Single-Cell RNA Sequencing with Spatial Transcriptomics of Cancer Tissues
Abstract
:1. Introduction
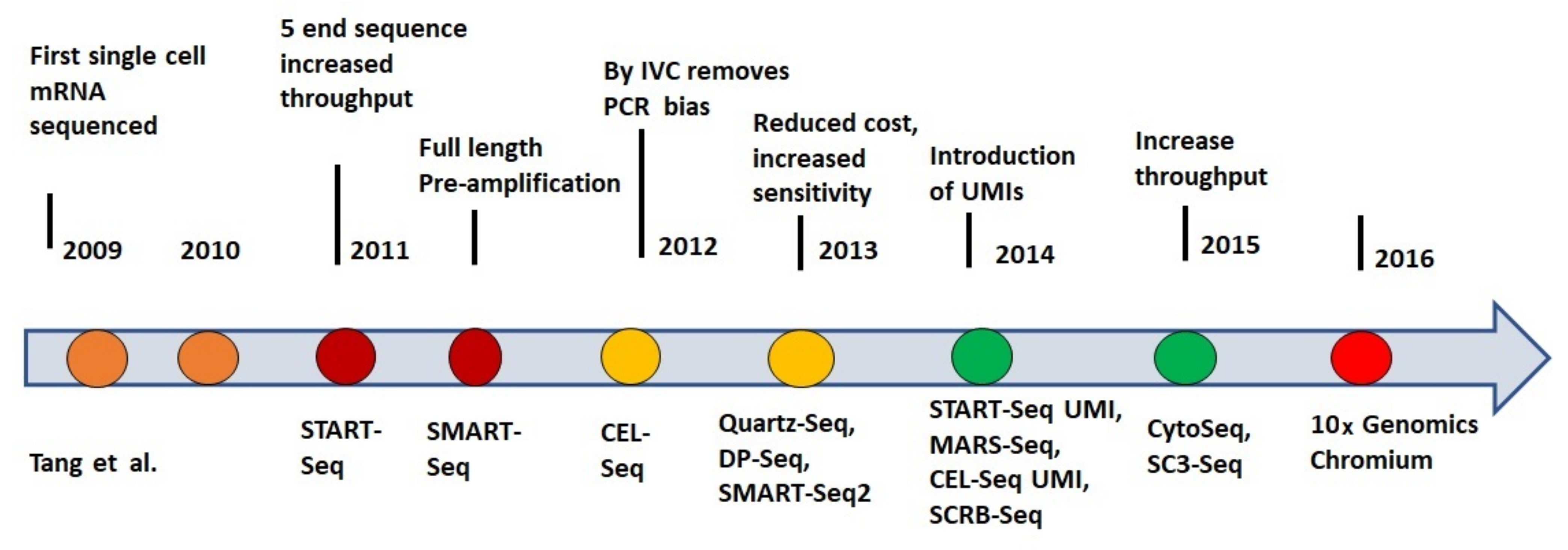
2. History of Single-Cell RNA-Seq Techniques
3. Methods of Single-Cell RNA-Seq Techniques
3.1. Cell Expression by Linear Amplification and Sequencing (CEL-Seq)
3.2. Single-Cell RNA Barcoding and Sequencing (SCRB-Seq)
3.3. Switching Mechanism at the End of the 5′-End of the RNA Transcript Sequencing (Smart-Seq)
3.4. Drop-Sequencing (Drop-Seq)
3.5. Massively Parallel RNA Single-Cell Sequencing Framework (MARS-Seq)
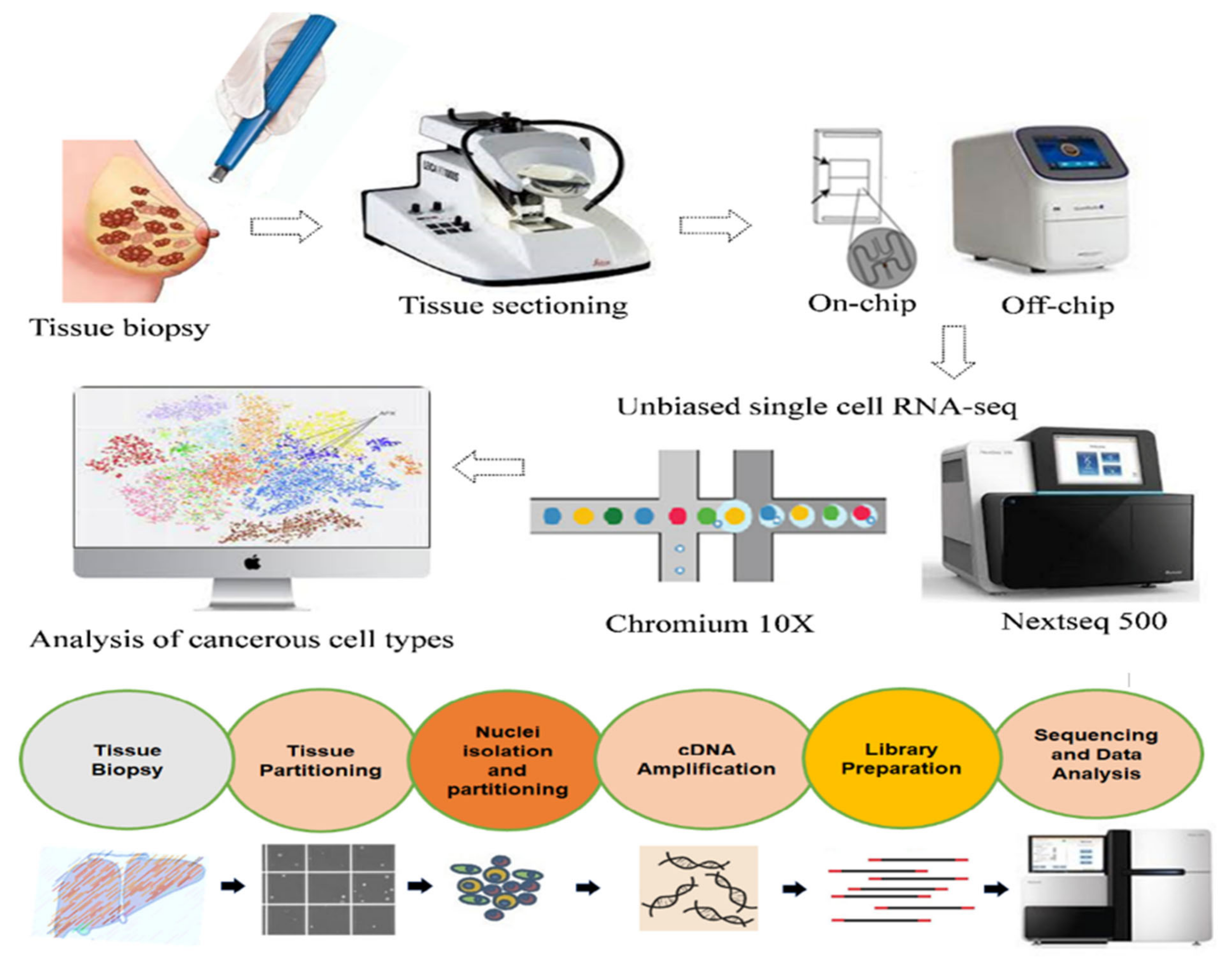
3.6. 10x Genomics Single-Cell RNA-Seq
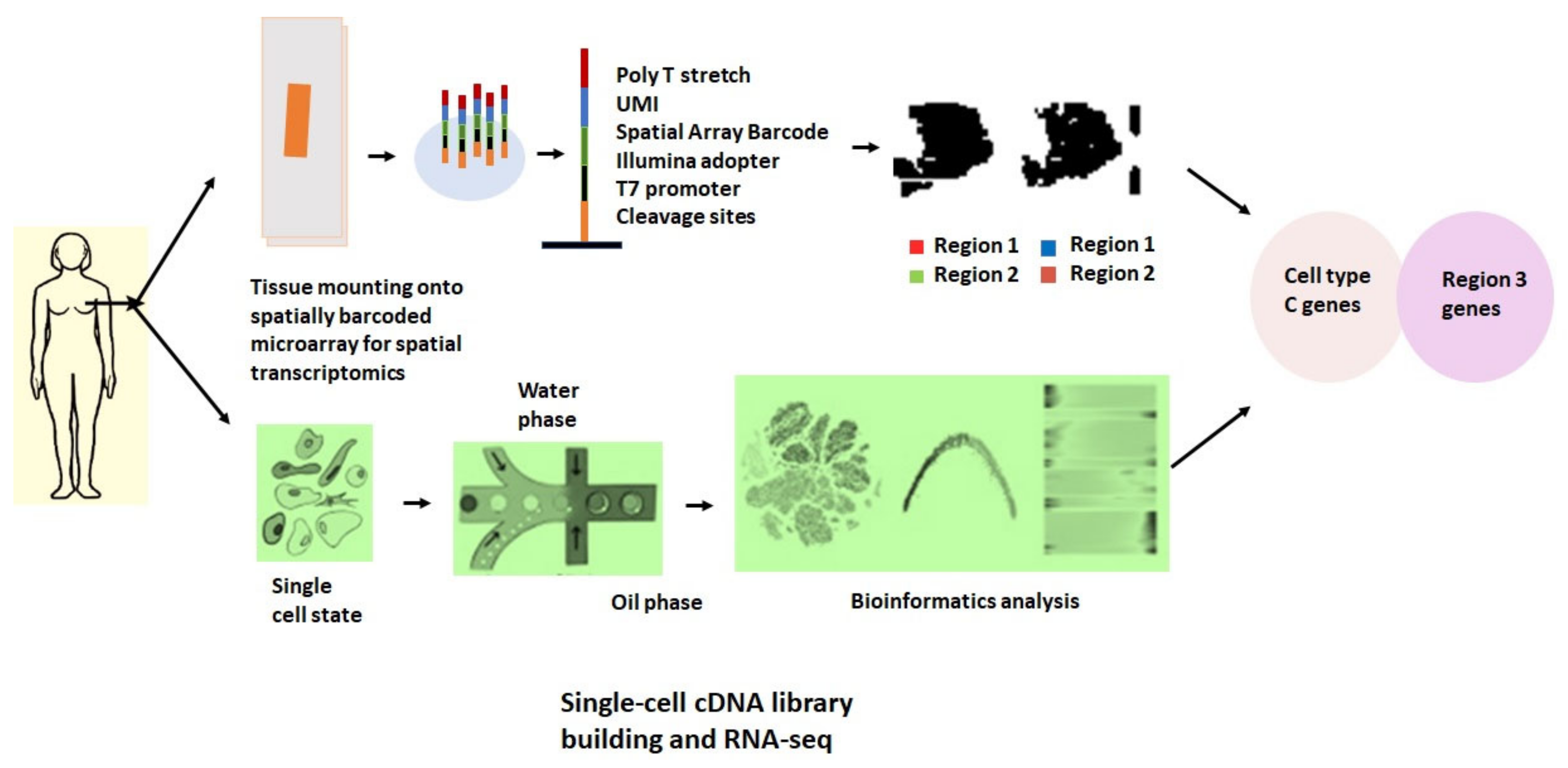
4. Spatial Transcriptomics
4.1. Next-Generation Sequencing (NGS)-Based Approaches
4.2. Imaging-Based Approaches
4.2.1. Multiplex Error Robust Fluorescent In Situ Hybridization (MERFISH)
4.2.2. Fourth-Generation RNA-Seq
4.2.3. Laser Capture Micro-Dissected RNA-Seq
5. Integration of Single-Cell RNA-Seq with Spatial Mapping Techniques
6. Clinical Applications of Single-Cell RNA-Seq Techniques
7. Existing Challenges and Prospects
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lee, J.; Hyeon, D.Y.; Hwang, D. Single-cell multiomics: Technologies and data analysis methods. Exp. Mol. Med. 2020, 52, 1428–1442. [Google Scholar] [CrossRef]
- Heng, H.H.; Stevens, J.B.; Bremer, S.W.; Liu, G.; Abdallah, B.Y.; Christine, J.Y. Evolutionary mechanisms and diversity in cancer. Adv. Cancer Res. 2011, 112, 217–253. [Google Scholar] [PubMed]
- Hong, M.; Tao, S.; Zhang, L.; Diao, L.-T.; Huang, X.; Huang, S.; Xie, S.-J.; Xiao, Z.-D.; Zhang, H. RNA sequencing: New technologies and applications in cancer research. J. Hematol. Oncol. 2020, 13, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Saltz, J.; Gupta, R.; Hou, L.; Kurc, T.; Singh, P.; Nguyen, V.; Samaras, D.; Shroyer, K.R.; Zhao, T.; Batiste, R. Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep. 2018, 23, 181–193.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asp, M.; Bergenstråhle, J.; Lundeberg, J. Spatially resolved transcriptomes—Next generation tools for tissue exploration. BioEssays 2020, 42, 1900221. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, R.; Augustine, R.; Valera, E.; Ganguli, A.; Mesaeli, N.; Ahmad, I.S.; Bashir, R.; Hasan, A. Spatial mapping of cancer tissues by OMICS technologies. Biochim. Biophys. Acta BBA Rev. Cancer 2022, 1877, 188663. [Google Scholar] [CrossRef]
- Ståhl, P.L.; Salmén, F.; Vickovic, S.; Lundmark, A.; Navarro, J.F.; Magnusson, J.; Giacomello, S.; Asp, M.; Westholm, J.O.; Huss, M. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 2016, 353, 78–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saiselet, M.; Rodrigues-Vitória, J.; Tourneur, A.; Craciun, L.; Spinette, A.; Larsimont, D.; Andry, G.; Lundeberg, J.; Maenhaut, C.; Detours, V. Transcriptional output, cell-type densities, and normalization in spatial transcriptomics. J. Mol. Cell Biol. 2020, 12, 906–908. [Google Scholar] [CrossRef]
- Tang, F.; Barbacioru, C.; Wang, Y.; Nordman, E.; Lee, C.; Xu, N.; Wang, X.; Bodeau, J.; Tuch, B.B.; Siddiqui, A. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 2009, 6, 377–382. [Google Scholar] [CrossRef]
- Eberwine, J.; Yeh, H.; Miyashiro, K.; Cao, Y.; Nair, S.; Finnell, R.; Zettel, M.; Coleman, P. Analysis of gene expression in single live neurons. Proc. Natl. Acad. Sci. USA 1992, 89, 3010–3014. [Google Scholar] [CrossRef] [Green Version]
- Brady, G.; Barbara, M.; Iscove, N.N. Representative in vitro cDNA amplification from individual hemopoietic cells and colonies. Methods Mol. Cell Biol. 1990, 2, 17–25. [Google Scholar]
- Islam, S.; Kjällquist, U.; Moliner, A.; Zajac, P.; Fan, J.-B.; Lönnerberg, P.; Linnarsson, S. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res. 2011, 21, 1160–1167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hashimshony, T.; Wagner, F.; Sher, N.; Yanai, I. CEL-Seq: Single-cell RNA-Seq by multiplexed linear amplification. Cell Rep. 2012, 2, 666–673. [Google Scholar] [CrossRef] [Green Version]
