Genome-Wide Identification of Petunia HSF Genes and Potential Function of PhHSF19 in Benzenoid/Phenylpropanoid Biosynthesis
Abstract
:1. Introduction
2. Results
2.1. Identification of HSF Gene Family Members in Petunia
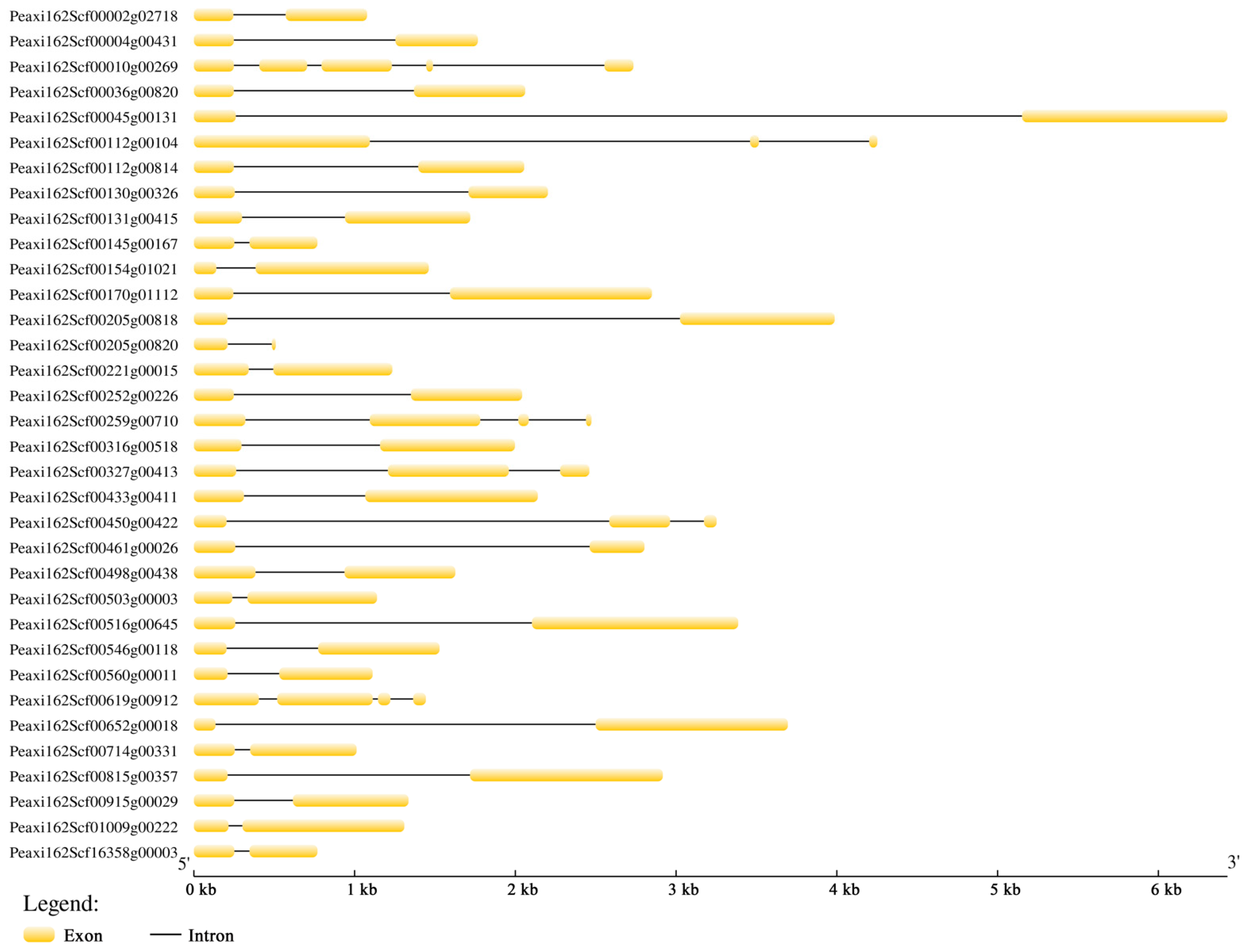
2.2. Gene Structure, Phylogeny Analysis, and Conserved Motifs of Petunia HSFs
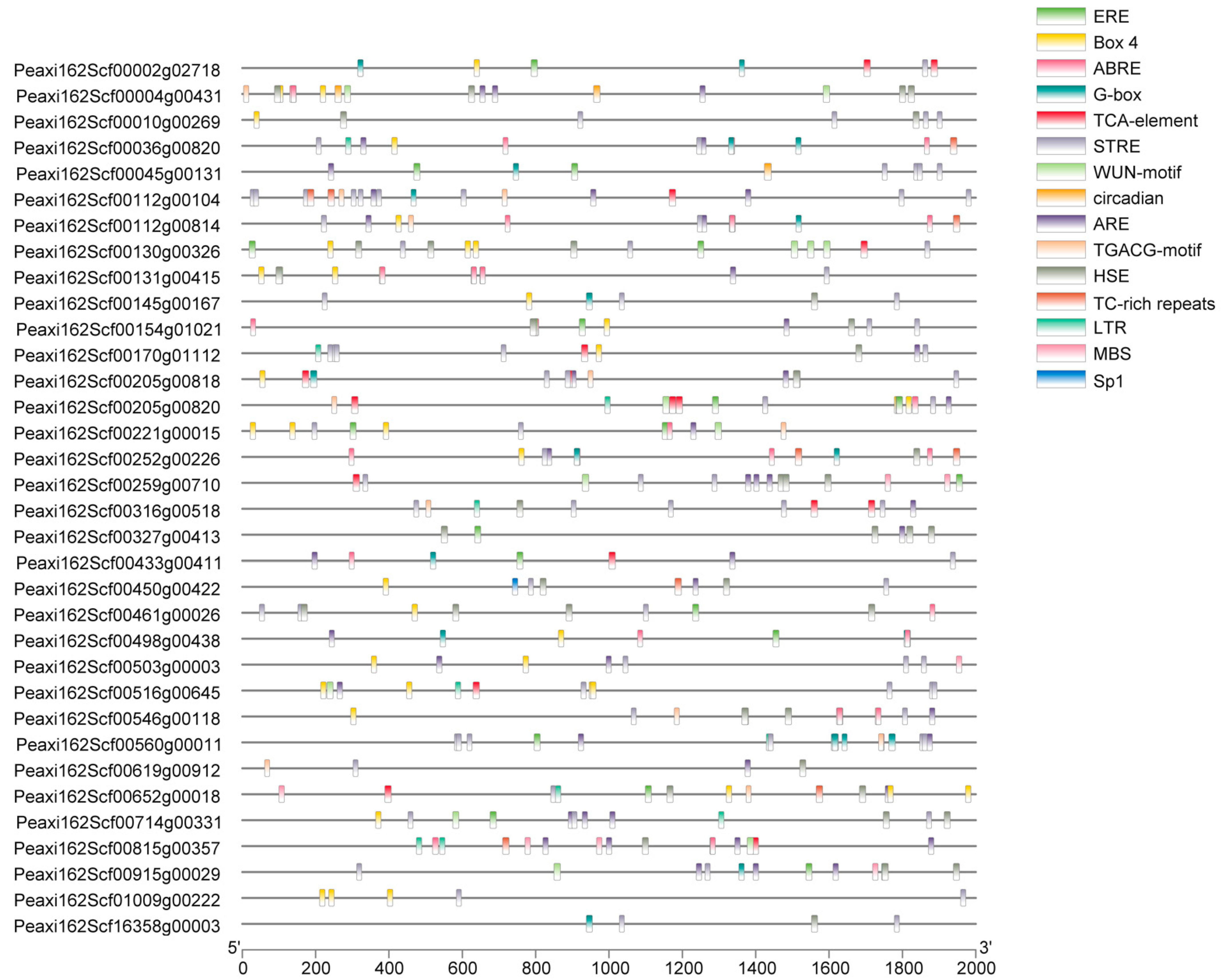
2.3. Cis-Acting Elements Analysis of Petunia HSFs
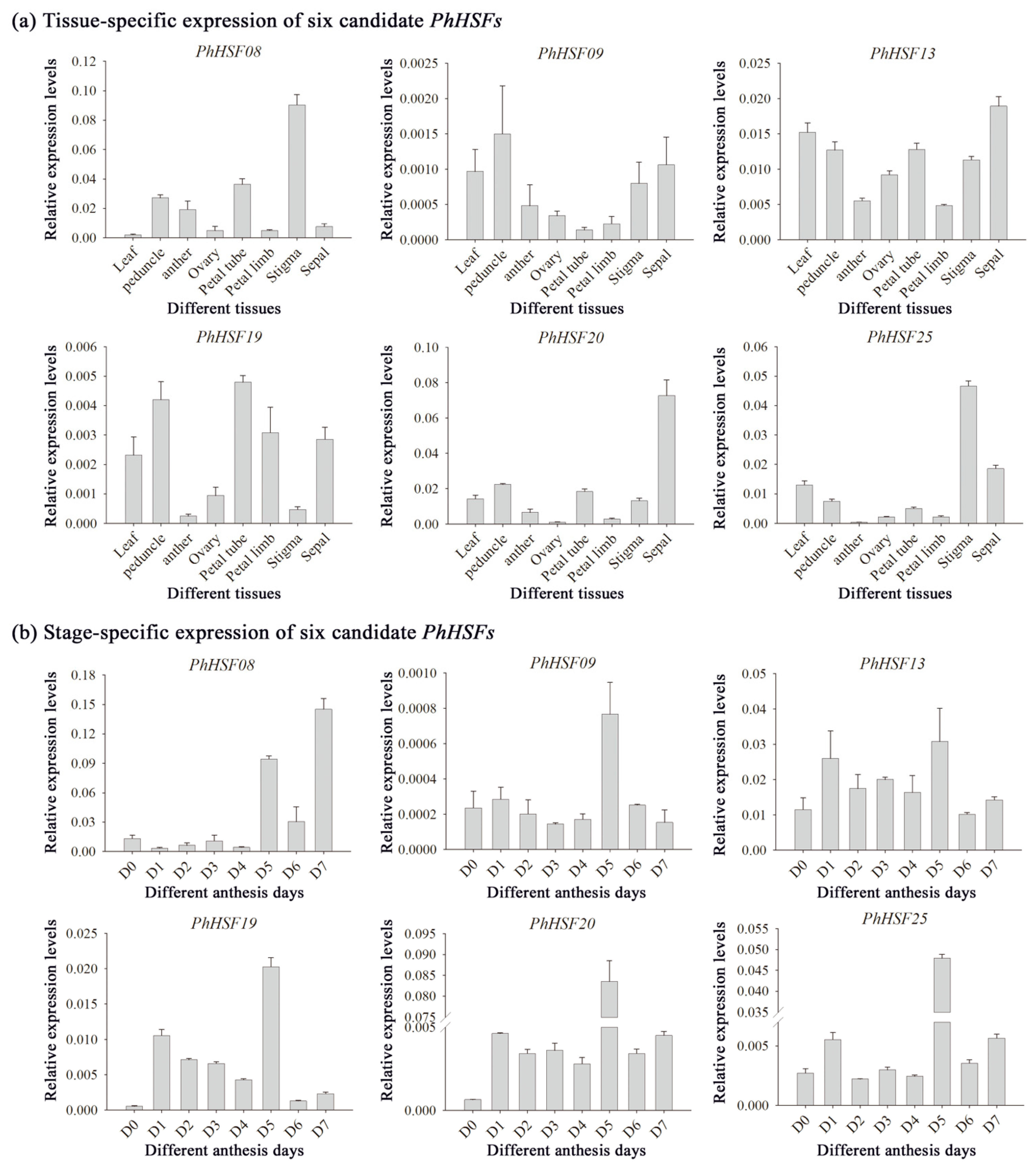
2.4. Selection of Potential Key Petunia HSF Genes Based on RNA Sequencing and qRT-PCR Analysis
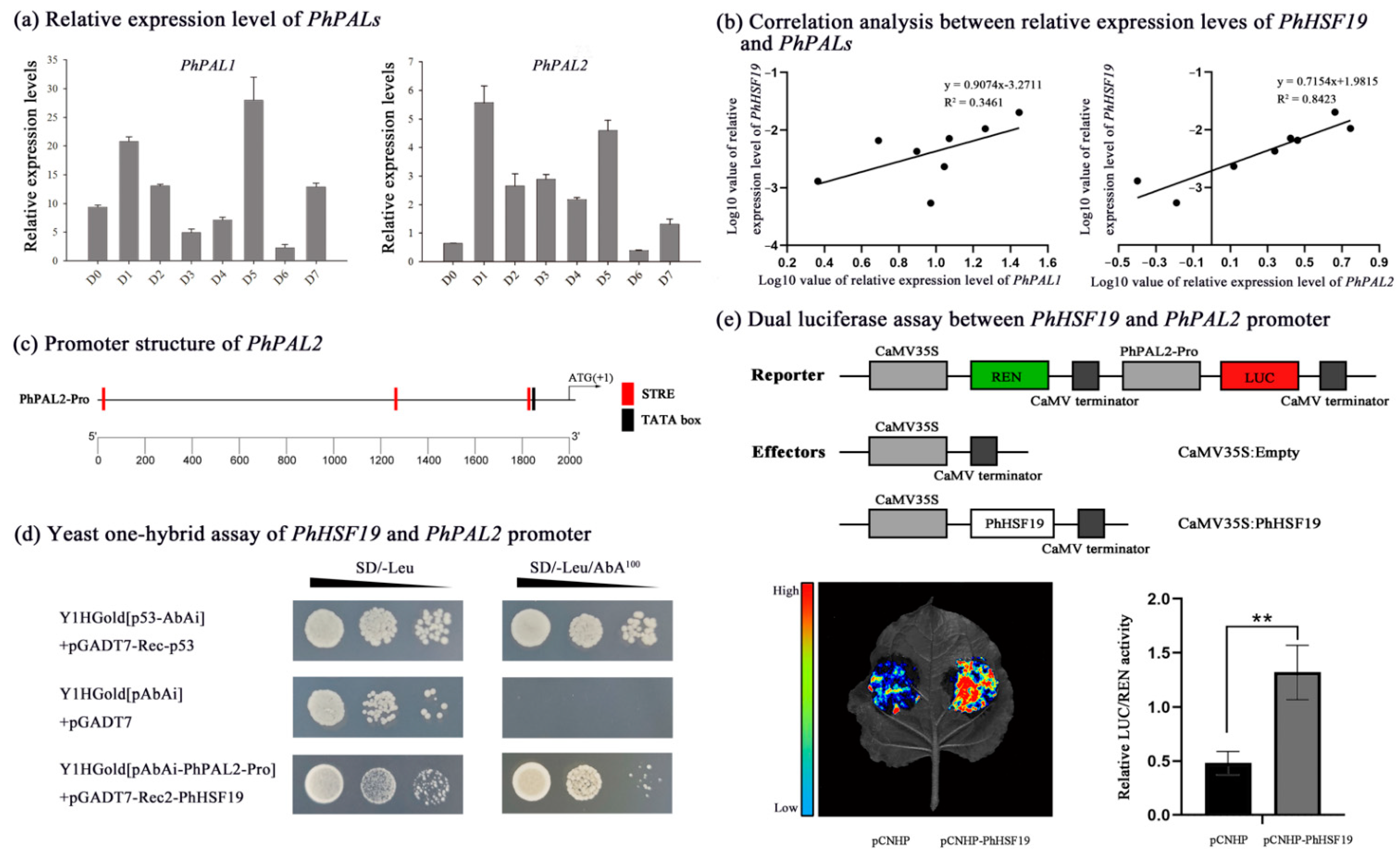
2.5. Y1H and Dual Luciferase Assays
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Identification HSF Genes in Petunia
4.3. Gene Structure and Phylogenetic Analysis of Petunia HSFs
4.4. Conserved Motifs and Cis-Acting Elements Analysis of Petunia HSFs
4.5. Selection of Potential Key PhHSF Genes Based on RNA Sequencing
4.6. Total RNA Extraction, cDNA Reverse Transcription and qRT-PCR Analysis
4.7. Y1H Assay
4.8. Dual Luciferase Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Scharf, K.D.; Berberich, T.; Ebersberger, I.; Nover, L. The plant heat stress transcription factor (Hsf) family: Structure, function and evolution. Biochim. Biophys. Acta 2012, 1819, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Mittal, D.; Chakrabarti, S.; Sarkar, A.; Singh, A.; Grover, A. Heat shock factor gene family in rice: Genomic organization and transcript expression profiling in response to high temperature, low temperature and oxidative stresses. Plant Physiol. Biochem. 2009, 47, 785–795. [Google Scholar] [CrossRef]
- Nover, N.; Bharti, K.; DRing, P.; Mishra, S.K.; Scharf, G. Arabidopsis and the heat stress transcription factor world: How many heat stress transcription factors do we need? Cell Stress Chaperones 2001, 6, 177–189. [Google Scholar] [CrossRef]
