433. [Google Scholar] [CrossRef]
- Cheng, X.; Du, J.; Shen, L.; Tan, Z.; Jiang, D.; Jiang, A.; Li, Q.; Tang, G.; Jiang, Y.; Wang, J.; et al. MiR-204-5p regulates C2C12 myoblast differentiation by targeting MEF2C and ERRγ. Biomed. Pharmacother. 2018, 101, 528–535. [Google Scholar] [CrossRef]
- Gagan, J.; Dey, B.K.; Layer, R.; Yan, Z.; Dutta, A. Notch3 and Mef2c Proteins Are Mutually Antagonistic via Mkp1 Protein and miR-1/206 MicroRNAs in Differentiating Myoblasts. J. Biol. Chem. 2012, 287, 40360–40370. [Google Scholar] [CrossRef]
- Tang, C.-M.; Liu, F.-Z.; Zhu, J.-N.; Fu, Y.-H.; Lin, Q.-X.; Deng, C.-Y.; Hu, Z.-Q.; Yang, H.; Zheng, X.-L.; Cheng, J.-D.; et al. Myocyte-specific enhancer factor 2C: A novel target gene of miR-214-3p in suppressing angiotensin II-induced cardiomyocyte hypertrophy. Sci. Rep. 2016, 6, 36146. [Google Scholar] [CrossRef]
- Zhang, R.; Sui, L.; Hong, X.; Yang, M.; Li, W. MiR-448 promotes vascular smooth muscle cell proliferation and migration in through directly targeting MEF2C. Environ. Sci. Pollut. Res. 2017, 24, 22294–22300. [Google Scholar] [CrossRef]
- Klattenhoff, C.A.; Scheuermann, J.C.; Surface, L.E.; Bradley, R.K.; Fields, P.A.; Steinhauser, M.L.; Ding, H.; Butty, V.L.; Torrey, L.; Haas, S.; et al. Braveheart, a Long Noncoding RNA Required for Cardiovascular Lineage Commitment. Cell 2013, 152, 570–583. [Google Scholar] [CrossRef]
- Hou, J.; Long, H.; Zhou, C.; Zheng, S.; Wu, H.; Guo, T.; Wu, Q.; Zhong, T.; Wang, T. Long noncoding RNA Braveheart promotes cardiogenic differentiation of mesenchymal stem cells in vitro. Stem Cell Res. Ther. 2017, 8, 4. [Google Scholar] [CrossRef]
- García-Padilla, C.; Domínguez, J.N.; Aránega, A.E.; Franco, D. Differential chamber-specific expression and regulation of long non-coding RNAs during cardiac development. Biochim. Biophys. Acta 2019, 1862, 194435. [Google Scholar] [CrossRef]
- Yin, A.; Feng, M.; Cheng, Z.; Zhang, Q.; Li, H.; Xu, J.; Zhang, H.; Li, Y.; Qian, L. Altered DNA Methylation of Long Noncoding RNA uc.167 Inhibits Cell Differentiation in Heart Development. BioMed Res. Int. 2018, 2018, 4658024. [Google Scholar] [CrossRef]
- Liu, H.; Hu, Y.; Yin, J.; Yan, X.; Chen, W.; Wang, X.; Han, S.; Yu, Z.; Li, M. Effects of long non-coding RNA uc.245 on cardiomyocyte-like differentiation in P19 cells via FOG2. Gene 2019, 694, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Song, G.; Shen, Y.; Ruan, Z.; Li, X.; Chen, Y.; Yuan, W.; Ding, X.; Zhu, L.; Qian, L. LncRNA-uc.167 influences cell proliferation, apoptosis and differentiation of P19 cells by regulating Mef2c. Gene 2016, 590, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.-H.; Chang, M.-W.; Pandey, P.R.; Tsitsipatis, D.; Yang, X.; Martindale, J.L.; Munk, R.; De, S.; Abdelmohsen, K.; Gorospe, M. Interaction of OIP5-AS1 with MEF2C mRNA promotes myogenic gene expression. Nucleic Acids Res. 2020, 48, 12943–12956. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.-J.; Lee, K.-H.; Son, Y.; Nam, A.-R.; Moon, E.-H.; Pyun, J.-H.; Park, J.; Kang, J.-S.; Lee, Y.J.; Cho, J.-Y. Spatiotemporal expression of long noncoding RNA Moshe modulates heart cell lineage commitment. RNA Biol. 2021, 18, 640–654. [Google Scholar] [CrossRef]
- Grote, P.; Wittler, L.; Hendrix, D.; Koch, F.; Währisch, S.; Beisaw, A.; Macura, K.; Bläss, G.; Kellis, M.; Werber, M.; et al. The Tissue-Specific lncRNA Fendrr Is an Essential Regulator of Heart and Body Wall Development in the Mouse. Dev. Cell 2013, 24, 206–214. [Google Scholar] [CrossRef]
- Zhu, H. Forkhead box transcription factors in embryonic heart development and congenital heart disease. Life Sci. 2016, 144, 194–201. [Google Scholar] [CrossRef]
- Watanabe, M.; Whitman, M. FAST-1 is a key maternal effector of mesoderm inducers in the early Xenopus embryo. Development 1999, 126, 5621–5634. [Google Scholar] [CrossRef]
- Weisberg, E.; Winnier, G.E.; Chen, X.; Farnsworth, C.L.; Hogan, B.L.; Whitman, M. A mouse homologue of FAST-1 transduces TGFβ superfamily signals and is expressed during early embryogenesis. Mech. Dev. 1998, 79, 17–27. [Google Scholar] [CrossRef]
- von Both, I.; Silvestri, C.; Erdemir, T.; Lickert, H.; Walls, J.R.; Henkelman, R.M.; Rossant, J.; Harvey, R.P.; Attisano, L.; Wrana, J.L. Foxh1 is essential for development of the anterior heart field. Dev. Cell 2004, 7, 331–345. [Google Scholar] [CrossRef]
- Huang, C.-X.; Chen, N.; Wu, X.-J.; He, Y.; Huang, C.-H.; Liu, H.; Wang, W.-M.; Wang, H.-L. Zebrafish let-7b acts downstream of hypoxia-inducible factor-1α to assist in hypoxia-mediated cell proliferation and cell cycle regulation. Life Sci. 2017, 171, 21–29. [Google Scholar] [CrossRef]
- Heigwer, J.; Kutzner, J.; Haeussler, M.; Burkhalter, M.D.; Draebing, T.; Juergensen, L.; Katus, H.A.; Philipp, M.; Westhoff, J.H.; Hassel, D. miR-103/107 regulates left-right asymmetry in zebrafish by modulating Kupffer’s vesicle development and ciliogenesis. Biochem. Biophys. Res. Commun. 2020, 527, 432–439. [Google Scholar] [CrossRef]
- Fischer, P.; Chen, H.; Pacho, F.; Rieder, D.; Kimmel, R.A.; Meyer, D. FoxH1 represses miR-430 during early embryonic development of zebrafish via non-canonical regulation. BMC Biol. 2019, 17, 61. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, L.; Meng, Y.; Huang, L. Benzyl isothiocyanate inhibits breast cancer cell tumorigenesis via repression of the FoxH1-Mediated Wnt/β-catenin pathway. Int. J. Clin. Exp. Med. 2015, 8, 17601–17611. [Google Scholar]
- Vincent, S.D.; Buckingham, M. Chapter One-How to Make a Heart: The Origin and Regulation of Cardiac Progenitor Cells. Curr. Top. Dev. Biol. 2010, 90, 1–41. [Google Scholar]
- Evans, S.M.; Yelon, D.; Conlon, F.L.; Kirby, M.L. Myocardial Lineage Development. Circ. Res. 2010, 107, 1428–1444. [Google Scholar] [CrossRef]
- Ren, J.; Miao, D.; Li, Y.; Gao, R. Spotlight on Isl1: A Key Player in Cardiovascular Development and Diseases. Front. Cell Dev. Biol. 2021, 9, 793605. [Google Scholar] [CrossRef]
- Wang, J.; Greene, S.B.; Bonilla-Claudio, M.; Tao, Y.; Zhang, J.; Huang, Z.; Black, B.L.; Wang, F.; Martin, J.F. Bmp-signaling regulates myocardial differentiation from cardiac progenitors through a micro RNA-mediated mechanism. Dev. Cell 2010, 19, 903–912. [Google Scholar] [CrossRef]
- Zlabinger, K.; Spannbauer, A.; Traxler, D.; Gugerell, A.; Lukovic, D.; Winkler, J.; Mester-Tonczar, J.; Podesser, B.; Gyöngyösi, M. MiR-21, MiR-29a, GATA4, and MEF2c Expression Changes in Endothelin-1 and Angiotensin II Cardiac Hypertrophy Stimulated Isl-1+Sca-1+c-kit+ Porcine Cardiac Progenitor Cells In Vitro. Cells 2019, 8, 1416. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, X.; Li, F.; Ji, Q. MiR-128-3p accelerates cardiovascular calcification and insulin resistance through ISL1-dependent Wnt pathway in type 2 diabetes mellitus rats. J. Cell. Physiol. 2018, 234, 4997–5010. [Google Scholar] [CrossRef]
- Steimle, J.D.; Moskowitz, I.P. Chapter Seven—TBX5: A Key Regulator of Heart Development. Curr. Top. Dev. Biol. 2017, 122, 195–221. [Google Scholar]
- D’Aurizio, R.; Russo, F.; Chiavacci, E.; Baumgart, M.; Groth, M.; D’Onofrio, M.; Arisi, I.; Rainaldi, G.; Pitto, L.; Pellegrini, M. Discovering miRNA Regulatory Networks in Holt–Oram Syndrome Using a Zebrafish Model. Front. Bioeng. Biotechnol. 2016, 4, 60. [Google Scholar] [CrossRef][Green Version]
- Chiavacci, E.; Daurizio, R.; Guzzolino, E.; Russo, F.; Baumgart, M.; Groth, M.; Mariani, L.; D’Onofrio, M.; Arisi, I.; Pellegrini, M.; et al. MicroRNA 19a replacement partially res-cues fin and cardiac defects in zebrafish model of Holt Oram syndrome. Sci. Rep. 2015, 5, 18240. [Google Scholar] [CrossRef]
- Boon, R.A.; Iekushi, K.; Lechner, S.; Seeger, T.; Fischer, A.; Heydt, S.; Kaluza, D.; Tréguer, K.; Carmona, G.; Bonauer, A.; et al. MicroRNA-34a regulates cardiac ageing and function. Nature 2013, 495, 107–110. [Google Scholar] [CrossRef]