- Jaitin, D.A.; Kenigsberg, E.; Keren-Shaul, H.; Elefant, N.; Paul, F.; Zaretsky, I.; Mildner, A.; Cohen, N.; Jung, S.; Tanay, A. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 2014, 343, 776–779. [Google Scholar] [CrossRef] [PubMed]
- Klein, A.M.; Mazutis, L.; Akartuna, I.; Tallapragada, N.; Veres, A.; Li, V.; Peshkin, L.; Weitz, D.A.; Kirschner, M.W. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 2015, 161, 1187–1201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tirosh, I.; Izar, B.; Prakadan, S.M.; Wadsworth, M.H.; Treacy, D.; Trombetta, J.J.; Rotem, A.; Rodman, C.; Lian, C.; Murphy, G. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016, 352, 189–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joost, S.; Zeisel, A.; Jacob, T.; Sun, X.; La Manno, G.; Lönnerberg, P.; Linnarsson, S.; Kasper, M. Single-cell transcriptomics reveals that differentiation and spatial signatures shape epidermal and hair follicle heterogeneity. Cell Syst. 2016, 3, 221–237.e229. [Google Scholar] [CrossRef] [Green Version]
- Shalek, A.K.; Benson, M. Single-cell analyses to tailor treatments. Sci. Transl. Med. 2017, 9, eaan4730. [Google Scholar] [CrossRef] [Green Version]
- Gierahn, T.M.; Wadsworth, M.H.; Hughes, T.K.; Bryson, B.D.; Butler, A.; Satija, R.; Fortune, S.; Love, J.C.; Shalek, A.K. Seq-Well: Portable, low-cost RNA sequencing of single cells at high throughput. Nat. Methods 2017, 14, 395–398. [Google Scholar] [CrossRef]
- Shnayder, M.; Nachshon, A.; Krishna, B.; Poole, E.; Boshkov, A.; Binyamin, A.; Maza, I.; Sinclair, J.; Schwartz, M.; Stern-Ginossar, N. Defining the transcriptional landscape during cytomegalovirus latency with single-cell RNA sequencing. mBio 2018, 9, e00013-18. [Google Scholar] [CrossRef] [Green Version]
- Yuan, J.; Sheng, J.; Sims, P.A. An optically decodable bead array for linking imaging and sequencing with single-cell resolution. bioRxiv 2018, 355677. [Google Scholar] [CrossRef]
- Slyper, M.; Porter, C.B.; Ashenberg, O.; Waldman, J.; Drokhlyansky, E.; Wakiro, I.; Smillie, C.; Smith-Rosario, G.; Wu, J.; Dionne, D. A single-cell and single-nucleus RNA-Seq toolbox for fresh and frozen human tumors. Nat. Med. 2020, 26, 792–802. [Google Scholar] [CrossRef] [PubMed]
- Macosko, E.Z.; Basu, A.; Satija, R.; Nemesh, J.; Shekhar, K.; Goldman, M.; Tirosh, I.; Bialas, A.R.; Kamitaki, N.; Martersteck, E.M. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 2015, 161, 1202–1214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, G.X.; Terry, J.M.; Belgrader, P.; Ryvkin, P.; Bent, Z.W.; Wilson, R.; Ziraldo, S.B.; Wheeler, T.D.; McDermott, G.P.; Zhu, J. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017, 8, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Sheng, K.; Cao, W.; Niu, Y.; Deng, Q.; Zong, C. Effective detection of variation in single-cell transcriptomes using MATQ-seq. Nat. Methods 2017, 14, 267–270. [Google Scholar] [CrossRef]
- Fan, X.; Zhang, X.; Wu, X.; Guo, H.; Hu, Y.; Tang, F.; Huang, Y. Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome Biol. 2015, 16, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Kivioja, T.; Vähärautio, A.; Karlsson, K.; Bonke, M.; Enge, M.; Linnarsson, S.; Taipale, J. Counting absolute numbers of molecules using unique molecular identifiers. Nat. Methods 2012, 9, 72–74. [Google Scholar] [CrossRef]
- Donati, G. The niche in single-cell technologies. Immunol. Cell Biol. 2016, 94, 250–255. [Google Scholar] [CrossRef]
- Van Dijk, E.L.; Auger, H.; Jaszczyszyn, Y.; Thermes, C. Ten years of next-generation sequencing technology. Trends Genet. 2014, 30, 418–426. [Google Scholar] [CrossRef]