- Pelham, H.R.B. A regulatory upstream promoter element in the Drosophila Hsp 70 heat-shock gene. Cell 1982, 30, 517–528. [Google Scholar] [CrossRef]
- Li, S.; Liu, S.; Lin, X.; Grierson, D.; Yin, X.; Chen, K. Citrus heat shock transcription factor CitHsfA7-mediated citric acid degradation in response to heat stress. Plant Cell Environ. 2021, 45, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Ning, K.; Han, Y.; Chen, Z.; Luo, C.; Wang, S.; Zhang, W.; Li, L.; Zhang, X.; Fan, S.; Wang, Q. Genome-wide analysis of MADS-box family genes during flower development in lettuce. Plant Cell Environ. 2019, 42, 1868–1881. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Chen, S.; Liu, K.; Liu, Y.; Ni, L.; Zhang, K.; Zhang, L. Isolation of heat shock factor HsfA1a-binding sites in vivo revealed variations of heat shock elements in Arabidopsis thaliana. Plant Cell Physiol. 2008, 9, 1306–1315. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Zheng, Q.; Chen, L.; Liang, Y.; Wu, J. Ectopic overexpression of maize heat shock transcription factor gene ZmHsf04 confers increased thermo and salt-stress tolerance in transgenic Arabidopsis. Acta Physiol. Plant. 2017, 40, 9–20. [Google Scholar] [CrossRef]
- Zhu, B.; Ye, C.; Lü, H.; Chen, X.; Chai, G.; Chen, J.; Wang, C. Identification and characterization of a novel heat shock transcription factor gene, GmHsfA1, in soybeans (Glycine max). J. Plant Res. 2006, 119, 247–256. [Google Scholar] [CrossRef]
