Cryoprotectants-Free Vitrification and Conventional Freezing of Human Spermatozoa: A Comparative Transcript Profiling
Abstract
:1. Introduction
2. Results
2.1. Overview Analysis of RNA Sequencing
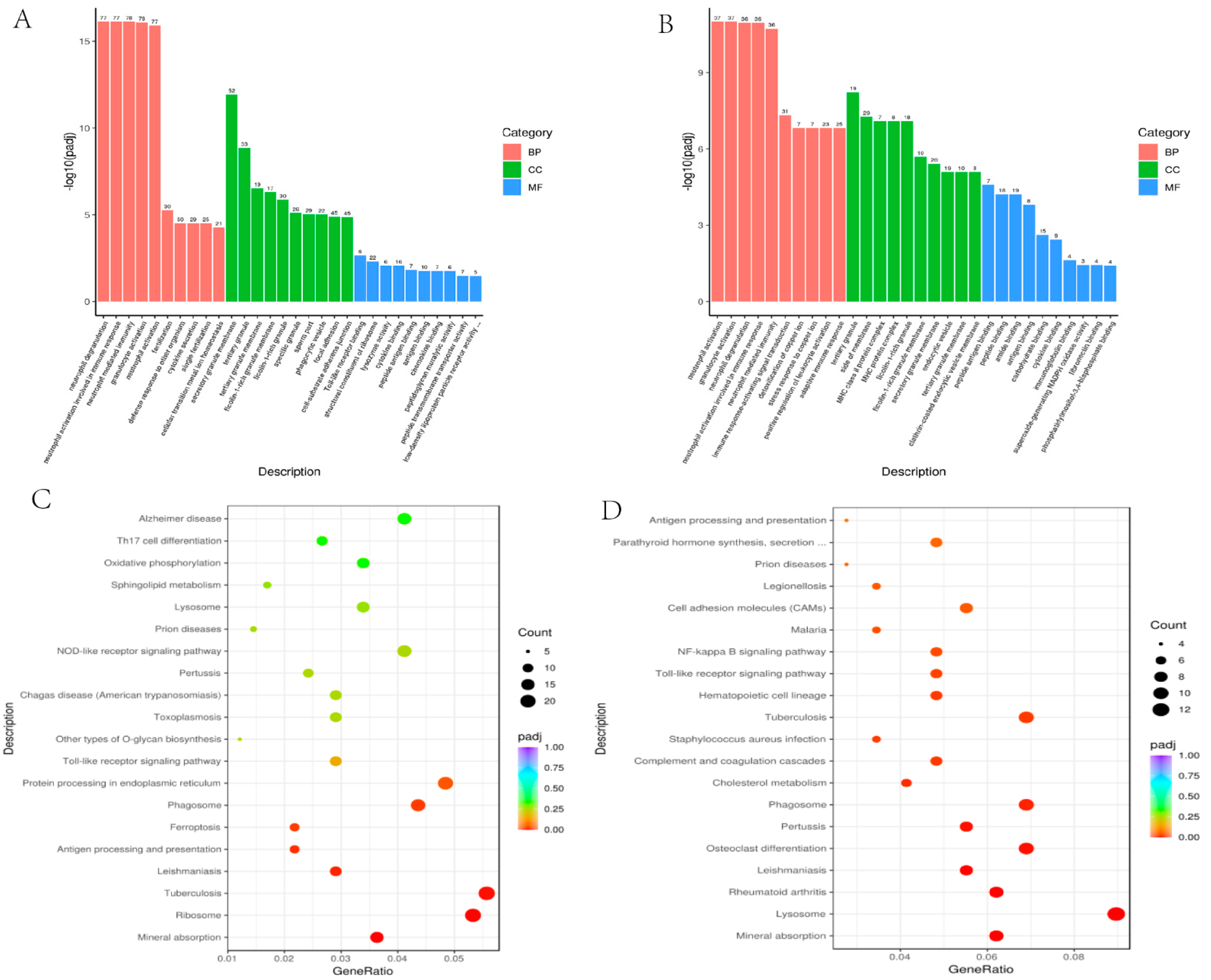
2.2. Gene Ontology Enrichment Analysis
2.3. Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Analysis
2.4. Protein–Protein Interaction (PPI) Construction and Hub Gens Identification
3. Discussion
3.1. Indication from DEGs
3.2. Findings in GO Terms
3.3. Interesting Results from KEGG Pathway Analysis
3.4. What the PPI Analysis Tell Us
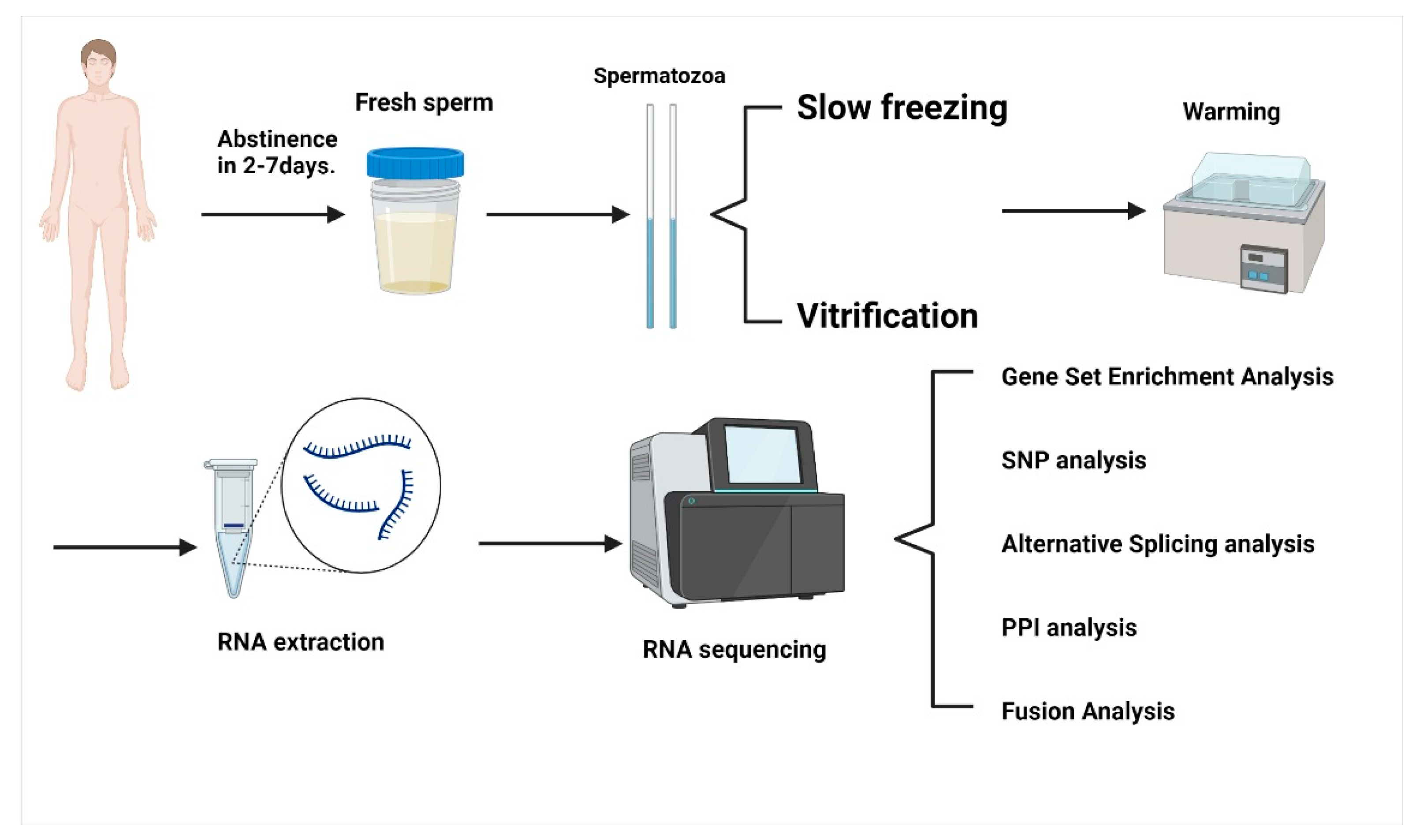
4. Materials and Methods
4.1. Sample Collection
4.2. Spermatozoa Cryopreservation
4.2.1. Spermatozoa Slow Freezing
4.2.2. Spermatozoa Vitrification
4.3. RNA Extraction and Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bunge, R.G.; Sherman, J.K. Fertilizing Capacity of Frozen Human Spermatozoa. Nature 1953, 172, 767–768. [Google Scholar] [CrossRef] [PubMed]
- Sanger, W.G.; Olson, J.H.; Sherman, J.K. Semen cryobanking for men with cancer—Criteria change. Fertil. Steril. 1992, 58, 1024–1027. [Google Scholar] [CrossRef]
- Li, Y.-X.; Zhou, L.; Lv, M.-Q.; Ge, P.; Liu, Y.-C.; Zhou, D.-X. Vitrification and conventional freezing methods in sperm cryopreservation: A systematic review and meta-analysis. Eur. J. Obstet. Gynecol. Reprod. Biol. 2019, 233, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Todorov, P.; Isachenko, E.; Rahimi, G.; Wang, W.; von Brandenstein, M.; Kumar, P.; Mallmann, P.; Isachenko, V. Aseptic capillary vitrification of human spermatozoa: Cryoprotectant-free vs. cryoprotectant-included technologies. Cryobiology 2021, 99, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Isachenko, V.; Maettner, R.; Petrunkina, A.; Sterzik, K.; Mallmann, P.; Rahimi, G.; Sanchez, R.; Risopatron, J.; Damjanoski, I.; Isachenko, E. Vitrification of human ICSI/IVF spermatozoa without cryoprotectants: New capillary technology. J. Androl. 2012, 33, 462–468. [Google Scholar] [CrossRef] [PubMed]