- Hashimshony, T.; Senderovich, N.; Avital, G.; Klochendler, A.; De Leeuw, Y.; Anavy, L.; Gennert, D.; Li, S.; Livak, K.J.; Rozenblatt-Rosen, O. CEL-Seq2: Sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol. 2016, 17, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.R.; Yong, K.W.; Choi, J.Y.; Cowie, A.C. Single-cell RNA sequencing and its combination with protein and DNA analyses. Cells 2020, 9, 1130. [Google Scholar] [CrossRef] [PubMed]
- Bagnoli, J.W.; Ziegenhain, C.; Janjic, A.; Wange, L.E.; Vieth, B.; Parekh, S.; Geuder, J.; Hellmann, I.; Enard, W. Sensitive and powerful single-cell RNA sequencing using mcSCRB-seq. Nat. Commun. 2018, 9, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shapiro, E.; Biezuner, T.; Linnarsson, S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat. Rev. Genet. 2013, 14, 618–630. [Google Scholar] [CrossRef] [PubMed]
- Ramsköld, D.; Luo, S.; Wang, Y.-C.; Li, R.; Deng, Q.; Faridani, O.R.; Daniels, G.A.; Khrebtukova, I.; Loring, J.F.; Laurent, L.C. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 2012, 30, 777–782. [Google Scholar] [CrossRef] [Green Version]
- Goetz, J.J.; Trimarchi, J.M. Transcriptome sequencing of single cells with Smart-Seq. Nat. Biotechnol. 2012, 30, 763–765. [Google Scholar] [CrossRef]
- Picelli, S.; Björklund, Å.K.; Faridani, O.R.; Sagasser, S.; Winberg, G.; Sandberg, R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 2013, 10, 1096–1098. [Google Scholar] [CrossRef]
- Hangauer, M.J.; Vaughn, I.W.; McManus, M.T. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 2013, 9, e1003569. [Google Scholar] [CrossRef]
- Picelli, S.; Faridani, O.R.; Björklund, Å.K.; Winberg, G.; Sagasser, S.; Sandberg, R. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014, 9, 171–181. [Google Scholar] [CrossRef]
- FA, V.B.; Miragaia, R.J. Tissue Handling and Dissociation for Single-Cell RNA-Seq. Methods Mol. Biol. 2019, 1979, 9–21. [Google Scholar]
- Picelli, S. Full-length single-cell RNA sequencing with smart-seq2. In Single Cell Methods; Humana: New York, NY, USA, 2019; pp. 25–44. [Google Scholar]
- Pollen, A.A.; Nowakowski, T.J.; Shuga, J.; Wang, X.; Leyrat, A.A.; Lui, J.H.; Li, N.; Szpankowski, L.; Fowler, B.; Chen, P. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat. Biotechnol. 2014, 32, 1053–1058. [Google Scholar] [CrossRef] [Green Version]
- Skelly, D.A.; Squiers, G.T.; McLellan, M.A.; Bolisetty, M.T.; Robson, P.; Rosenthal, N.A.; Pinto, A.R. Single-cell transcriptional profiling reveals cellular diversity and intercommunication in the mouse heart. Cell Rep. 2018, 22, 600–610. [Google Scholar] [CrossRef] [Green Version]
- Cochain, C.; Vafadarnejad, E.; Arampatzi, P.; Pelisek, J.; Winkels, H.; Ley, K.; Wolf, D.; Saliba, A.-E.; Zernecke, A. Single-cell RNA-seq reveals the transcriptional landscape and heterogeneity of aortic macrophages in murine atherosclerosis. Circ. Res. 2018, 122, 1661–1674. [Google Scholar] [CrossRef]
- Terabayashi, A.; Germino, G.G.; Menezes, L.F. Pathway identification through transcriptome analysis. Cell. Signal. 2020, 74, 109701. [Google Scholar] [CrossRef] [PubMed]
- Doyle, M.D.; Pescitelli, M.J., Jr.; Williams, B.S.; Michaels, G.S. Method and System for the Multidimensional Morphological Reconstruction of Genome Expression Activity. U.S. Patent US7613571B2, 3 November 2009. [Google Scholar]
- Zhuang, X. Spatially resolved single-cell genomics and transcriptomics by imaging. Nat. Methods 2021, 18, 18–22. [Google Scholar] [CrossRef]
- Larsson, L.; Frisén, J.; Lundeberg, J. Spatially resolved transcriptomics adds a new dimension to genomics. Nat. Methods 2021, 18, 15–18. [Google Scholar] [CrossRef]
- Crosetto, N.; Bienko, M.; Van Oudenaarden, A. Spatially resolved transcriptomics and beyond. Nat. Rev. Genet. 2015, 16, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Moor, A.E.; Itzkovitz, S. Spatial transcriptomics: Paving the way for tissue-level systems biology. Curr. Opin. Biotechnol. 2017, 46, 126–133. [Google Scholar] [CrossRef] [Green Version]