- Hu, Y.; Han, Y.T.; Wei, W.; Li, Y.J.; Zhang, K.; Gao, Y.R.; Zhao, F.L.; Feng, J.Y. Identification, isolation, and expression analysis of heat shock transcription factors in the diploid woodland strawberry Fragaria vesca. Front. Plant Sci. 2015, 6, 736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, L.; Li, X.; Hou, D.; Cheng, Z.; Gao, J. Genome-Wide Analysis and Expression Profiling of the Heat Shock Factor Gene Family in Phyllostachys edulis during Development and in Response to Abiotic Stresses. Forests 2019, 10, 100. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Wan, X.L.; Yu, J.Y.; Wang, K.L.; Zhang, J. Genome-Wide Identification, Classification, and Expression Analysis of the Hsf Gene Family in Carnation (Dianthus caryophyllus). Int. J. Mol. Sci. 2019, 20, 5233. [Google Scholar] [CrossRef] [Green Version]
- Zinsmeister, J.; Berriri, S.; Basso, D.; Vu, B.L.; Buitink, J. The seed-specific heat shock factor A9 regulates the depth of dormancy in Medicago truncatula seeds via ABA signaling. Plant Cell Environ. 2020, 43, 2508–2522. [Google Scholar] [CrossRef]
- Shang, J.; Tian, J.; Cheng, H.; Yan, Q.; Chen, L. The chromosome-level wintersweet (Chimonanthus praecox) genome provides insights into floral scent biosynthesis and flowering in winter. Genome Biol. 2020, 21, 200–227. [Google Scholar] [CrossRef]
- Yang, J.W.; Ren, Y.J.; Zhang, D.Y.; Chen, X.W.; Huang, J.Z.; Xu, Y. Transcriptome-Based WGCNA Analysis Reveals Regulated Metabolite Fluxes between Floral Color and Scent in Narcissus tazetta Flower. Int. J. Mol. Sci. 2021, 15, 8249. [Google Scholar] [CrossRef] [PubMed]
- Verdonk, J.C.; Haring, M.; van Tunen, A.J.; Schuurink, R.C. ODORANT1 Regulates Fragrance Biosynthesis in Petunia Flowers. Plant Cell 2005, 17, 1612–1624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boatright, J.; Negre, F.; Chen, X.; Kish, C.M. Understanding in Vivo Benzenoid Metabolism in Petunia Petal Tissue. Plant Physiol. 2004, 135, 1993–2011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verdonk, J.C.; Vos, C.; Verhoeven, H.A.; Haring, M.A.; Schuurink, R.C. Regulation of floral scent production in Petunia revealed by targeted metabolomics. Phytochemistry 2003, 62, 997–1008. [Google Scholar] [CrossRef]