- Isachenko, E.; Isachenko, V.; Weiss, J.M.; Kreienberg, R.; Katkov, I.I.; Schulz, M.; Lulat, A.G.; Risopatrón, M.J.; Sánchez, R. Acrosomal status and mitochondrial activity of human spermatozoa vitrified with sucrose. Reproduction 2008, 136, 167–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Bansal, S.K.; Gupta, N.; Sankhwar, S.N.; Rajender, S. Differential Genes Expression between Fertile and Infertile Spermatozoa Revealed by Transcriptome Analysis. PLoS ONE 2015, 10, e0127007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ostermeier, G.C.; Goodrich, R.J.; Diamond, M.P.; Dix, D.J.; Krawetz, S.A. Toward using stable spermatozoal RNAs for prognostic assessment of male factor fertility. Fertil. Steril. 2005, 83, 1687–1694. [Google Scholar] [CrossRef]
- Dai, D.H.; Qazi, I.H.; Ran, M.X.; Liang, K.; Zhang, Y.; Zhang, M.; Zhou, G.B.; Angel, C.; Zeng, C.J. Exploration of miRNA and mRNA Profiles in Fresh and Frozen-Thawed Boar Sperm by Transcriptome and Small RNA Sequencing. Int. J. Mol. Sci. 2019, 20, 802. [Google Scholar] [CrossRef] [Green Version]
- Fraser, L.; Brym, P.; Pareek, C.S.; Mogielnicka-Brzozowska, M.; Paukszto, Ł.; Jastrzębski, J.P.; Wasilewska-Sakowska, K.; Mańkowska, A.; Sobiech, P.; Żukowski, K. Transcriptome analysis of boar spermatozoa with different freezability using RNA-Seq. Theriogenology 2020, 142, 400–413. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, Y.; Zhu, H.; Hao, H.; Zhao, X.; Qin, T.; Wang, D. Comparative transcript profiling of gene expression of fresh and frozen-thawed bull sperm. Theriogenology 2015, 83, 504–511. [Google Scholar] [CrossRef] [PubMed]
- Gur, Y.; Breitbart, H. Mammalian sperm translate nuclear-encoded proteins by mitochondrial-type ribosomes. Genes Dev. 2006, 20, 411–416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Z.; Umehara, T.; Okazaki, T.; Goto, M.; Fujita, Y.; Hoque, S.A.M.; Kawai, T.; Zeng, W.; Shimada, M. Gene Expression and Protein Synthesis in Mitochondria Enhance the Duration of High-Speed Linear Motility in Boar Sperm. Front. Physiol. 2019, 10, 252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, P.; Wang, M.; Isachenko, E.; Rahimi, G.; Mallmann, P.; Wang, W.; von Brandenstein, M.; Isachenko, V. Unraveling Subcellular and Ultrastructural Changes During Vitrification of Human Spermatozoa: Effect of a Mitochondria-Targeted Antioxidant and a Permeable Cryoprotectant. Front. Cell Dev. Biol. 2021, 9, 672862. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wang, W.; Xu, Y.; Tang, M.; Fang, J.; Sun, H.; Sun, Y.; Gu, M.; Liu, Z.; Zhang, Z.; et al. Proteomic characteristics of human sperm cryopreservation. Proteomics 2014, 14, 298–310. [Google Scholar] [CrossRef] [PubMed]
- Bogle, O.A.; Kumar, K.; Attardo-Parrinello, C.; Lewis, S.E.; Estanyol, J.M.; Ballescà, J.L.; Oliva, R. Identification of protein changes in human spermatozoa throughout the cryopreservation process. Andrology 2017, 5, 10–22. [Google Scholar] [CrossRef]
- Li, S.; Ao, L.; Yan, Y.; Jiang, J.; Chen, B.; Duan, Y.; Shen, F.; Chen, J.; Inglis, B.; Ni, R.; et al. Differential motility parameters and identification of proteomic profiles of human sperm cryopreserved with cryostraw and cryovial. Clin. Proteom. 2019, 16, 24. [Google Scholar] [CrossRef]
- Zhang, Y.; Dai, D.; Chang, Y.; Li, Y.; Zhang, M.; Zhou, G.; Peng, Z.; Zeng, C. Cryopreservation of boar sperm induces differential microRNAs expression. Cryobiology 2017, 76, 24–33. [Google Scholar] [CrossRef]
- Valcarce, D.G.; Cartón-García, F.; Herráez, M.P.; Robles, V. Effect of cryopreservation on human sperm messenger RNAs crucial for fertilization and early embryo development. Cryobiology 2013, 67, 84–90. [Google Scholar] [CrossRef]
- Zeng, C.; Peng, W.; Ding, L.; He, L.; Zhang, Y.; Fang, D.; Tang, K. A preliminary study on epigenetic changes during boar spermatozoa cryopreservation. Cryobiology 2014, 69, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Card, C.J.; Anderson, E.J.; Zamberlan, S.; Krieger, K.E.; Kaproth, M.; Sartini, B.L. Cryopreserved bovine spermatozoal transcript profile as revealed by high-throughput ribonucleic acid sequencing. Biol. Reprod. 2013, 88, 49. [Google Scholar] [CrossRef]
- Jodar, M.; Selvaraju, S.; Sendler, E.; Diamond, M.P.; Krawetz, S.A. The presence, role and clinical use of spermatozoal RNAs. Hum. Reprod. Update 2013, 19, 604–624. [Google Scholar] [CrossRef]