- Augustine, R.; Al Mamun, A.; Hasan, A.; Salam, S.A.; Chandrasekaran, R.; Ahmed, R.; Thakor, A.S. Imaging cancer cells with nanostructures: Prospects of nanotechnology driven non-invasive cancer diagnosis. Adv. Colloid Interface Sci. 2021, 294, 102457. [Google Scholar] [CrossRef] [PubMed]
- Rao, A.; Barkley, D.; França, G.S.; Yanai, I. Exploring tissue architecture using spatial transcriptomics. Nature 2021, 596, 211–220. [Google Scholar] [CrossRef]
- Thrane, K.; Eriksson, H.; Maaskola, J.; Hansson, J.; Lundeberg, J. Spatially resolved transcriptomics enables dissection of genetic heterogeneity in stage III cutaneous malignant melanoma. Cancer Res. 2018, 78, 5970–5979. [Google Scholar] [CrossRef] [Green Version]
- Moncada, R.; Barkley, D.; Wagner, F.; Chiodin, M.; Devlin, J.C.; Baron, M.; Hajdu, C.H.; Simeone, D.M.; Yanai, I. Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat. Biotechnol. 2020, 38, 333–342. [Google Scholar] [CrossRef]
- Ji, A.L.; Rubin, A.J.; Thrane, K.; Jiang, S.; Reynolds, D.L.; Meyers, R.M.; Guo, M.G.; George, B.M.; Mollbrink, A.; Bergenstråhle, J. Multimodal analysis of composition and spatial architecture in human squamous cell carcinoma. Cell 2020, 182, 497–514.e22. [Google Scholar] [CrossRef] [PubMed]
- Rodriques, S.G.; Stickels, R.R.; Goeva, A.; Martin, C.A.; Murray, E.; Vanderburg, C.R.; Welch, J.; Chen, L.M.; Chen, F.; Macosko, E.Z. Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. Science 2019, 363, 1463–1467. [Google Scholar] [CrossRef]
- Stickels, R.R.; Murray, E.; Kumar, P.; Li, J.; Marshall, J.L.; Di Bella, D.J.; Arlotta, P.; Macosko, E.Z.; Chen, F. Highly sensitive spatial transcriptomics at near-cellular resolution with Slide-seqV2. Nat. Biotechnol. 2021, 39, 313–319. [Google Scholar] [CrossRef]
- Vickovic, S.; Eraslan, G.; Salmén, F.; Klughammer, J.; Stenbeck, L.; Schapiro, D.; Äijö, T.; Bonneau, R.; Bergenstråhle, L.; Navarro, J.F. High-definition spatial transcriptomics for in situ tissue profiling. Nat. Methods 2019, 16, 987–990. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yang, M.; Deng, Y.; Su, G.; Enninful, A.; Guo, C.C.; Tebaldi, T.; Zhang, D.; Kim, D.; Bai, Z. High-spatial-resolution multi-omics sequencing via deterministic barcoding in tissue. Cell 2020, 183, 1665–1681. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, S.; Sonnhammer, E.L. Fusion transcript detection using spatial transcriptomics. BMC Med. Genom. 2020, 13, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Cho, C.-S.; Xi, J.; Park, S.-R.; Hsu, J.-E.; Kim, M.; Jun, G.; Kang, H.M.; Lee, J.H. Seq-Scope: Submicrometer-resolution spatial transcriptomics for single cell and subcellular studies. bioRxiv 2021. [Google Scholar] [CrossRef]
- Fu, X.; Sun, L.; Chen, J.; Dong, R.; Lin, Y.; Palmiter, R.; Lin, S.; Gu, L. Continuous Polony Gels for Tissue Mapping with High Resolution and RNA Capture Efficiency. bioRxiv 2021. [Google Scholar] [CrossRef]
- Chen, K.H.; Boettiger, A.N.; Moffitt, J.R.; Wang, S.; Zhuang, X. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 2015, 348, aaa6090. [Google Scholar] [CrossRef] [Green Version]
- Moffitt, J.R.; Hao, J.; Wang, G.; Chen, K.H.; Babcock, H.P.; Zhuang, X. High-throughput single-cell gene-expression profiling with multiplexed error-robust fluorescence in situ hybridization. Proc. Natl. Acad. Sci. USA 2016, 113, 11046–11051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Femino, A.M.; Fay, F.S.; Fogarty, K.; Singer, R.H. Visualization of single RNA transcripts in situ. Science 1998, 280, 585–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raj, A.; Van Den Bogaard, P.; Rifkin, S.A.; Van Oudenaarden, A.; Tyagi, S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods 2008, 5, 877–879. [Google Scholar] [CrossRef] [Green Version]