- Fish, J.E.; Wythe, J.D.; Xiao, T.; Bruneau, B.G.; Stainier, D.Y.R.; Srivastava, D.; Woo, S. A Slit/miR-218/Robo regulatory loop is required during heart tube formation in zebrafish. Development 2011, 138, 1409–1419. [Google Scholar] [CrossRef]
- Qu, X.; Jia, H.; Garrity, D.M.; Tompkins, K.; Batts, L.; Appel, B.; Zhong, T.P.; Baldwin, H.S. ndrg4 is required for normal myocyte proliferation during early cardiac development in zebrafish. Dev. Biol. 2008, 317, 486–496. [Google Scholar] [CrossRef]
- Chiavacci, E.; Dolfi, L.; Verduci, L.; Meghini, F.; Gestri, G.; Evangelista, M.; Wilson, S.W.; Cremisi, F.; Pitto, L. MicroRNA 218 Mediates the Effects of Tbx5a Over-Expression on Zebrafish Heart Development. PLoS ONE 2012, 7, e50536. [Google Scholar] [CrossRef]
- Alzein, M.; Lozano-Velasco, E.; Hernández-Torres, F.; García-Padilla, C.; Domínguez, J.N.; Aránega, A.; Franco, D. Differential Spatio-Temporal Regulation of T-Box Gene Expression by microRNAs during Cardiac Development. J. Cardiovasc. Dev. Dis. 2021, 8, 56. [Google Scholar] [CrossRef]
- Ma, J.; Chen, S.; Hao, L.; Sheng, W.; Chen, W.; Ma, X.; Zhang, B.; Ma, D.; Huang, G. Hypermethylation-mediated down-regulation of lncRNA TBX5-AS1:2 in Tetralogy of Fallot inhibits cell proliferation by reducingTBX5expression. J. Cell. Mol. Med. 2020, 24, 6472–6484. [Google Scholar] [CrossRef]
- Hori, Y.; Tanimoto, Y.; Takahashi, S.; Furukawa, T.; Koshiba-Takeuchi, K.; Takeuchi, J.K. Important cardiac transcription factor genes are accompanied by bidirectional long non-coding RNAs. BMC Genom. 2018, 19, 967. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.H.; Nadadur, R.D.; Hilvering, C.; Bianchi, V.; Werner, M.; Mazurek, S.R.; Gadek, M.; Shen, K.M.; Goldman, J.A.; Tyan, L.; et al. Transcription-factor-dependent enhancer transcription defines a gene regulatory network for cardiac rhythm. eLife 2017, 6, e31683. [Google Scholar] [CrossRef] [PubMed]
- Shiratori, H.; Hamada, H. TGFβ signaling in establishing left–right asymmetry. Semin. Cell Dev. Biol. 2014, 32, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Hamada, H.; Meno, C.; Watanabe, D.; Saijoh, Y. Establishment of vertebrate left–right asymmetry. Nat. Rev. Genet. 2002, 3, 103–113. [Google Scholar] [CrossRef]
- Franco, D. The Role of Pitx2 during Cardiac Development Linking Left–Right Signaling and Congenital Heart Diseases. Trends Cardiovasc. Med. 2003, 13, 157–163. [Google Scholar] [CrossRef]
- Grimes, D.T.; Burdine, R.D. Left–Right Patterning: Breaking Symmetry to Asymmetric Morphogenesis. Trends Genet. 2017, 33, 616–628. [Google Scholar] [CrossRef]
- Franco, D.; Sedmera, D.; Lozano-Velasco, E. Multiple Roles of Pitx2 in Cardiac Development and Disease. J. Cardiovasc. Dev. Dis. 2017, 4, 16. [Google Scholar] [CrossRef]
- Franco, D.; Christoffels, V.M.; Campione, M. Homeobox transcription factor Pitx2: The rise of an asymmetry gene in cardiogenesis and arrhythmogenesis. Trends Cardiovasc. Med. 2014, 24, 23–31. [Google Scholar] [CrossRef]
- Gage, P.; Suh, H.; Camper, S. Dosage requirement of Pitx2 for development of multiple organs. Development 1999, 126, 4643–4651. [Google Scholar] [CrossRef]
- Lu, M.-F.; Pressman, C.L.; Dyer, R.; Johnson, R.L.; Martin, J.F. Function of Rieger syndrome gene in left–right asymmetry and craniofacial development. Nature 1999, 401, 276–278. [Google Scholar] [CrossRef]
- Barroso-Deljesus, A.; Lucena-Aguilar, G.; Sanchez, L.; Ligero, G.; Gutierrez-Aranda, I.; Menendez, P. The Nodal inhibitor Lefty is negatively modulated by the microRNA miR-302 in human embryonic stem cells. FASEB J. 2011, 25, 1497–1508. [Google Scholar] [CrossRef]
- Rosa, A.; Spagnoli, F.M.; Brivanlou, A.H. The miR-430/427/302 Family Controls Mesendodermal Fate Specification via Species-Specific Target Selection. Dev. Cell 2009, 16, 517–527. [Google Scholar] [CrossRef]
- Rosa, A.; Papaioannou, M.D.; Krzyspiak, J.E.; Brivanlou, A.H. miR-373 is regulated by TGFβ signaling and promotes mesendoderm differentiation in human Embryonic Stem Cells. Dev. Biol. 2014, 391, 81–88. [Google Scholar] [CrossRef]
- Choi, W.-Y.; Giraldez, A.J.; Schier, A.F. Target Protectors Reveal Dampening and Balancing of Nodal Agonist and Antagonist by miR-430. Science 2007, 318, 271–274. [Google Scholar] [CrossRef]
- Ma, H.; Lin, Y.; Zhao, Z.-A.; Lu, X.; Yu, Y.; Zhang, X.; Wang, Q.; Li, L. MicroRNA-127 Promotes Mesendoderm Differentiation of Mouse Embryonic Stem Cells by Targeting Left-Right Determination Factor 2. J. Biol. Chem. 2016, 291, 12126–12135. [Google Scholar] [CrossRef]
- Yang, F.; Qi, J. miR-430a regulates the development of left–right asymmetry by targeting sqt in the teleost. Gene 2020, 745, 144628. [Google Scholar] [CrossRef]
- Li, M.; Hu, X.; Zhu, J.; Zhu, C.; Zhu, S.; Liu, X.; Xu, J.; Han, S.; Yu, Z. Overexpression of miR-19b Impairs Cardiac Development in Zebrafish by Targeting ctnnb1. Cell. Physiol. Biochem. 2014, 33, 1988–2002. [Google Scholar] [CrossRef]
- Ryan, A.K.; Blumberg, B.; Rodriguez-Esteban, C.; Yonei-Tamura, S.; Tamura, K.; Tsukui, T.; De La Peña, J.; Sabbagh, W.; Greenwald, J.; Choe, S.; et al. Pitx2 determines left–right asymmetry of internal organs in vertebrates. Nature 1998, 394, 545–551. [Google Scholar] [CrossRef]
- Piedra, M.E.; Icardo, J.M.; Albajar, M.; Rodriguez-Rey, J.C.; Ros, M.A. Pitx2 participates in the late phase of the pathway controlling left-right asymmetry. Cell 1998, 94, 319–324. [Google Scholar] [CrossRef]
- Logan, M.; Pagán-Westphal, S.M.; Smith, D.M.; Paganessi, L.; Tabin, C.J. The Transcription Factor Pitx2 Mediates Situs-Specific Morphogenesis in Response to Left-Right Asymmetric Signals. Cell 1998, 94, 307–317. [Google Scholar] [CrossRef]
- Campione, M.; Steinbeisser, H.; Schweickert, A.; Deissler, K.; van Bebber, F.; Lowe, L.A.; Nowotschin, S.; Viebahn, C.; Haffter, P.; Kuehn, M.R.; et al. The homeobox gene Pitx2: Mediator of asymmetric left-right signaling in vertebrate heart and gut looping. Development 1999, 126, 1225–1234. [Google Scholar] [CrossRef]
- Bisgrove, B.W.; Essner, J.J.; Yost, H.J. Multiple pathways in the midline regulate concordant brain, heart and gut left-right asymmetry. Development 2000, 127, 3567–3579. [Google Scholar] [CrossRef]
- Bisgrove, B.W.; Yost, H.J. Classification of left-right patterning defects in zebrafish, mice, and humans. Am. J. Med Genet. 2001, 101, 315–323. [Google Scholar] [CrossRef]
- Branford, W.W.; Essner, J.; Yost, H. Regulation of Gut and Heart Left–Right Asymmetry by Context-Dependent Interactions between Xenopus Lefty and BMP4 Signaling. Dev. Biol. 2000, 223, 291–306. [Google Scholar] [CrossRef]
- Chinchilla, A.; Daimi, H.; Lozano-Velasco, E.; Dominguez, J.N.; Caballero, R.; Delpo, E.; Tamargo, J.; Cinca, J.; Hove-Madsenet, L.; Aranega, A.E.; et al. PITX2 insufficiency leads to atrial electrical and structural remodeling linked to arrhythmogenesis. Circ. Cardiovasc. Genet. 2011, 4, 269–279. [Google Scholar] [CrossRef]
- Wang, J.; Klysik, E.; Sood, S.; Johnson, R.L.; Wehrens, X.H.T.; Martin, J.F. Pitx2 prevents susceptibility to atrial arrhythmias by inhibiting left-sided pacemaker specification. Proc. Natl. Acad. Sci. USA 2010, 107, 9753–9758. [Google Scholar] [CrossRef]
- Kirchhof, P.; Kahr, P.C.; Kaese, S.; Piccini, I.; Vokshi, I.; Scheld, H.-H.; Rotering, H.; Fortmueller, L.; Laakmann, S.; Verheule, S.; et al. PITX2c Is Expressed in the Adult Left Atrium, and Reducing Pitx2c Expression Promotes Atrial Fibrillation Inducibility and Complex Changes in Gene Expression. Circ. Cardiovasc. Genet. 2011, 4, 123–133. [Google Scholar] [CrossRef]
- Lozano-Velasco, E.; Hernández-Torres, F.; Daimi, H.; Serra, S.A.; Herraiz, A.; Hove-Madsen, L.; Aránega, A.; Franco, D. Pitx2 impairs calcium handling in a dose-dependent manner by modulating Wnt signalling. Cardiovasc. Res. 2016, 109, 55–66. [Google Scholar] [CrossRef]