- Aureliano, B.; Michel, M.; Avichai, A.; Manzhu, B.; Laure, B.; Cornelius, S.B.; Mattijs, B. Insight into the evolution of the Solanaceae from the parental genomes of Petunia hybrida. Nat. Plants 2016, 2, 16074. [Google Scholar]
- Spitzer-Rimon, B.; Marhevka, E.; Barkai, O.; Marton, I.; Edelbaum, O.; Masci, T.; Prathapani, N.K.; Shklarman, E.; Vainstein, O.A. EOBII, a Gene Encoding a Flower-Specific Regulator of Phenylpropanoid Volatiles’ Biosynthesis in Petunia. Plant Cell 2010, 22, 1961–1976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spitzer-Rimon, B.; Farhi, M.; Albo, B.; Cna’Ani, A.; Zvi, M.M.B. The R2R3-MYB–Like Regulatory Factor EOBI, Acting Downstream of EOBII, Regulates Scent Production by Activating ODO1 and Structural Scent-Related Genes in Petunia. Plant Cell 2012, 24, 5089–5105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, J.; Jian, W.; Qian, J.; Chao, W.; Lei, L.; Yi, Y.; Wang, Y. Genome-wide analysis of heat shock transcription factor families in rice and Arabidopsis. J. Genet. Genom. 2008, 035, 105–118. [Google Scholar] [CrossRef]
- Widhalm, J.R.; Gutensohn, M.; Yoo, H.; Adebesin, F.; Qian, Y.; Guo, L.; Jaini, R.; Lynch, J.H.; Mccoy, R.M.; Shreve, J.T. Identification of a plastidial phenylalanine exporter that influences flux distribution through the phenylalanine biosynthetic network. Nat. Commun. 2015, 6, 8142. [Google Scholar] [CrossRef] [PubMed]