- Levsky, J.M.; Shenoy, S.M.; Pezo, R.C.; Singer, R.H. Single-cell gene expression profiling. Science 2002, 297, 836–840. [Google Scholar] [CrossRef] [PubMed]
- Lubeck, E.; Coskun, A.F.; Zhiyentayev, T.; Ahmad, M.; Cai, L. Single-cell in situ RNA profiling by sequential hybridization. Nat. Methods 2014, 11, 360–361. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Allen, W.E.; Wright, M.A.; Sylwestrak, E.L.; Samusik, N.; Vesuna, S.; Evans, K.; Liu, C.; Ramakrishnan, C.; Liu, J. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 2018, 361, eaat5691. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.H.; Daugharthy, E.R.; Scheiman, J.; Kalhor, R.; Yang, J.L.; Ferrante, T.C.; Terry, R.; Jeanty, S.S.; Li, C.; Amamoto, R. Highly multiplexed subcellular RNA sequencing in situ. Science 2014, 343, 1360–1363. [Google Scholar] [CrossRef] [Green Version]
- Shah, S.; Lubeck, E.; Zhou, W.; Cai, L. In situ transcription profiling of single cells reveals spatial organization of cells in the mouse hippocampus. Neuron 2016, 92, 342–357. [Google Scholar] [CrossRef] [Green Version]
- Moffitt, J.R.; Bambah-Mukku, D.; Eichhorn, S.W.; Vaughn, E.; Shekhar, K.; Perez, J.D.; Rubinstein, N.D.; Hao, J.; Regev, A.; Dulac, C. Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science 2018, 362, 6400. [Google Scholar] [CrossRef] [Green Version]
- Moffitt, J.R.; Hao, J.; Bambah-Mukku, D.; Lu, T.; Dulac, C.; Zhuang, X. High-performance multiplexed fluorescence in situ hybridization in culture and tissue with matrix imprinting and clearing. Proc. Natl. Acad. Sci. USA 2016, 113, 14456–14461. [Google Scholar] [CrossRef] [Green Version]
- Di Guardo, G. Lipofuscin, lipofuscin-like pigments and autofluorescence. Eur. J. Histochem. EJH 2015, 59, 2485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wotton, J.M.; Peterson, E.; Anderson, L.; Murray, S.A.; Braun, R.E.; Chesler, E.J.; White, J.K.; Kumar, V. Machine learning-based automated phenotyping of inflammatory nocifensive behavior in mice. Mol. Pain 2020, 16, 1744806920958596. [Google Scholar] [CrossRef] [PubMed]
- Ke, R.; Mignardi, M.; Hauling, T.; Nilsson, M. Fourth Generation of Next-Generation Sequencing Technologies: Promise and Consequences. Hum. Mutat. 2016, 37, 1363–1367. [Google Scholar] [CrossRef] [PubMed]
- Lein, E.; Borm, L.E.; Linnarsson, S. The promise of spatial transcriptomics for neuroscience in the era of molecular cell typing. Science 2017, 358, 64–69. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.H.; Daugharthy, E.R.; Scheiman, J.; Kalhor, R.; Ferrante, T.C.; Terry, R.; Turczyk, B.M.; Yang, J.L.; Lee, H.S.; Aach, J. Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nat. Protoc. 2015, 10, 442–458. [Google Scholar] [CrossRef] [Green Version]
- Farris, S.; Wang, Y.; Ward, J.M.; Dudek, S.M. Optimized method for robust transcriptome profiling of minute tissues using laser capture microdissection and low-input RNA-Seq. Front. Mol. Neurosci. 2017, 10, 185. [Google Scholar] [CrossRef]
- Singh, S.; Wang, L.; Schaff, D.L.; Sutcliffe, M.D.; Koeppel, A.F.; Kim, J.; Onengut-Gumuscu, S.; Park, K.-S.; Zong, H.; Janes, K.A. In situ 10-cell RNA sequencing in tissue and tumor biopsy samples. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Foley, J.W.; Zhu, C.; Jolivet, P.; Zhu, S.X.; Lu, P.; Meaney, M.J.; West, R.B. Gene expression profiling of single cells from archival tissue with laser-capture microdissection and Smart-3SEQ. Genome Res. 2019, 29, 1816–1825. [Google Scholar] [CrossRef] [Green Version]
- Vahrenkamp, J.M.; Szczotka, K.; Dodson, M.K.; Jarboe, E.A.; Soisson, A.P.; Gertz, J. FFPEcap-seq: A method for sequencing capped RNAs in formalin-fixed paraffin-embedded samples. Genome Res. 2019, 29, 1826–1835. [Google Scholar] [CrossRef]