- Lozano-Velasco, E.; Wangensteen, R.; Quesada, A.; Garcia-Padilla, C.; Osorio, J.A.; Ruiz-Torres, M.D.; Aranega, A.; Franco, D. Hyperthyroidism, but not hypertension, impairs PITX2 expression leading to Wnt-microRNA-ion channel remodeling. PLoS ONE 2017, 12, e0188473. [Google Scholar] [CrossRef]
- Franco, D.; Aranega, A.; Dominguez, J.N. Non-coding RNAs and Atrial Fibrillation. In Non-coding RNAs in Cardiovascular Diseases; Springer: Berlin/Heidelberg, Germany, 2020; Volume 1229, pp. 311–325. [Google Scholar] [CrossRef]
- Petkova, M.; Atkinson, A.J.; Yanni, J.; Stuart, L.; Aminu, A.J.; Ivanova, A.D.; Pustovit, K.B.; Geragthy, C.; Feather, A.; Li, N.; et al. Identification of Key Small Non-Coding MicroRNAs Controlling Pacemaker Mechanisms in the Human Sinus Node. J. Am. Heart Assoc. 2020, 9, e016590. [Google Scholar] [CrossRef]
- García-Padilla, C.; Aránega, A.; Franco, D. The role of long non-coding RNAs in cardiac development and disease. AIMS Genet. 2018, 5, 124–140. [Google Scholar] [CrossRef] [PubMed]
- Ocaña, O.H.; Córcoles, R.; Fabra, Á.; Moreno-Bueno, G.; Acloque, H.; Vega, S.; Barrallo-Gimeno, A.; Cano, A.; Nieto, M.A. Metastatic Colonization Requires the Re-pression of the Epithelial-Mesenchymal Transition Inducer Prrx1. Cancer Cell. 2012, 22, 709–724. [Google Scholar] [CrossRef] [PubMed]
- Fazilaty, H.; Rago, L.; Youssef, K.K.; Ocaña, O.H.; Garcia-Asencio, F.; Arcas, A.; Galceran, J.; Nieto, M.A. A gene regulatory network to control EMT programs in development and disease. Nat. Commun. 2019, 10, 5115. [Google Scholar] [CrossRef] [PubMed]
- Rago, L.; Castroviejo, N.; Fazilaty, H.; Garcia-Asencio, F.; Ocaña, O.H.; Galceran, J.; Nieto, M.A. MicroRNAs Establish the Right-Handed Dominance of the Heart Laterality Pathway in Vertebrates. Dev. Cell 2019, 51, 446–459. [Google Scholar] [CrossRef]
- Welsh, I.C.; Kwak, H.; Chen, F.L.; Werner, M.; Shopland, L.S.; Danko, C.G.; Lis, J.T.; Zhang, M.; Martin, J.F.; Kurpios, N.A. Chromatin Architecture of the Pitx2 Locus Requires CTCF- and Pitx2-Dependent Asymmetry that Mirrors Embryonic Gut Laterality. Cell Rep. 2015, 13, 337–349. [Google Scholar] [CrossRef]
- Gore-Panter, S.R.; Hsu, J.; Barnard, J.; Moravec, C.S.; Van Wagoner, D.R.; Chung, M.K.; Smith, J.D. PANCR, the PITX2 Adjacent Noncoding RNA, Is Expressed in Human Left Atria and Regulates PITX2c Expression. Circ. Arrhythmia Electrophysiol. 2016, 9, e003197. [Google Scholar] [CrossRef]
- Cao, Y.; Duca, S.; Cao, J. Epicardium in Heart Development. Cold Spring Harb. Perspect. Biol. 2019, 12, a037192. [Google Scholar] [CrossRef]
- Quijada, P.; Trembley, M.A.; Small, E.M. The Role of the Epicardium During Heart Development and Repair. Circ. Res. 2020, 126, 377–394. [Google Scholar] [CrossRef]
- Pomares, J.M.P.; De La Pompa, J.L. Signaling During Epicardium and Coronary Vessel Development. Circ. Res. 2011, 109, 1429–1442. [Google Scholar] [CrossRef]
- Dueñas, A.; Aranega, A.E.; Franco, D. More than Just a Simple Cardiac Envelope; Cellular Contributions of the Epicardium. Front. Cell Dev. Biol. 2017, 5, 44. [Google Scholar] [CrossRef]
- Sridurongrit, S.; Larsson, J.; Schwartz, R.; Ruiz-Lozano, P.; Kaartinen, V. Signaling via the Tgf-β type I receptor Alk5 in heart development. Dev. Biol. 2008, 322, 208–218. [Google Scholar] [CrossRef]
- Azhar, M.; Schultz, J.E.J.; Grupp, I.; Dorn, G.W.; Meneton, P.; Molin, D.G.; Groot, A.C.G.-D.; Doetschman, T. Transforming growth factor beta in cardiovascular development and function. Cytokine Growth Factor Rev. 2003, 14, 391–407. [Google Scholar] [CrossRef]
- Allison, P.; Espiritu, D.; Barnett, J.V.; Camenisch, T.D. Type III TGFβ receptor and Src direct hyaluronan-mediated invasive cell motility. Cell. Signal. 2014, 27, 453–459. [Google Scholar] [CrossRef]
- Austin, A.F.; Compton, L.A.; Love, J.D.; Brown, C.B.; Barnett, J.V. Primary and immortalized mouse epicardial cells undergo differentiation in response to TGFβ. Dev. Dyn. 2008, 237, 366–376. [Google Scholar] [CrossRef]
- Clark, C.R.; Robinson, J.Y.; Sanchez, N.S.; Townsend, T.A.; Arrieta, J.A.; Merryman, W.D.; Trykall, D.Z.; Olivey, H.E.; Hong, C.C.; Barnett, J.V. Common pathways regulate Type III TGFβ receptor-dependent cell invasion in epicardial and endocardial cells. Cell. Signal. 2016, 28, 688–698. [Google Scholar] [CrossRef]
- Bax, N.A.M.; Van Oorschot, A.A.M.; Maas, S.; Braun, J.; Van Tuyn, J.; De Vries, A.A.F.; Groot, A.C.G.-D.; Goumans, M.-J. In vitro epithelial-to-mesenchymal transformation in human adult epicardial cells is regulated by TGFβ-signaling and WT1. Basic Res. Cardiol. 2011, 106, 829–847. [Google Scholar] [CrossRef]
- Hill, C.R.; Sanchez, N.S.; Love, J.D.; Arrieta, J.A.; Hong, C.C.; Brown, C.B.; Austin, A.F.; Barnett, J.V. BMP2 signals loss of epithelial character in epicardial cells but requires the Type III TGFβ receptor to promote invasion. Cell. Signal. 2012, 24, 1012–1022. [Google Scholar] [CrossRef][Green Version]
- Morabito, C.J.; Dettman, R.; Kattan, J.; Collier, J.M.; Bristow, J. Positive and Negative Regulation of Epicardial–Mesenchymal Transformation during Avian Heart Development. Dev. Biol. 2001, 234, 204–215. [Google Scholar] [CrossRef]
- Sánchez, N.S.; Barnett, J.V. TGFβ and BMP-2 regulate epicardial cell invasion via TGFβR3 activation of the Par6/Smurf1/RhoA pathway. Cell. Signal. 2012, 24, 539–548. [Google Scholar] [CrossRef]
- Brønnum, H.; Andersen, D.C.; Schneider, M.; Sandberg, M.B.; Eskildsen, T.; Nielsen, S.B.; Kalluri, R.; Sheikh, S.P. miR-21 Promotes Fibrogenic Epithelial-to-Mesenchymal Transition of Epicardial Mesothelial Cells Involving Programmed Cell Death 4 and Sprouty-1. PLoS ONE 2013, 8, e56280. [Google Scholar] [CrossRef]
- Pontemezzo, E.; Foglio, E.; Vernucci, E.; Magenta, A.; D’Agostino, M.; Sileno, S.; Astanina, E.; Bussolino, F.; Pellegrini, L.; Germani, A.; et al. miR-200c-3p Regulates Epitelial-to-Mesenchymal Transition in Epicardial Mesothelial Cells by Targeting Epicardial Follistatin-Related Protein 1. Int. J. Mol. Sci. 2021, 22, 4971. [Google Scholar] [CrossRef]
- Tandon, P.; Miteva, Y.V.; Kuchenbrod, L.M.; Cristea, I.M.; Conlon, F.L. Tcf21 regulates the specification and maturation of proepicardial cells. Development 2013, 140, 2409–2421. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Lin, S.; Wang, S.; Chen, X. The Role of Transcription Factor 21 in Epicardial Cell Differentiation and the Development of Coronary Heart Disease. Front Cell Dev. Biol. 2020, 8, 457. [Google Scholar] [CrossRef] [PubMed]
- Seeger, T.; Xu, Q.-F.; Muhly-Reinholz, M.; Fischer, A.; Kremp, E.-M.; Zeiher, A.M.; Dimmeler, S. Inhibition of let-7 augments the recruitment of epicardial cells and improves cardiac function after myocardial infarction. J. Mol. Cell. Cardiol. 2016, 94, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.M.; Dykeman, C.A.; King, B.L.; Yin, V.P. Modulation of TNFα Activity by the microRNA Let-7 Coordi-nates Zebrafish Heart Regeneration. iScience 2019, 15, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.L.; Haas, U.; Diaz, R.; Leeper, N.J.; Kundu, R.K.; Patlolla, B.; Assimes, T.L.; Kaiser, F.J.; Perisic, L.; Hedin, U.; et al. Coronary Heart Disease-Associated Variation in TCF21 Disrupts a miR-224 Binding Site and miRNA-Mediated Regulation. PLoS Genet. 2014, 10, e1004263. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Gao, X.; Luo, H.; Huang, Q.; Su, D.; Tan, X.; Lu, C. TCF21 rs12190287 Polymorphisms Are Associated with Ventricu-lar Septal Defects in a Chinese Population. Genet. Test. Mol. Biomark. 2017, 21, 312–315. [Google Scholar] [CrossRef]
- Dechamethakun, S.; Ikeda, S.; Arai, T.; Sato, N.; Sawabe, M.; Muramatsu, M. Associations between the CDKN2A/B, ADTRP and PDGFD Polymorphisms and the Development of Coronary Atherosclerosis in Japanese Patients. J. Atheroscler. Thromb. 2014, 21, 680–690. [Google Scholar] [CrossRef]
- Fujimaki, T.; Oguri, M.; Horibe, H.; Kato, K.; Matsuoka, R.; Abe, S.; Tokoro, F.; Arai, M.; Noda, T.; Watanabe, S.; et al. Association of a transcription factor 21 gene polymorphism with hypertension. Biomed. Rep. 2015, 3, 118–122. [Google Scholar] [CrossRef]