- Underwood, B.A.; Tieman, D.M.; Shibuya, K.; Dexter, R.J.; Loucas, H.M.; Simkin, A.J.; Clark, D.G. Ethylene-Regulated Floral Volatile Synthesis in Petunia Corollas. Plant Physiol. 2005, 138, 255–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colquhoun, T.A.; Verdonk, J.C.; Schimmel, B.C.J.; Tieman, D.M.; Underwood, B.A.; Clark, D.G. Petunia floral volatile benzenoid/phenylpropanoid genes are regulated in a similar manner. Phytochemistry 2010, 71, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Dong, Q.; Jiang, H.; Zhu, S.; Chen, B.; Xiang, Y. Genome-wide analysis of the heat shock transcription factors in Populus trichocarpa and Medicago truncatula. Mol. Biol. Rep. 2012, 39, 1877–1886. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Huang, Q.; Sun, W.; Ma, Z.; Huang, L.; Wu, Q.; Tang, Z.; Bu, T.; Li, C.; Chen, H. Genome-wide investigation of the heat shock transcription factor (Hsf) gene family in Tartary buckwheat (Fagopyrum tataricum). BMC Genom. 2019, 20, 871–887. [Google Scholar] [CrossRef] [PubMed]
- Lohani, N.; Golicz, A.A.; Singh, M.B.; Bhalla, P.L. Genome-wide analysis of the Hsf gene family in Brassica oleracea and a comparative analysis of the Hsf gene family in B. oleracea, B. rapa and B. napus. Funct. Integr. Genom. 2019, 18, 649–665. [Google Scholar] [CrossRef]
- Guo, M.; Lu, J.P.; Zhai, Y.F.; Chai, W.G.; Gong, Z.H.; Lu, M.H. Genome-wide analysis, expression profile of heat shock factor gene family (CaHsfs) and characterisation of CaHsfA2 in pepper (Capsicum annuum L.). BMC Plant Biol. 2015, 15, 151. [Google Scholar] [CrossRef] [Green Version]
- Sun, T.; Wang, C.; Liu, R.; Zhang, Y.; Wang, Y.; Wang, L. Thhsfa1 Confers Sa1t Stress Tolerance through Modulation of Reactive Oxygen Species Scavenging by Directly Regulating ThWRKY4. Int. J. Mol. Sci. 2021, 22, 5048. [Google Scholar] [CrossRef]
- Rao, S.; Das, J.R.; Mathur, S. Exploring the master regulator heat stress transcription factor HSFA1a-mediated transcriptional cascade of HSFs in the heat stress response of tomato. J. Plant Biochem. Biot. 2021, 30, 878–888. [Google Scholar] [CrossRef]