- Morton, M.L.; Bai, X.; Merry, C.R.; Linden, P.A.; Khalil, A.M.; Leidner, R.S.; Thompson, C.L. Identification of mRNAs and lincRNAs associated with lung cancer progression using next-generation RNA sequencing from laser micro-dissected archival FFPE tissue specimens. Lung Cancer 2014, 85, 31–39. [Google Scholar] [CrossRef] [Green Version]
- Civita, P.; Franceschi, S.; Aretini, P.; Ortenzi, V.; Menicagli, M.; Lessi, F.; Pasqualetti, F.; Naccarato, A.G.; Mazzanti, C.M. Laser capture microdissection and RNA-seq analysis: High sensitivity approaches to explain histopathological heterogeneity in human glioblastoma FFPE archived tissues. Front. Oncol. 2019, 9, 482. [Google Scholar] [CrossRef] [PubMed]
- Wels, J.; Kaplan, R.N.; Rafii, S.; Lyden, D. Migratory neighbors and distant invaders: Tumor-associated niche cells. Genes Dev. 2008, 22, 559–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Travaglini, K.J.; Nabhan, A.N.; Penland, L.; Sinha, R.; Gillich, A.; Sit, R.V.; Chang, S.; Conley, S.D.; Mori, Y.; Seita, J. A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature 2020, 587, 619–625. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Ning, B.; Shi, T. Single-cell RNA-seq technologies and related computational data analysis. Front. Genet. 2019, 10, 317. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Qing, T.; Zheng, Y.; Jin, L.; Shi, L. Advances in single-cell RNA sequencing and its applications in cancer research. Oncotarget 2017, 8, 53763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rantalainen, M. Application of single-cell sequencing in human cancer. Brief. Funct. Genom. 2018, 17, 273–282. [Google Scholar] [CrossRef] [Green Version]
- Andersson, A.; Bergenstråhle, J.; Asp, M.; Bergenstråhle, L.; Jurek, A.; Navarro, J.F.; Lundeberg, J. Single-cell and spatial transcriptomics enables probabilistic inference of cell type topography. Commun. Biol. 2020, 3, 1–8. [Google Scholar] [CrossRef]
- Abdelaal, T.; Mourragui, S.; Mahfouz, A.; Reinders, M.J. SpaGE: Spatial gene enhancement using scRNA-seq. Nucleic Acids Res. 2020, 48, e107. [Google Scholar] [CrossRef]
- Ren, X.; Zhong, G.; Zhang, Q.; Zhang, L.; Sun, Y.; Zhang, Z. Reconstruction of cell spatial organization from single-cell RNA sequencing data based on ligand-receptor mediated self-assembly. Cell Res. 2020, 30, 763–778. [Google Scholar] [CrossRef]
- Gyllborg, D.; Langseth, C.M.; Qian, X.; Choi, E.; Salas, S.M.; Hilscher, M.M.; Lein, E.S.; Nilsson, M. Hybridization-based in situ sequencing (HybISS) for spatially resolved transcriptomics in human and mouse brain tissue. Nucleic Acids Res. 2020, 48, e112. [Google Scholar] [CrossRef]
- Asp, M.; Giacomello, S.; Larsson, L.; Wu, C.; Fürth, D.; Qian, X.; Wärdell, E.; Custodio, J.; Reimegård, J.; Salmén, F. A spatiotemporal organ-wide gene expression and cell atlas of the developing human heart. Cell 2019, 179, 1647–1660.e19. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Suo, S.; Tam, P.P.; Han, J.-D.J.; Peng, G.; Jing, N. Spatial transcriptomic analysis of cryosectioned tissue samples with Geo-seq. Nat. Protoc. 2017, 12, 566–580. [Google Scholar] [CrossRef] [PubMed]
- Saviano, A.; Henderson, N.C.; Baumert, T.F. Single-cell genomics and spatial transcriptomics: Discovery of novel cell states and cellular interactions in liver physiology and disease biology. J. Hepatol. 2020, 73, 1219–1230. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Sun, B.-F.; Chen, C.-Y.; Zhou, J.-Y.; Chen, Y.-S.; Chen, H.; Liu, L.; Huang, D.; Jiang, J.; Cui, G.-S. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 2019, 29, 725–738. [Google Scholar] [CrossRef]