- Wirtwein, M.; Melander, O.; Sjőgren, M.; Hoffmann, M.; Narkiewicz, K.; Gruchala, M.; Sobiczewski, W. Relationship between selected DNA polymorphisms and coronary artery disease complications. Int. J. Cardiol. 2016, 228, 814–820. [Google Scholar] [CrossRef]
- Zhao, Q.; Wirka, R.; Nguyen, T.; Nagao, M.; Cheng, P.; Miller, C.L.; Kim, J.B.; Pjanic, M.; Quertermous, T. TCF21 and AP-1 interact through epigenetic modifi-cations to regulate coronary artery disease gene expression. Genome Med. 2019, 11, 23. [Google Scholar] [CrossRef]
- Nagao, M.; Lyu, Q.; Zhao, Q.; Wirka, R.C.; Bagga, J.; Nguyen, T.; Cheng, P.; Kim, J.; Pjanic, M.; Miano, J.M.; et al. Coronary Disease-Associated Gene TCF21 Inhibits Smooth Muscle Cell Differentiation by Blocking the Myocardin-Serum Response Factor Pathway. Circ. Res. 2020, 126, 517–529. [Google Scholar] [CrossRef]
- Bastami, M.; Ghaderian, S.M.; Omrani, M.D.; Mirfakhraie, R.; Vakili, H.; Parsa, S.A.; Nariman-Saleh-Fam, Z.; Masotti, A. MiRNA-Related Polymorphisms in miR-146a and TCF21 Are Associated with Increased Susceptibility to Coronary Artery Disease in an Iranian Population. Genet. Test. Mol. Biomark. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Aminu, A.J.; Petkova, M.; Chen, W.; Yin, Z.; Kuzmin, V.S.; Atkinson, A.J.; Dobrzynski, H. MiR-486-3p and MiR-938—Important Inhibitors of Pacemaking Ion Channels and/or Markers of Immune Cells. Appl. Sci. 2021, 11, 11366. [Google Scholar] [CrossRef]
- Yanni, J.; D’Souza, A.; Wang, Y.; Li, N.; Hansen, B.J.; Zakharkin, S.O.; Smith, M.; Hayward, C.; Whitson, B.A.; Mohler, P.J.; et al. Silencing miR-370-3p rescues funny current and sinus node function in heart failure. Sci. Rep. 2020, 10, 11279. [Google Scholar] [CrossRef]
- Carmona, R.; Guadix, J.A.; Cano, E.; Ruiz-Villalba, A.; Portillo-Sánchez, V.; Pérez-Pomares, J.M.; Muñoz-Chápuli, R. The embryonic epicardium: An essential element of cardiac development. J. Cell. Mol. Med. 2010, 14, 2066–2072. [Google Scholar] [CrossRef]
- Martínez-Estrada, O.M.; Lettice, A.L.; Essafi, A.; Guadix, J.A.; Slight, J.; Velecela, V.; Hall, E.; Reichmann, J.; Devenney, P.S.; Hohenstein, P.; et al. Wt1 is required for cardiovascular progenitor cell formation through transcriptional control of Snail and E-cadherin. Nat. Genet. 2009, 42, 89–93. [Google Scholar] [CrossRef]
- von Gise, A.; Zhou, B.; Honor, L.B.; Ma, Q.; Petryk, A.; Pu, W.T. WT1 regulates epicardial epithelial to mesenchymal transition through β-catenin and retinoic acid signaling pathways. Dev. Biol. 2011, 356, 421–431. [Google Scholar] [CrossRef]
- Bakker, M.L.; Christoffels, V.M.; Moorman, A.F.M. The Cardiac Pacemaker and Conduction System Develops From Embryonic Myocardium that Retains Its Primitive Phenotype. J. Cardiovasc. Pharmacol. 2010, 56, 6–15. [Google Scholar] [CrossRef]
- Lv, L.; Chen, G.; Zhou, J.; Li, J.; Gong, J. WT1-AS promotes cell apoptosis in hepatocellular carcinoma through down-regulating of WT1. J. Exp. Clin. Cancer Res. 2015, 34, 119. [Google Scholar] [CrossRef]
- Zhang, Y.; Na, R.; Wang, X. LncRNA WT1-AS up-regulates p53 to inhibit the proliferation of cervical squamous carcino-ma cells. BMC Cancer 2019, 19, 1052. [Google Scholar] [CrossRef]
- Wang, Q.; Ge, X.; Zhang, J.; Chen, L. Effect of lncRNA WT1-AS regulating WT1 on oxidative stress injury and apoptosis of neurons in Alzheimer’s disease via inhibition of the miR-375/SIX4 axis. Aging 2020, 12, 23974–23995. [Google Scholar] [CrossRef]
- Christoffels, V.M.; Smits, G.J.; Kispert, A.; Moorman, A.F.M. Development of the Pacemaker Tissues of the Heart. Circ. Res. 2010, 106, 240–254. [Google Scholar] [CrossRef]
- Hoogaars, W.M.H.; Barnett, P.; Moorman, A.F.M.; Christoffels, V.M. Cardiovascular development: Towards biomedical applicability. Cell. Mol. Life Sci. 2007, 64, 646. [Google Scholar] [CrossRef]
- Singh, R.; Hoogaars, W.M.; Barnett, P.; Grieskamp, T.; Rana, M.S.; Buermans, H.; Farin, H.F.; Petry, M.; Heallen, T.; Martin, J.F.; et al. Tbx2 and Tbx3 induce atrioventricular myocardial development and endocardial cushion formation. Cell. Mol. Life Sci. 2012, 69, 1377–1389. [Google Scholar] [CrossRef]
- Wang, J.; Bai, Y.; Li, N.; Ye, W.; Zhang, M.; Greene, S.B.; Tao, Y.; Chen, Y.; Wehrens, X.H.; Martin, J.F. Pitx2-microRNA pathway that delimits sinoatrial node development and inhibits predisposition to atrial fibrillation. Proc. Natl. Acad. Sci. USA 2014, 111, 9181–9186. [Google Scholar] [CrossRef]
- Benzoni, P.; Nava, L.; Giannetti, F.; Guerini, G.; Gualdoni, A.; Bazzini, C.; Milanesi, R.; Bucchi, A.; Baruscotti, M.; Barbuti, A. Dual role of miR-1 in the development and function of sinoatrial cells. J. Mol. Cell. Cardiol. 2021, 157, 104–112. [Google Scholar] [CrossRef]
- D’Souza, A.; Bucchi, A.; Johnsen, A.B.; Logantha, S.J.; Monfredi, O.; Yanni, J.; Prehar, S.; Hart, G.; Cartwright, E.; Wisløff, U.; et al. Exercise training reduces resting heart rate via downregulation of the funny channel HCN4. Nat. Commun. 2014, 5, 3775. [Google Scholar] [CrossRef]
- Zhao, H.; Wang, F.; Zhang, W.; Yang, M.; Tang, Y.; Wang, X.; Zhao, Q.; Huang, C. Overexpression of TBX3 in human induced pluripotent stem cells (hiPSCs) increases their differentiation into cardiac pacemaker-like cells. Biomed. Pharmacother. 2020, 130, 110612. [Google Scholar] [CrossRef]
- Jiang, K.; Ren, C.; Nair, V. MicroRNA-137 represses Klf4 and Tbx3 during differentiation of mouse embryonic stem cells. Stem Cell Res. 2013, 11, 1299–1313. [Google Scholar] [CrossRef]
- Han, J.; Yuan, P.; Yang, H.; Zhang, J.; Soh, B.S.; Li, P.; Lim, S.L.; Cao, S.; Tay, J.; Orlov, Y.; et al. Tbx3 improves the germ-line competency of induced pluripotent stem cells. Nature 2010, 463, 1096–1100. [Google Scholar] [CrossRef]
- Russell, R.; Ilg, M.; Lin, Q.; Wu, G.; Lechel, A.; Bergmann, W.; Eiseler, T.; Linta, L.; Kumar, P.P.; Klingenstein, M.; et al. A Dynamic Role of TBX3 in the Pluripotency Circuitry. Stem Cell Rep. 2015, 5, 1155–1170. [Google Scholar] [CrossRef] [PubMed]
- Niwa, H.; Ogawa, K.; Shimosato, D.; Adachi, K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature 2009, 460, 118–122. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Liu, M.; Zhao, C.; Chen, C.; Kong, Q.; Cai, Z.; Li, D. MiR-1/133 attenuates cardiomyocyte apoptosis and electrical remodeling in mice with viral myocarditis. Cardiol. J. 2020, 27, 285–294. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, J.; Huo, C.; Ding, N.; Li, J.; Xiao, J.; Lin, X.; Cai, B.; Zhang, Y.; Xu, J. Dynamic Organization of lncRNA and Circular RNA Regulators Collectively Controlled Cardiac Differentiation in Humans. EBioMedicine 2017, 24, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Salguero-Jiménez, A.; Grego-Bessa, J.; D’Amato, G.; Jiménez-Borreguero, L.J.; De La Pompa, J.L. Myocardial Notch1-Rbpj deletion does not affect NOTCH signaling, heart development or function. PLoS ONE 2018, 13, e0203100. [Google Scholar] [CrossRef]
- MacGrogan, D.; Münch, J.; De La Pompa, J.L. Notch and interacting signalling pathways in cardiac development, disease, and regeneration. Nat. Rev. Cardiol. 2018, 15, 685–704. [Google Scholar] [CrossRef] [PubMed]
- Luxán, G.; D’Amato, G.; MacGrogan, D.; De La Pompa, J.L. Endocardial Notch Signaling in Cardiac Development and Disease. Circ. Res. 2016, 118, e1–e18. [Google Scholar] [CrossRef]
- Gassmann, M.; Casagranda, F.; Orioli, D.; Simon, H.; Lai, C.; Klein, R.; Lemke, G. Aberrant neural and cardiac development in mice lacking the ErbB4 neuregulin receptor. Nature 1995, 378, 390–394. [Google Scholar] [CrossRef]
- Lee, K.-F.; Simon, H.; Chen, H.; Bates, B.; Hung, M.-C.; Hauser, C. Requirement for neuregulin receptor erbB2 in neural and cardiac development. Nature 1995, 378, 394–398. [Google Scholar] [CrossRef]