- Mishra, S.K.; Poonia, A.; Chaudhary, B.; Baranwal, V.K.; Arora, D.; Kumar, R.; Chauhan, H. Genome-wide identification, phylogeny and expression analysis of HSF gene family in barley during abiotic stress response and reproductive development. Plant Gene 2020, 23, 100231. [Google Scholar] [CrossRef]
- Bharti, K.; Schmidt, E.; Lyck, R.; Heerklotz, D.; Scharf, K.D. Isolation and characterization of HsfA3, a new heat stress transcription factor of Lycopersicon peruvianum. Plant J. 2010, 22, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Patrick, R.M.; Huang, X.Q.; Natalia, D.; Li, Y. Dynamic histone acetylation in floral volatile synthesis and emission in petunia flowers. J. Exp. Bot. 2021, 10, 3704–3722. [Google Scholar] [CrossRef] [PubMed]
- Noe, F.P.; Naama, M.; Edwards, J.D.; Surya, S.; Tecle, I.Y.; Strickler, S.R.; Aureliano, B.; Thomas, F.Y.; Anuradha, P.; Hartmut, F. The Sol Genomics Network (SGN)—From genotype to phenotype to breeding. Nucleic Acids Res. 2015, 10, 1036–1041. [Google Scholar]


| Loci | Protein Length (aa) | Molecular Weight (kD) | Theoretical pI | Instability Index | Hydrophilic Index | Aliphatic Index | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| Peaxi162Scf00002g02718 | 250 | 28.83 | 5.77 | 55.15 | −0.868 | 64.40 | nucleus |
| Peaxi162Scf00004g00431 | 253 | 29.36 | 6.56 | 41.27 | −0.745 | 84.31 | nucleus |
| Peaxi162Scf00010g00269 | 403 | 45.91 | 6.34 | 60.36 | −0.734 | 70.89 | nucleus |
| Peaxi162Scf00036g00820 | 314 | 35.82 | 6.12 | 47.69 | −0.805 | 72.07 | nucleus |
| Peaxi162Scf00045g00131 | 512 | 56.56 | 4.71 | 68.62 | −0.636 | 68.57 | nucleus |
| Peaxi162Scf00112g00104 | 400 | 45.40 | 5.48 | 47.29 | −0.649 | 79.20 | nucleus |