- Lee, M.-C.W.; Lopez-Diaz, F.J.; Khan, S.Y.; Tariq, M.A.; Dayn, Y.; Vaske, C.J.; Radenbaugh, A.J.; Kim, H.J.; Emerson, B.M.; Pourmand, N. Single-cell analyses of transcriptional heterogeneity during drug tolerance transition in cancer cells by RNA sequencing. Proc. Natl. Acad. Sci. USA 2014, 111, E4726–E4735. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.-T.; Lee, H.W.; Lee, H.-O.; Song, H.J.; Shin, S.; Kim, H.; Shin, Y.; Nam, D.-H.; Jeong, B.C.; Kirsch, D.G. Application of single-cell RNA sequencing in optimizing a combinatorial therapeutic strategy in metastatic renal cell carcinoma. Genome Biol. 2016, 17, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Villani, A.-C.; Satija, R.; Reynolds, G.; Sarkizova, S.; Shekhar, K.; Fletcher, J.; Griesbeck, M.; Butler, A.; Zheng, S.; Lazo, S. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 2017, 356, eaah4573. [Google Scholar] [CrossRef] [Green Version]
- Zheng, C.; Zheng, L.; Yoo, J.-K.; Guo, H.; Zhang, Y.; Guo, X.; Kang, B.; Hu, R.; Huang, J.Y.; Zhang, Q. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell 2017, 169, 1342–1356. e1316. [Google Scholar] [CrossRef] [Green Version]
- Chung, W.; Eum, H.H.; Lee, H.-O.; Lee, K.-M.; Lee, H.-B.; Kim, K.-T.; Ryu, H.S.; Kim, S.; Lee, J.E.; Park, Y.H. Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat. Commun. 2017, 8, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Sharma, P.; Allison, J.P. Immune checkpoint targeting in cancer therapy: Toward combination strategies with curative potential. Cell 2015, 161, 205–214. [Google Scholar] [CrossRef] [Green Version]
- Simmons, A.J.; Lau, K.S. Deciphering tumor heterogeneity from FFPE tissues: Its promise and challenges. Mol. Cell. Oncol. 2017, 4, e1260191. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Mashock, M.; Tong, Z.; Mu, X.; Chen, H.; Zhou, X.; Zhang, H.; Zhao, G.; Liu, B.; Li, X. Changing technologies of RNA sequencing and their applications in clinical oncology. Front. Oncol. 2020, 10, 447. [Google Scholar] [CrossRef] [Green Version]
- Hagemann-Jensen, M.; Abdullayev, I.; Sandberg, R.; Faridani, O.R. Small-seq for single-cell small-RNA sequencing. Nat. Protoc. 2018, 13, 2407–2424. [Google Scholar] [CrossRef]
- Hayashi, T.; Ozaki, H.; Sasagawa, Y.; Umeda, M.; Danno, H.; Nikaido, I. Single-cell full-length total RNA sequencing uncovers dynamics of recursive splicing and enhancer RNAs. Nat. Commun. 2018, 9, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Armour, C.D.; Castle, J.C.; Chen, R.; Babak, T.; Loerch, P.; Jackson, S.; Shah, J.K.; Dey, J.; Rohl, C.A.; Johnson, J.M. Digital transcriptome profiling using selective hexamer priming for cDNA synthesis. Nat. Methods 2009, 6, 647–649. [Google Scholar] [CrossRef] [PubMed]
- Loi, D.S.; Yu, L.; Wu, A.R. Effective ribosomal RNA depletion for single-cell total RNA-seq by scDASH. PeerJ 2021, 9, e10717. [Google Scholar] [CrossRef] [PubMed]
- Brennecke, P.; Anders, S.; Kim, J.K.; Kołodziejczyk, A.A.; Zhang, X.; Proserpio, V.; Baying, B.; Benes, V.; Teichmann, S.A.; Marioni, J.C. Accounting for technical noise in single-cell RNA-seq experiments. Nat. Methods 2013, 10, 1093–1095. [Google Scholar] [CrossRef]
- Islam, S.; Zeisel, A.; Joost, S.; La Manno, G.; Zajac, P.; Kasper, M.; Lönnerberg, P.; Linnarsson, S. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat. Methods 2014, 11, 163–166. [Google Scholar] [CrossRef]
- Stylianopoulou, E.; Lykidis, D.; Ypsilantis, P.; Simopoulos, C.; Skavdis, G.; Grigoriou, M. A rapid and highly sensitive method of non radioactive colorimetric in situ hybridization for the detection of mRNA on tissue sections. PLoS ONE 2012, 7, e33898. [Google Scholar] [CrossRef] [Green Version]
- Zaghlool, A.; Ameur, A.; Wu, C.; Westholm, J.O.; Niazi, A.; Manivannan, M.; Bramlett, K.; Nilsson, M.; Feuk, L. Expression profiling and in situ screening of circular RNAs in human tissues. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Berglund, E.; Maaskola, J.; Schultz, N.; Friedrich, S.; Marklund, M.; Bergenstråhle, J.; Tarish, F.; Tanoglidi, A.; Vickovic, S.; Larsson, L. Spatial maps of prostate cancer transcriptomes reveal an unexplored landscape of heterogeneity. Nat. Commun. 2018, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Yoosuf, N.; Navarro, J.F.; Salmén, F.; Ståhl, P.L.; Daub, C.O. Identification and transfer of spatial transcriptomics signatures for cancer diagnosis. Breast Cancer Res. 2020, 22, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Technique | UMI | mRNA Priming | cDNA Preamplification | Library Generation | Transcript Coverage | Strand Specificity | Positional Bias | Costs | Reference |