- Rupert, C.; Coulombe, K.L. The Roles of Neuregulin-1 in Cardiac Development, Homeostasis, and Disease. Biomark. Insights 2015, 10s1, BMI-S20061. [Google Scholar] [CrossRef]
- Kirabo, A.; Ryzhov, S.; Gupte, M.; Sengsayadeth, S.; Gumina, R.J.; Sawyer, D.B.; Galindo, C.L. Neuregulin-1β induces proliferation, survival and paracrine signaling in normal human cardiac ventricular fibroblasts. J. Mol. Cell. Cardiol. 2017, 105, 59–69. [Google Scholar] [CrossRef]
- Sun, M.; Yan, X.; Bian, Y.; Caggiano, A.O.; Morgan, J.P. Improving murine embryonic stem cell differentiation into cardiomyocytes with neuregulin-1: Differential expression of microRNA. Am. J. Physiol. Physiol. 2011, 301, C21–C30. [Google Scholar] [CrossRef]
- von Gise, A.; Lin, Z.; Schlegelmilch, K.; Honor, L.B.; Pan, G.M.; Buck, J.N.; Ma, Q.; Ishiwata, T.; Zhou, B.; Camargo, F.D.; et al. YAP1, the nuclear target of Hippo signaling, stimulates heart growth through cardiomyocyte proliferation but not hypertrophy. Proc. Natl. Acad. Sci. USA 2012, 109, 2394–2399. [Google Scholar] [CrossRef]
- Xiao, Y.; Leach, J.; Wang, J.; Martin, J.F. Hippo/Yap Signaling in Cardiac Development and Regeneration. Curr. Treat. Opt. Cardiovasc. Med. 2016, 18, 38. [Google Scholar] [CrossRef]
- Moya, I.M.; Halder, G. Hippo–YAP/TAZ signalling in organ regeneration and regenerative medicine. Nat. Rev. Mol. Cell Biol. 2018, 20, 211–226. [Google Scholar] [CrossRef]
- Mia, M.M.; Singh, M.K. The Hippo Signaling Pathway in Cardiac Development and Diseases. Front. Cell Dev. Biol. 2019, 7, 211. [Google Scholar] [CrossRef]
- Xie, Y.; Wang, Q.; Gao, N.; Wu, F.; Lan, F.; Zhang, F.; Jin, L.; Huang, Z.; Ge, J.; Wang, H.; et al. MircroRNA-10b Promotes Human Embryonic Stem Cell-Derived Cardiomyocyte Proliferation via Novel Target Gene LATS1. Mol. Ther. Nucleic Acids 2019, 19, 437–445. [Google Scholar] [CrossRef]
- Wu, K.-H.; Xiao, Q.-R.; Yang, Y.; Xu, J.-L.; Zhang, F.; Liu, C.-M.; Zhang, Z.-M.; Lu, Y.-Q.; Huang, N.-P. MicroRNA-34a modulates the Notch signaling pathway in mice with congenital heart disease and its role in heart development. J. Mol. Cell. Cardiol. 2017, 114, 300–308. [Google Scholar] [CrossRef]
- Han, C.; Song, Y.; Lian, C. MiR-769 Inhibits Colorectal Cancer Cell Proliferation and Invasion by Targeting HEY1. Med. Sci. Monit. 2018, 24, 9232–9239. [Google Scholar] [CrossRef]
- Wei, B.; Liu, Y.; Guan, H. MicroRNA-145-5p attenuates high glucose-induced apoptosis by targeting the Notch signaling pathway in podocytes. Exp. Ther. Med. 2020, 19, 1915–1924. [Google Scholar] [CrossRef]
- Lina, S.; Lihong, Q.; Di, Y.; Bo, Y.; Xiaolin, L.; Jing, M. microRNA-146a and Hey2 form a mutual negative feedback loop to regulate the inflammatory response in chronic apical periodontitis. J. Cell. Biochem. 2018, 120, 645–657. [Google Scholar] [CrossRef]
- Wu, D.-C.; Zhang, M.-F.; Su, S.-G.; Fang, H.-Y.; Wang, X.-H.; He, D.; Xie, Y.-Y.; Liu, X.-H. HEY2, a target of miR-137, indicates poor outcomes and promotes cell proliferation and migration in hepatocellular carcinoma. Oncotarget 2016, 7, 38052–38063. [Google Scholar] [CrossRef]
- Bao, Y.; Lu, Y.; Feng, W.; Yu, H.; Tao, Y.; Shi, Q.; Chen, W.; Wang, X. COUP-TFII promotes epithelial-mesenchymal transition by inhibiting miR-34a expression in colorectal cancer. Int. J. Oncol. 2019, 54, 1337–1344. [Google Scholar] [CrossRef] [PubMed]
- Jeong, B.-C.; Kang, I.-H.; Hwang, Y.-C.; Kim, S.-H.; Koh, J.-T. MicroRNA-194 reciprocally stimulates osteogenesis and inhibits adipogenesis via regulating COUP-TFII expression. Cell Death Dis. 2014, 5, e1532. [Google Scholar] [CrossRef] [PubMed]
- Kang, I.-H.; Jeong, B.-C.; Hur, S.-W.; Choi, H.; Choi, S.-H.; Ryu, J.-H.; Hwang, Y.-C.; Koh, J.-T. MicroRNA-302a Stimulates Osteoblastic Differentiation by Repressing COUP-TFII Expression. J. Cell. Physiol. 2014, 230, 911–921. [Google Scholar] [CrossRef] [PubMed]
- Lu, F.; Langenbacher, A.; Chen, J.-N. Tbx20 drives cardiac progenitor formation and cardiomyocyte proliferation in zebrafish. Dev. Biol. 2016, 421, 139–148. [Google Scholar] [CrossRef]
- Gupta, M.K.; Rao, T.N. Hearty miR-363 controls HAND1 in cardiac cell specification. Stem Cell Res. Ther. 2014, 5, 89. [Google Scholar] [CrossRef]
- Wagh, V.; Pomorski, A.; Wilschut, K.J.; Piombo, S.; Bernstein, H.S. MicroRNA-363 negatively regulates the left ventricular determining transcription factor HAND1 in human embryonic stem cell-derived cardiomyocytes. Stem Cell Res. Ther. 2014, 5, 75. [Google Scholar] [CrossRef]
- Cao, L.; Kong, L.-P.; Yu, Z.-B.; Han, S.-P.; Bai, Y.-F.; Zhu, J.; Hu, X.; Zhu, C.; Zhu, S.; Guo, X.-R. microRNA expression profiling of the developing mouse heart. Int. J. Mol. Med. 2012, 30, 1095–1104. [Google Scholar] [CrossRef]
- Zhao, Y.; Samal, E.; Srivastava, D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 2005, 436, 214–220. [Google Scholar] [CrossRef]
- Vo, N.K.; Dalton, R.P.; Liu, N.; Olson, E.N.; Goodman, R.H. Affinity purification of microRNA-133a with the cardiac transcription factor, Hand2. Proc. Natl. Acad. Sci. USA 2010, 107, 19231–19236. [Google Scholar] [CrossRef]
- Liu, L.; Yuan, Y.; He, X.; Xia, X.; Mo, X. MicroRNA-1 upregulation promotes myocardiocyte proliferation and suppresses apoptosis during heart development. Mol. Med. Rep. 2017, 15, 2837–2842. [Google Scholar] [CrossRef][Green Version]
- Lavallée, G.; Andelfinger, G.; Nadeau, M.; Lefebvre, C.; Nemer, G.; Horb, M.E.; Nemer, M. The Kruppel-like transcription factor KLF13 is a novel regulator of heart development. EMBO J. 2006, 25, 5201–5213. [Google Scholar] [CrossRef]
- Gu, M.; Wang, J.; Wang, Y.; Xu, Y.; Zhang, Y.; Wu, W.; Liao, S. MiR-147b inhibits cell viability and promotes apoptosis of rat H9c2 cardiomyocytes via down-regulating KLF13 expression. Acta Biochim. Biophys. Sin. 2018, 50, 288–297. [Google Scholar] [CrossRef]
- Bayoumi, A.S.; Park, K.-M.; Wang, Y.; Teoh, J.-P.; Aonuma, T.; Tang, Y.; Su, H.; Weintraub, N.L.; Kim, I.-M. A carvedilol-responsive microRNA, miR-125b-5p protects the heart from acute myocardial infarction by repressing pro-apoptotic bak1 and klf13 in cardiomyocytes. J. Mol. Cell. Cardiol. 2017, 114, 72–82. [Google Scholar] [CrossRef]
- Mikhailov, A.T.; Torrado, M. Myocardial transcription factors in diastolic dysfunction: Clues for model systems and disease. Heart Fail. Rev. 2016, 21, 783–794. [Google Scholar] [CrossRef]
- Kong, L.; Hu, N.; Du, X.; Wang, W.; Chen, H.; Li, W.; Wei, S.; Zhuang, H.; Li, X.; Li, C. Upregulation of miR-483-3p contributes to endothelial progenitor cells dysfunction in deep vein thrombosis patients via SRF. J. Transl. Med. 2016, 14, 23. [Google Scholar] [CrossRef]
- Xu, D.; Guo, Y.; Liu, T.; Li, S.; Sun, Y. miR-22 contributes to endosulfan-induced endothelial dysfunction by targeting SRF in HUVECs. Toxicol. Lett. 2017, 269, 33–40. [Google Scholar] [CrossRef]
- You, G.; Zu, B.; Wang, B.; Fu, Q.; Li, F. Identification of miRNA–mRNA–TFs Regulatory Network and Crucial Pathways Involved in Tetralogy of Fallot. Front. Genet. 2020, 11, 552. [Google Scholar] [CrossRef]
- Wei, X.; Hou, X.; Li, J.; Liu, Y. miRNA-181a/b Regulates Phenotypes of Vessel Smooth Muscle Cells Through Serum Response Factor. DNA Cell Biol. 2017, 36, 127–135. [Google Scholar] [CrossRef]
- Bolte, C.; Zhang, Y.; Wang, I.-C.; Kalin, T.V.; Molkentin, J.D.; Kalinichenko, V.V. Expression of Foxm1 Transcription Factor in Cardiomyocytes Is Required for Myocardial Development. PLoS ONE 2011, 6, e22217. [Google Scholar] [CrossRef]
- Chen, F.; Kook, H.; Milewski, R.; Gitler, A.D.; Lu, M.M.; Li, J.; Nazarian, R.; Schnepp, R.; Jen, K.-Y.; Biben, C.; et al. Hop Is an Unusual Homeobox Gene that Modulates Cardiac Development. Cell 2002, 110, 713–723. [Google Scholar] [CrossRef]
- Hadji, F.; Boulanger, M.-C.; Guay, S.-P.; Gaudreault, N.; Amellah, S.; Mkannez, G.; Bouchareb, R.; Marchand, J.T.; Nsaibia, M.J.; Guauque-Olarte, S.; et al. Altered DNA Methylation of Long Noncoding RNA H19 in Calcific Aortic Valve Disease Promotes Mineralization by Silencing NOTCH1. Circulation 2016, 134, 1848–1862. [Google Scholar] [CrossRef]