| Peaxi162Scf00112g00814 | 302 | 34.25 | 5.99 | 42.22 | −0.728 | 75.56 | nucleus |
| Peaxi162Scf00130g00326 | 249 | 28.91 | 6.68 | 54.38 | −0.865 | 65.30 | nucleus |
| Peaxi162Scf00131g00415 | 359 | 41.74 | 5.00 | 51.96 | −0.835 | 64.32 | nucleus |
| Peaxi162Scf00145g00167 | 224 | 25.98 | 9.18 | 41.12 | −0.728 | 74.06 | nucleus |
| Peaxi162Scf00154g01021 | 405 | 46.04 | 5.39 | 54.92 | −0.820 | 71.23 | nucleus |
| Peaxi162Scf00170g01112 | 500 | 55.63 | 4.75 | 67.61 | −0.647 | 66.08 | nucleus |
| Peaxi162Scf00205g00818 | 390 | 45.15 | 4.63 | 54.33 | −0.673 | 73.46 | nucleus |
| Peaxi162Scf00205g00820 | 69 | 8.02 | 4.32 | 42.50 | 0.333 | 94.49 | nucleus |
| Peaxi162Scf00221g00015 | 360 | 41.84 | 6.64 | 57.17 | −0.759 | 70.92 | nucleus |
| Peaxi162Scf00252g00226 | 314 | 35.81 | 5.35 | 48.60 | −0.803 | 71.43 | nucleus |
| Peaxi162Scf00259g00710 | 369 | 42.10 | 5.42 | 51.46 | −0.751 | 65.77 | nucleus |
| Peaxi162Scf00316g00518 | 378 | 44.14 | 5.08 | 49.36 | −0.883 | 67.57 | nucleus |
| Peaxi162Scf00327g00413 | 399 | 45.58 | 5.19 | 53.32 | −0.406 | 87.39 | nucleus |
| Peaxi162Scf00433g00411 | 461 | 51.58 | 4.90 | 54.28 | −0.526 | 66.18 | nucleus |
| Peaxi162Scf00450g00422 | 221 | 24.71 | 5.85 | 41.11 | −0.765 | 54.71 | nucleus |
| Peaxi162Scf00461g00026 | 199 | 23.31 | 9.42 | 49.99 | −0.735 | 69.10 | nucleus |
| Peaxi162Scf00498g00438 | 357 | 41.59 | 5.08 | 55.61 | −0.777 | 70.73 | nucleus |
| Peaxi162Scf00503g00003 | 348 | 39.75 | 7.75 | 52.24 | −0.664 | 64.91 | nucleus |
| Peaxi162Scf00516g00645 | 503 | 55.67 | 5.67 | 47.46 | −0.604 | 74.77 | nucleus |
| Peaxi162Scf00546g00118 | 319 | 36.05 | 5.35 | 69.94 | −0.600 | 63.86 | nucleus |
| Peaxi162Scf00560g00011 | 263 | 30.25 | 6.62 | 47.95 | −0.651 | 64.52 | nucleus |
| Peaxi162Scf00619g00912 | 385 | 45.37 | 5.43 | 42.70 | −0.907 | 60.08 | nucleus |