|---|---|---|---|---|---|---|---|---|---|
| CEL-seq2 | Yes | Poly T | In vitro transcription | Transposon tagmentation | 3′-only | No | Weakley 3′ | High | [30] |
| SCRB-seq | Yes | Poly T | PCR | RNA fragmentation and adapter ligation | Nearly full length | No | Strongly 3′ | High | [9] |
| Smart-Seq | No | Poly T | PCR | Transposon tagmentation | Full length | No | Medium 3′ | High | [38] |
| Drop-seq | Yes | Poly T | PCR | Transposon tagmentation | 3′-only | Yes | 3′ only | Low | [41] |
| MARS-seq | Yes | Poly T | In vitro transcription | RNA fragmentation and adapter ligation | 3′-only | Yes | 3′ only | Low | [14] |
| 10×Genomics | Yes | Poly T | PCR | cDNA fragmentation, adapter ligation, and library amp | 3′-only | Yes | 3′ only | Low | [42] |
| Type | Strength | Weaknesses | Suitable Applications |
|---|---|---|---|
| Bulk RNA-seq | Well-developed, cost-effective, and high throughput technique | Unable to determine spatial content; gene expression profiling is average | Whole transcriptome-based biomarker discovery, targeted RNA-seq panel for gene fusion |
| MERFISH | High-throughput, high-sensitivity, high-multiplex power | Reduced specificity and off-target binding | Spatial organization of the transcriptome inside the cells, 3D organization of the chromatin and chromosome, spatial atlases of cells in complex tissues |
| LCM-RNAseq | Performs cell-specific gene expression analysis | Low-quality data, time-consuming, unable to perform spatial profiling | Applied for tumor heterogeneity to the specific population of cells |
| Single-cell RNA-Seq | Capable to perform >10,000 single-cell gene expression analysis | Applicable to a limited number of unique transcripts, unable to reveal spatial content, high cost | Characterization and discovery of cell type tumor heterogeneity |
| Digital Spatial Profiling | Useful for FFPE materials, spatial profiling | Unable to reveal sequence information, restricted to a small number of gene panels only | Biomarker discovery, tumor microenvironments |
| Spatial transcriptomics | Spatial profiling, whole transcriptome analysis, sequence information | Time-consuming, the early phase of development | Tumor microenvironments, tumor heterogeneity |
| Fourth-generation RNA-seq | Potential of in situ sequencing | Not properly well developed | Great future potential but not demonstrated yet |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmed, R.; Zaman, T.; Chowdhury, F.; Mraiche, F.; Tariq, M.; Ahmad, I.S.; Hasan, A. Single-Cell RNA Sequencing with Spatial Transcriptomics of Cancer Tissues. Int. J. Mol. Sci. 2022, 23, 3042. https://doi.org/10.3390/ijms23063042
Ahmed R, Zaman T, Chowdhury F, Mraiche F, Tariq M, Ahmad IS, Hasan A. Single-Cell RNA Sequencing with Spatial Transcriptomics of Cancer Tissues. International Journal of Molecular Sciences. 2022; 23(6):3042. https://doi.org/10.3390/ijms23063042
Chicago/Turabian StyleAhmed, Rashid, Tariq Zaman, Farhan Chowdhury, Fatima Mraiche, Muhammad Tariq, Irfan S. Ahmad, and Anwarul Hasan. 2022. "Single-Cell RNA Sequencing with Spatial Transcriptomics of Cancer Tissues" International Journal of Molecular Sciences 23, no. 6: 3042. https://doi.org/10.3390/ijms23063042
APA StyleAhmed, R., Zaman, T., Chowdhury, F., Mraiche, F., Tariq, M., Ahmad, I. S., & Hasan, A. (2022). Single-Cell RNA Sequencing with Spatial Transcriptomics of Cancer Tissues. International Journal of Molecular Sciences, 23(6), 3042. https://doi.org/10.3390/ijms23063042