- Anderson, K.M.; Anderson, D.M.; McAnally, J.R.; Shelton, J.M.; Bassel-Duby, R.; Olson, E.N. Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature 2016, 539, 433–436. [Google Scholar] [CrossRef]
- Han, X.; Zhang, J.; Liu, Y.; Fan, X.; Ai, S.; Luo, Y.; Li, X.; Jin, H.; Luo, S.; Zheng, H.; et al. The lncRNA Hand2os1/Uph locus orchestrates heart development through regulation of precise expression of Hand2. Development 2019, 146, dev176198. [Google Scholar] [CrossRef]
- Anderson, D.M.; Anderson, K.M.; Nelson, B.R.; McAnally, J.R.; Bezprozvannaya, S.; Shelton, J.M.; Bassel-Duby, R.; Olson, E.N. A myocardin-adjacent lncRNA balances SRF-dependent gene transcription in the heart. Genes Dev. 2021, 35, 835–840. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, W.; Lin, M.; Wu, W.; Jiang, P.; Tou, E.; Xue, M.; Richards, A.; Jourd’Heuil, D.; Asif, A.; et al. MYOSLID Is a Novel Serum Response Factor–Dependent Long Noncoding RNA That Amplifies the Vascular Smooth Muscle Differentiation Program. Arter. Thromb. Vasc. Biol. 2016, 36, 2088–2099. [Google Scholar] [CrossRef]
- Liu, J.; Li, Y.; Lin, B.; Sheng, Y.; Yang, L. HBL1 Is a Human Long Noncoding RNA that Modulates Cardiomyocyte Development from Pluripotent Stem Cells by Counteracting MIR1. Dev. Cell 2017, 42, 333–348.e5. [Google Scholar] [CrossRef]
- Xiao, X.; Zhou, T.; Guo, S.; Guo, C.; Zhang, Q.; Dong, N.; Wang, Y. LncRNA MALAT1 sponges miR-204 to promote osteoblast differentiation of human aortic valve interstitial cells through up-regulating Smad4. Int. J. Cardiol. 2017, 243, 404–412. [Google Scholar] [CrossRef]
- Leisegang, M.S.; Bibli, S.-I.; Günther, S.; Pflüger-Müller, B.; Oo, J.A.; Höper, C.; Seredinski, S.; Yekelchyk, M.; Schmitz-Rixen, T.; Schürmann, C.; et al. Pleiotropic effects of laminar flow and statins depend on the Krüppel-like factor-induced lncRNA MANTIS. Eur. Heart J. 2019, 40, 2523–2533. [Google Scholar] [CrossRef] [PubMed]
- Shyu, K.; Wang, B.; Fang, W.; Pan, C.; Lin, C. Hyperbaric oxygen-induced long non-coding RNA MALAT1 exosomes suppress MicroRNA-92a expression in a rat model of acute myocardial infarction. J. Cell. Mol. Med. 2020, 24, 12945–12954. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Wang, J.; Morikawa, Y.; Bonilla-Claudio, M.; Klysik, E.; Martin, J.F. Bmp signaling represses Vegfa to promote outflow tract cushion development. Development 2013, 140, 3395–3402. [Google Scholar] [CrossRef] [PubMed]
- Stottmann, R.W.; Choi, M.; Mishina, Y.; Meyers, E.N.; Klingensmith, J. BMP receptor IA is required in mammalian neural crest cells for development of the cardiac outflow tract and ventricular myocardium. Development 2004, 131, 2205–2218. [Google Scholar] [CrossRef]
- Cai, C.-L.; Liang, X.; Shi, Y.; Chu, P.-H.; Pfaff, S.L.; Chen, J.; Evans, S. Isl1 Identifies a Cardiac Progenitor Population that Proliferates Prior to Differentiation and Contributes a Majority of Cells to the Heart. Dev. Cell 2003, 5, 877–889. [Google Scholar] [CrossRef]
- Engleka, K.A.; Manderfield, L.J.; Brust, R.D.; Li, L.; Cohen, A.; Dymecki, S.M.; Epstein, J.A. Islet1Derivatives in the Heart Are of Both Neural Crest and Second Heart Field Origin. Circ. Res. 2012, 110, 922–926. [Google Scholar] [CrossRef]
- Liu, W.; Selever, J.; Wang, D.; Lu, M.-F.; Moses, K.A.; Schwartz, R.J.; Martin, J.F. Bmp4 signaling is required for outflow-tract septation and branchial-arch artery remodeling. Proc. Natl. Acad. Sci. USA 2004, 101, 4489–4494. [Google Scholar] [CrossRef]
- Solloway, M.J.; Robertson, E.J. Early embryonic lethality in Bmp5; Bmp7 double mutant mice suggests functional redundancy within the 60A subgroup. Development 1999, 126, 1753–1768. [Google Scholar] [CrossRef]
- Dobaczewski, M.; Chen, W.; Frangogiannis, N.G. Transforming growth factor (TGF)-β signaling in cardiac remodeling. J. Mol. Cell. Cardiol. 2011, 51, 600–606. [Google Scholar] [CrossRef]
- Hurlstone, A.; Haramis, A.-P.G.; Wienholds, E.; Begthel, H.; Korving, J.; Van Eeden, F.; Cuppen, E.; Zivkovic, D.; Plasterk, R.H.A.; Clevers, H. The Wnt/β-catenin pathway regulates cardiac valve formation. Nature 2003, 425, 633–637. [Google Scholar] [CrossRef]
- Jarrett, M.J.; Houk, A.K.; McCuistion, P.E.; Weyant, M.J.; Reece, T.B.; Meng, X.; Fullerton, D.A. Wnt Signaling Mediates Pro-Fibrogenic Activity in Human Aortic Valve Interstitial Cells. Ann. Thorac. Surg. 2021, 112, 519–525. [Google Scholar] [CrossRef]
- Cai, X.; Zhang, W.; Hu, J.; Zhang, L.; Sultana, N.; Wu, B.; Cai, W.; Zhou, B.; Cai, C.-L. Tbx20 acts upstream of Wnt signaling to regulate endocardial cushion formation and valve remodeling during mouse cardiogenesis. Development 2013, 140, 3176–3187. [Google Scholar] [CrossRef]
- Kim, Y.-S.; Kim, M.-J.; Koo, T.-H.; Kim, J.-D.; Koun, S.; Ham, H.J.; Lee, Y.M.; Rhee, M.; Yeo, S.-Y.; Huh, T.-L. Histone deacetylase is required for the activation of Wnt/β-catenin signaling crucial for heart valve formation in zebrafish embryos. Biochem. Biophys. Res. Commun. 2012, 423, 140–146. [Google Scholar] [CrossRef]
- Alfieri, C.M.; Cheek, J.; Chakraborty, S.; Yutzey, K.E. Wnt signaling in heart valve development and osteogenic gene induction. Dev. Biol. 2010, 338, 127–135. [Google Scholar] [CrossRef]
- Verhoeven, M.C.; Haase, C.; Christoffels, V.M.; Weidinger, G.; Bakkers, J. Wnt signaling regulates atrioventricular canal formation upstream of BMP and Tbx2. Birth Defects Res. Part A: Clin. Mol. Teratol. 2011, 91, 435–440. [Google Scholar] [CrossRef]
- Bosada, F.M.; Devasthali, V.; Jones, K.A.; Stankunas, K. Wnt/β-catenin signaling enables developmental transitions during valvulogenesis. Development 2016, 143, 1041–1054. [Google Scholar] [CrossRef]
- Fang, M.; Wang, C.; Zheng, C.; Luo, J.; Hou, S.; Liu, K.; Li, X. Mir-29b promotes human aortic valve interstitial cell calcification via inhibiting TGF-β3 through activation of wnt3/β-catenin/Smad3 signaling. J. Cell. Biochem. 2017, 119, 5175–5185. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, Z.; Zhang, L.; Wang, J.; Zhang, M.; Zhu, B. miR-27a protects human mitral valve interstitial cell from TNF-α-induced inflammatory injury via up-regulation of NELL-1. Braz. J. Med Biol. Res. 2018, 51. [Google Scholar] [CrossRef]
- Lv, X.; Li, J.; Hu, Y.; Wang, S.; Yang, C.; Li, C.; Zhong, G. Overexpression of miR-27b-3p Targeting Wnt3a Regulates the Signaling Pathway of Wnt/β-Catenin and Attenuates Atrial Fibrosis in Rats with Atrial Fibrillation. Oxid. Med. Cell. Longev. 2019, 2019, 1–13. [Google Scholar] [CrossRef]
- Yang, M.; Kong, D.; Chen, J. Inhibition of miR-148b ameliorates myocardial ischemia/reperfusion injury via regulation of Wnt/β-catenin signaling pathway. J. Cell. Physiol. 2019, 234, 17757–17766. [Google Scholar] [CrossRef]
- Nakajima, Y.; Miyazono, K.; Kato, M.; Takase, M.; Yamagishi, T.; Nakamura, H. Extracellular Fibrillar Structure of Latent TGFβ Binding Protein-1: Role in TGFβ-dependent Endothelial-Mesenchymal Transformation during Endocardial Cushion Tissue Formation in Mouse Embryonic Heart. J. Cell Biol. 1997, 136, 193–204. [Google Scholar] [CrossRef]
- Brown, C.B.; Boyer, A.S.; Runyan, R.B.; Barnett, J.V. Requirement of Type III TGF-β Receptor for Endocardial Cell Transformation in the Heart. Science 1999, 283, 2080–2082. [Google Scholar] [CrossRef]
- Todorovic, V.; Finnegan, E.; Freyer, L.; Zilberberg, L.; Ota, M.; Rifkin, D.B. Long form of latent TGF-β binding protein 1 (Ltbp1L) regulates cardiac valve development. Dev. Dyn. 2010, 240, 176–187. [Google Scholar] [CrossRef]
- Ten Dijke, P.; Egorova, A.D.; Goumans, M.-J.T.H.; Poelmann, R.E.; Hierck, B.P. TGF-β Signaling in Endothelial-to-Mesenchymal Transition: The Role of Shear Stress and Primary Cilia. Sci. Signal. 2012, 5, pt2. [Google Scholar] [CrossRef]