| Peaxi162Scf00652g00018 | 443 | 49.37 | 5.39 | 57.72 | −0.794 | 65.80 | nucleus |
| Peaxi162Scf00714g00331 | 305 | 33.23 | 5.24 | 56.40 | −0.341 | 80.89 | nucleus |
| Peaxi162Scf00815g00357 | 469 | 52.79 | 5.40 | 49.77 | −0.550 | 72.69 | nucleus |
| Peaxi162Scf00915g00029 | 323 | 35.67 | 4.94 | 53.00 | −0.667 | 68.85 | nucleus |
| Peaxi162Scf01009g00222 | 407 | 46.63 | 5.20 | 48.51 | −0.754 | 70.42 | nucleus |
| Peaxi162Scf16358g00003 | 224 | 25.90 | 9.19 | 41.46 | −0.729 | 74.06 | nucleus |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, J.; Huang, S.; Qian, J.; Qing, H.; Wan, Z.; Cheng, H.; Zhang, C. Genome-Wide Identification of Petunia HSF Genes and Potential Function of PhHSF19 in Benzenoid/Phenylpropanoid Biosynthesis. Int. J. Mol. Sci. 2022, 23, 2974. https://doi.org/10.3390/ijms23062974
Fu J, Huang S, Qian J, Qing H, Wan Z, Cheng H, Zhang C. Genome-Wide Identification of Petunia HSF Genes and Potential Function of PhHSF19 in Benzenoid/Phenylpropanoid Biosynthesis. International Journal of Molecular Sciences. 2022; 23(6):2974. https://doi.org/10.3390/ijms23062974
Chicago/Turabian StyleFu, Jianxin, Shuying Huang, Jieyu Qian, Hongsheng Qing, Ziyun Wan, Hefeng Cheng, and Chao Zhang. 2022. "Genome-Wide Identification of Petunia HSF Genes and Potential Function of PhHSF19 in Benzenoid/Phenylpropanoid Biosynthesis" International Journal of Molecular Sciences 23, no. 6: 2974. https://doi.org/10.3390/ijms23062974
APA StyleFu, J., Huang, S., Qian, J., Qing, H., Wan, Z., Cheng, H., & Zhang, C. (2022). Genome-Wide Identification of Petunia HSF Genes and Potential Function of PhHSF19 in Benzenoid/Phenylpropanoid Biosynthesis. International Journal of Molecular Sciences, 23(6), 2974. https://doi.org/10.3390/ijms23062974