- Wang, J.; Sridurongrit, S.; Dudas, M.; Thomas, P.; Nagy, A.; Schneider, M.; Epstein, J.A.; Kaartinen, V. Atrioventricular cushion transformation is mediated by ALK2 in the developing mouse heart. Dev. Biol. 2005, 286, 299–310. [Google Scholar] [CrossRef]
- Ramsdell, A.F.; Markwald, R.R. Induction of Endocardial Cushion Tissue in the Avian Heart is Regulated, in Part, by TGFβ-3-Mediated Autocrine Signaling. Dev. Biol. 1997, 188, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Townsend, T.A.; Robinson, J.Y.; How, T.; DeLaughter, D.M.; Blobe, G.C.; Barnett, J.V. Endocardial cell epithelial-mesenchymal transformation requires Type III TGFβ receptor interaction with GIPC. Cell. Signal. 2012, 24, 247–256. [Google Scholar] [CrossRef]
- Lagendijk, A.K.; Goumans, M.J.; Burkhard, S.B.; Bakkers, J. MicroRNA-23 Restricts Cardiac Valve Formation by Inhibiting Has2 and Extracellular Hyaluronic Acid Production. Circ. Res. 2011, 109, 649–657. [Google Scholar] [CrossRef]
- Bonet, F.; Dueñas, Á.; López-Sánchez, C.; García-Martínez, V.; Aránega, A.E.; Franco, D. MiR-23b and miR-199a impair epithelial-to-mesenchymal transition during atrioventricular endocardial cushion formation. Dev. Dyn. 2015, 244, 1259–1275. [Google Scholar] [CrossRef]
- Koenig, S.N.; Bosse, K.; Majumdar, U.; Bonachea, E.M.; Radtke, F.; Garg, V. Endothelial Notch1 Is Required for Proper Development of the Semilunar Valves and Cardiac Outflow Tract. J. Am. Heart Assoc. 2016, 5, e003075. [Google Scholar] [CrossRef]
- Yang, J.-H.; Wylie-Sears, J.; Bischoff, J. Opposing actions of Notch1 and VEGF in post-natal cardiac valve endothelial cells. Biochem. Biophys. Res. Commun. 2008, 374, 512–516. [Google Scholar] [CrossRef] [PubMed]
- Nigam, V.; Srivastava, D. Notch1 represses osteogenic pathways in aortic valve cells. J. Mol. Cell. Cardiol. 2009, 47, 828–834. [Google Scholar] [CrossRef] [PubMed]
- Bosse, K.; Hans, C.P.; Zhao, N.; Koenig, S.N.; Huang, N.; Guggilam, A.; LaHaye, S.; Tao, G.; Lucchesi, P.A.; Lincoln, J.; et al. Endothelial nitric oxide signaling regulates Notch1 in aortic valve disease. J. Mol. Cell. Cardiol. 2013, 60, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Timmerman, L.A.; Grego-Bessa, J.; Raya, A.; Bertrán, E.; Pérez-Pomares, J.M.; Díez, J.; Aranda, S.; Palomo, S.; McCormick, F.; Izpisúa-Belmonte, J.C.; et al. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 2003, 18, 99–115. [Google Scholar] [CrossRef]
- Torregrosa-Carrión, R.; Zurita, L.L.; García-Marqués, F.; D’Amato, G.; Piñeiro-Sabarís, R.; Bonzón-Kulichenko, E.; Vázquez, J.; de la Pompa, J.L. NOTCH Activation Promotes Valve Formation by Regulating the Endocardial Secretome. Mol. Cell. Proteom. 2019, 18, 1782–1795. [Google Scholar] [CrossRef]
- Toshima, T.; Watanabe, T.; Narumi, T.; Otaki, Y.; Shishido, T.; Aono, T.; Goto, J.; Watanabe, K.; Sugai, T.; Takahashi, T.; et al. Therapeutic inhibition of microRNA-34a ameliorates aortic valve calcification via modulation of Notch1-Runx2 signaling. Cardiovasc. Res. 2020, 116, 983–994. [Google Scholar] [CrossRef]
- Raddatz, A.M.; Roest, M.J.V.; Merryman, W.D. Notch1 suppression by microRNA-34a: A new mechanism of calcific aortic valve disease. Cardiovasc. Res. 2019, 116, 871–873. [Google Scholar] [CrossRef]
- Wang, L.; Tang, R.; Zhang, Y.; Chen, S.; Guo, Y.; Wang, X.; Liu, Z.; Liu, H.; Zhang, X.; Liu, B.C. PTH-induced EndMT via miR-29a-5p/GSAP/Notch1 pathway contributed to valvular calcification in rats with CKD. Cell Prolif. 2021, 54, e13018. [Google Scholar] [CrossRef]
- Dai, Y.; Yan, T.; Gao, Y. Silence of miR-32-5p promotes endothelial cell viability by targeting KLF2 and serves as a diagnostic biomarker of acute myocardial infarction. Diagn. Pathol. 2020, 15, 19. [Google Scholar] [CrossRef]
- Natarelli, L.; Schober, A. Janus-Faced Role of Krüppel-Like Factor 2–Dependent Regulation of MicroRNAs in Endothelial Proliferation. Arter. Thromb. Vasc. Biol. 2014, 34, 1605–1606. [Google Scholar] [CrossRef][Green Version]
- Liu, H.; Li, G.; Zhao, W.; Hu, Y. Inhibition of MiR-92a may protect endothelial cells after acute myocardial infarction in rats: Role of KLF2/4. Med. Sci. Monit. 2016, 22, 2451–2462. [Google Scholar] [CrossRef]
- Demolli, S.; Doebele, C.; Doddaballapur, A.; Lang, V.; Fisslthaler, B.; Chavakis, E.; Vinciguerra, M.; Sciacca, S.; Henschler, R.; Hecker, M.; et al. MicroRNA-30 mediates anti-inflammatory effects of shear stress and KLF2 via repression of angiopoietin 2. J. Mol. Cell. Cardiol. 2015, 88, 111–119. [Google Scholar] [CrossRef]
- Hergenreider, E.; Heydt, S.; Tréguer, K.; Boettger, T.; Horrevoets, A.J.G.; Zeiher, A.M.; Scheffer, M.P.; Frangakis, A.S.; Yin, X.; Mayr, M.; et al. Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat. Cell Biol. 2012, 14, 249–256. [Google Scholar] [CrossRef]
- Sindi, H.A.; Russomanno, G.; Satta, S.; Salam, V.A.; Jo, K.B.; Qazi-Chaudhry, B.; Ainscough, A.J.; Szulcek, R.; Bogaard, H.J.; Morgan, C.C.; et al. Therapeutic potential of KLF2-induced exosomal microRNAs in pulmonary hypertension. Nat. Commun. 2020, 11, 1185. [Google Scholar] [CrossRef]
- Cui, S.; Liu, Z.; Tao, B.; Fan, S.; Pu, Y.; Meng, X.; Li, D.; Xia, H.; Xu, L. MiR-145 attenuates cardiac fibrosis through the AKT/GSK-3β/β-catenin signaling pathway by directly targeting SOX9 in fibroblasts. J. Cell Physiol. 2020, 122, 209–221. [Google Scholar] [CrossRef]
- Li, J.; Cao, Y.; Ma, X.-J.; Wang, H.-J.; Zhang, J.; Luo, X.; Chen, W.; Wu, Y.; Meng, Y.; Yuan, Y.; et al. Roles of miR-1-1 and miR-181c in ventricular septal defects. Int. J. Cardiol. 2013, 168, 1441–1446. [Google Scholar] [CrossRef]
- Cheng, N.; Li, L.; Wu, Y.; Wang, M.; Yang, M.; Wei, S.; Wang, R. microRNA-30e up-regulation alleviates myocardial ischemia-reperfusion injury and promotes ventricular remodeling via SOX9 repression. Mol. Immunol. 2020, 130, 96–103. [Google Scholar] [CrossRef]
- Zhang, Y.; Tian, C.; Liu, X.; Zhang, H. Identification of Genetic Biomarkers for Diagnosis of Myocardial Infarction Compared with Angina Patients. Cardiovasc. Ther. 2020, 2020, 8535314. [Google Scholar] [CrossRef]
- Saadat, S.; Noureddini, M.; Mahjoubin-Tehran, M.; Nazemi, S.; Shojaie, L.; Aschner, M.; Maleki, B.; Abbasi-Kolli, M.; Rajabi Moghadam, H.; Alani, B.; et al. Pivot-al Role of TGF-β/Smad Signaling in Cardiac Fibrosis: Non-coding RNAs as Effectual Players. Front. Cardiovasc. Med. 2021, 7, 256. [Google Scholar] [CrossRef]
- Milano, G.; Biemmi, V.; Lazzarini, E.; Balbi, C.; Ciullo, A.; Bolis, S.; Ameri, P.; Di Silvestre, D.; Mauri, P.; Barile, L.; et al. Intravenous administration of cardiac progenitor cell-derived exosomes protects against doxorubicin/trastuzumab-induced cardiac toxicity. Cardiovasc. Res. 2020, 116, 383–392. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, Q.; Feng, Y.; Chen, X.; Yang, L.; Xu, M.; Wang, X.; Li, W.; Niu, X.; Gao, D. MicroRNA-26a Protects the Heart Against Hypertension-Induced Myocardial Fibrosis. J. Am. Heart Assoc. 2020, 9, e017970. [Google Scholar] [CrossRef]
- Dong, D.; Zhang, Y.; Reece, E.A.; Wang, L.; Harman, C.R.; Yang, P. microRNA expression profiling and functional annotation analysis of their targets modulated by oxidative stress during embryonic heart development in diabetic mice. Reprod. Toxicol. 2016, 65, 365–374. [Google Scholar] [CrossRef]
- Baldini, A.; Fulcoli, F.; Illingworth, E. Chapter Eight—Tbx1: Transcriptional and Developmental Functions. Curr. Top. Dev. Biol. 2017, 122, 223–243. [Google Scholar] [CrossRef]
- Pan, Y.; Wang, Z.-G.; Liu, X.-Y.; Zhao, H.; Zhou, N.; Zheng, G.-F.; Qiu, X.-B.; Li, R.-G.; Yuan, F.; Shi, H.-Y.; et al. A Novel TBX1 Loss-of-Function Mutation Associated with Congenital Heart Disease. Pediatr. Cardiol. 2015, 36, 1400–1410. [Google Scholar] [CrossRef]
- Cao, M.-L.; Zhu, B.-L.; Sun, Y.-Y.; Qiu, G.-R.; Fu, W.-N.; Jiang, H.-K. MicroRNA-144 Regulates Cardiomyocyte Proliferation and Apoptosis by Targeting TBX1 through the JAK2/STAT1 Pathway. Cytogenet. Genome Res. 2019, 159, 190–200. [Google Scholar] [CrossRef]
- Aravalli, R.N.; Greten, T.F. FoxC1: Novel Regulator of Inflammation-Induced Metastasis in Hepatocellular Carcinoma. Gastroenterology 2015, 149, 861–863. [Google Scholar] [CrossRef] [PubMed]
- Prasitsak, T.; Nandar, M.; Okuhara, S.; Ichinose, S.; Ota, M.S.; Iseki, S. Foxc1is required for early stage telencephalic vascular development. Dev. Dyn. 2015, 244, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Lambers, E.; Arnone, B.; Fatima, A.; Qin, G.; Wasserstrom, J.A.; Kume, T. Foxc1 Regulates Early Cardiomyogenesis and Functional Properties of Embryonic Stem Cell Derived Cardiomyocytes. Stem Cells 2016, 34, 1487–1500. [Google Scholar] [CrossRef] [PubMed]
- Henn, D.; Abu-Halima, M.; Wermke, D.; Falkner, F.; Thomas, B.; Köpple, C.; Ludwig, N.; Schulte, M.; Brockmann, M.A.; Kim, Y.-J.; et al. MicroRNA-regulated pathways of flow-stimulated angiogenesis and vascular remodeling in vivo. J. Transl. Med. 2019, 17, 22. [Google Scholar] [CrossRef]
- Mellor, R.H.; Brice, G.; Stanton, A.W.; French, J.; Smith, A.; Jeffery, S.; Levick, J.R.; Burnand, K.G.; Mortimer, P.S. Mutations in FOXC2 Are Strongly Associated With Primary Valve Failure in Veins of the Lower Limb. Circulation 2007, 115, 1912–1920. [Google Scholar] [CrossRef]
- Zeng, N.; Huang, Y.-Q.; Yan, Y.-M.; Hu, Z.-Q.; Zhang, Z.; Feng, J.-X.; Guo, J.-S.; Zhu, J.-N.; Fu, Y.-H.; Wang, X.-P.; et al. Diverging targets mediate the pathological roleof miR-199a-5p and miR-199a-3p by promoting cardiac hypertrophy and fibrosis. Mol. Ther. Nucleic Acids 2021, 26, 1035–1050. [Google Scholar] [CrossRef]
- Cao, Y.; Cao, Z.; Wang, W.; Jie, X.; Li, L. MicroRNA-199a-5p regulates FOXC2 to control human vascular smooth muscle cell phenotypic switch. Mol. Med. Rep. 2021, 24, 627. [Google Scholar] [CrossRef]
- Du, B.; Cawthorn, W.P.; Su, A.; Doucette, C.R.; Yao, Y.; Hemati, N.; Kampert, S.; McCoin, C.; Broome, D.T.; Rosen, C.J.; et al. The Transcription Factor Paired-Related Homeobox 1 (Prrx1) Inhibits Adipogenesis by Activating Transforming Growth Factor-β (TGFβ) Signaling. J. Biol. Chem. 2013, 288, 3036–3047. [Google Scholar] [CrossRef]
- Zhong, X.; Yu, X.; Wen, X.; Chen, L.; Gu, N. Activation of the LINC00242/miR-141/FOXC1 axis underpins the development of gastric cancer. Cancer Cell Int. 2020, 20, 272. [Google Scholar] [CrossRef]
- Jiang, Y.; Mo, H.; Luo, J.; Zhao, S.; Liang, S.; Zhang, M.; Yuan, J. HOTAIR Is a Potential Novel Biomarker in Patients with Congenital Heart Diseases. BioMed Res. Int. 2018, 2018, 850657. [Google Scholar] [CrossRef]
- Moorman, A.F.M.; Christoffels, V.M. Cardiac Chamber Formation: Development, Genes, and Evolution. Physiol. Rev. 2003, 83, 1223–1267. [Google Scholar] [CrossRef]
- Kelly, R.G.; Buckingham, M.E.; Moorman, A.F. Heart Fields and Cardiac Morphogenesis. Cold Spring Harb. Perspect. Med. 2014, 4, a015750. [Google Scholar] [CrossRef]
- Kelly, R.G.; Buckingham, M.E. The anterior heart-forming field: Voyage to the arterial pole of the heart. Trends Genet. 2002, 18, 210–216. [Google Scholar] [CrossRef]
- Erhardt, S.; Zheng, M.; Zhao, X.; Le, T.; Findley, T.; Wang, J. The Cardiac Neural Crest Cells in Heart Development and Congenital Heart Defects. J. Cardiovasc. Dev. Dis. 2021, 8, 89. [Google Scholar] [CrossRef]
- Saretzki, E.; Pankratz, F.; Engert, B.; Grundmann, S.; Bode, C.; Moser, M.; Zhou, Q.; Esser, J.S. Bone morphogenetic protein 4 regulates microRNAs miR-494 and miR-126–5p in control of endothelial cell function in angiogenesis. Thromb. Haemost. 2017, 117, 734–749. [Google Scholar] [CrossRef]
- Yanagawa, B.; Lovren, F.; Pan, Y.; Garg, V.; Quan, A.; Tang, G.; Singh, K.K.; Shukla, P.C.; Kalra, N.P.; Peterson, M.D.; et al. miRNA-141 is a novel regulator of BMP-2–mediated calcification in aortic stenosis. J. Thorac. Cardiovasc. Surg. 2012, 144, 256–262.e2. [Google Scholar] [CrossRef]
- Balderman, J.A.F.; Lee, H.; Mahoney, C.E.; Handy, D.E.; White, K.; Annis, S.; Lebeche, D.; Hajjar, R.J.; Loscalzo, J.; Leopold, J.A. Bone Morphogenetic Protein-2 Decreases MicroRNA-30b and MicroRNA-30c to Promote Vascular Smooth Muscle Cell Calcification. J. Am. Heart Assoc. 2012, 1, e003905. [Google Scholar] [CrossRef]
- Song, R.; Fullerton, D.A.; Ao, L.; Zhao, K.-S.; Meng, X. An epigenetic regulatory loop controls pro-osteogenic activation by TGF-β1 or bone morphogenetic protein 2 in human aortic valve interstitial cells. J. Biol. Chem. 2017, 292, 8657–8666. [Google Scholar] [CrossRef]
- Yousefi, F.; Shabaninejad, Z.; Vakili, S.; Derakhshan, M.; Movahedpour, A.; Dabiri, H.; Ghasemi, Y.; Mahjoubin-Tehran, M.; Nikoozadeh, A.; Savardashtaki, A.; et al. TGF-β and WNT signaling pathways in cardiac fibrosis: Non-coding RNAs come into focus. Cell Commun. Signal. 2020, 18, 87. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, Y.; Zhu, H.; Hu, J.; Xie, Z. MiR-34a/miR-93 target c-Ski to modulate the proliferaton of rat cardiac fibroblasts and extracellular matrix deposition in vivo and in vitro. Cell. Signal. 2018, 46, 145–153. [Google Scholar] [CrossRef]
- He, X.; Zhang, K.; Gao, X.; Li, L.; Tan, H.; Chen, J.; Zhou, Y. Rapid atrial pacing induces myocardial fibrosis by down-regulating Smad7 via microRNA-21 in rabbit. Heart Vessel. 2016, 31, 1696–1708. [Google Scholar] [CrossRef]
- Zhang, D.; Cui, Y.; Li, B.; Luo, X.; Li, B.; Tang, Y. MiR-155 regulates high glucose-induced cardiac fibrosis via the TGF-β signaling pathway. Mol. BioSyst. 2016, 13, 215–224. [Google Scholar] [CrossRef]
- Wang, Y.; Cai, H.; Li, H.; Gao, Z.; Song, K. Atrial overexpression of microRNA-27b attenuates angiotensin II-induced atrial fibrosis and fibrillation by targeting ALK5. Hum. Cell 2018, 31, 251–260. [Google Scholar] [CrossRef]
- Hong, Y.; Cao, H.; Wang, Q.; Ye, J.; Sui, L.; Feng, J.; Cai, X.; Song, H.; Zhang, X.; Chen, X. MiR-22 may Suppress Fibrogenesis by Targeting TGFβR I in Cardiac Fibroblasts. Cell. Physiol. Biochem. 2016, 40, 1345–1353. [Google Scholar] [CrossRef]
- Yu, R.-B.; Li, K.; Wang, G.; Gao, G.-M.; Du, J.-X. MiR-23 enhances cardiac fibroblast proliferation and suppresses fibroblast apoptosis via targeting TGF-β1 in atrial fibrillation. Eur. Rev. Med Pharmacol. Sci. 2019, 23, 4419–4424. [Google Scholar]
- Xu, J.; Wu, H.; Chen, S.; Qi, B.; Zhou, G.; Cai, L.; Zhao, L.; Wei, Y.; Liu, S. Micro RNA -30c suppresses the pro-fibrogenic effects of cardiac fibroblasts induced by TGF -β1 and prevents atrial fibrosis by targeting TGF β RII. J. Cell. Mol. Med. 2018, 22, 3045–3057. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.J.Y.; Zhuang, J.Z.J.; Lin, Y.L.Y.; Wang, X.W.X.; Chen, J.C.J.; Han, F.H.F. Long noncoding RNA SNHG6 contributes to ventricular septal defect formation via negative regulation of miR-101 and activation of Wnt/β-catenin pathway. Die Pharm. -Int. J. Pharm. Sci. 2019, 74, 23–28. [Google Scholar] [CrossRef]
- Zhao, X.; Wei, X.; Wang, X.; Qi, G. Long non-coding RNA NORAD regulates angiogenesis of human umbilical vein endothelial cells via miR-590-3p under hypoxic conditions. Mol. Med. Rep. 2020, 21, 2560–2570. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Luo, M.; Gao, Z.; Han, X.; Yan, Z.; Xie, S.; Zhao, H.; Sun, H. TUG1 knockdown suppresses cardiac fibrosis after myocardial infarction. Mamm. Genome 2021, 32, 435–442. [Google Scholar] [CrossRef] [PubMed]
- García-Padilla, C.; Domínguez, J.N.; Lodde, V.; Munk, R.; Abdelmohsen, K.; Gorospe, M.; Jiménez-Sábado, V.; Ginel, A.; Hove-Madsen, L.; Aránega, A.E.; et al. Identification of atrial-enriched lncRNA Walras linked to cardiomyocyte cytoarchitecture and atrial fibrillation. FASEB J. 2021, 36, e22051. [Google Scholar] [CrossRef]
- Das, A.; Shyamal, S.; Sinha, T.; Mishra, S.S.; Panda, A.C. Identification of Potential circRNA-microRNA-mRNA Regulatory Network in Skeletal Muscle. Front. Mol. Biosci. 2021, 8, 762185. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).