- Bissonnette, N.; Lévesque-Sergerie, J.P.; Thibault, C.; Boissonneault, G. Spermatozoal transcriptome profiling for bull sperm motility: A potential tool to evaluate semen quality. Reproduction 2009, 138, 65–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cappallo-Obermann, H.; Schulze, W.; Jastrow, H.; Baukloh, V.; Spiess, A.N. Highly purified spermatozoal RNA obtained by a novel method indicates an unusual 28S/18S rRNA ratio and suggests impaired ribosome assembly. Mol. Hum. Reprod. 2011, 17, 669–678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuster, A.; Tang, C.; Xie, Y.; Ortogero, N.; Yuan, S.; Yan, W. SpermBase: A Database for Sperm-Borne RNA Contents. Biol. Reprod. 2016, 95, 99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selders, G.S.; Fetz, A.E.; Radic, M.Z.; Bowlin, G.L. An overview of the role of neutrophils in innate immunity, inflammation and host-biomaterial integration. Regen. Biomater. 2017, 4, 55–68. [Google Scholar] [CrossRef] [PubMed]
- Perobelli, S.M.; Galvani, R.G.; Gonçalves-Silva, T.; Xavier, C.R.; Nóbrega, A.; Bonomo, A. Plasticity of neutrophils reveals modulatory capacity. Braz. J. Med. Biol. Res. Rev. 2015, 48, 665–675. [Google Scholar] [CrossRef] [Green Version]
- Björnsdottir, H.; Welin, A.; Michaëlsson, E.; Osla, V.; Berg, S.; Christenson, K.; Sundqvist, M.; Dahlgren, C.; Karlsson, A.; Bylund, J. Neutrophil NET formation is regulated from the inside by myeloperoxidase-processed reactive oxygen species. Free. Radic. Biol. Med. 2015, 89, 1024–1035. [Google Scholar] [CrossRef]
- Chistiakov, D.A.; Bobryshev, Y.V.; Orekhov, A.N. Neutrophil’s weapons in atherosclerosis. Exp. Mol. Pathol. 2015, 99, 663–671. [Google Scholar] [CrossRef]
- Smith, J.A. Neutrophils, host defense, and inflammation: A double-edged sword. J. Leukoc. Biol. 1994, 56, 672–686. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.; Dicker, B.L.; Jayasinghe, S.N.; De Gregorio, F.; Tian, H.; Han, D.Y.; Hudson, K.R. Strong inhibition of neutrophil–sperm interaction in cattle by selective phosphatidylinositol 3-kinase inhibitors. Biol. Reprod. 2017, 97, 671–687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaikwad, A.S.; Anderson, A.L.; Merriner, D.J.; O’Connor, A.E.; Houston, B.J.; Aitken, R.J.; O’Bryan, M.K.; Nixon, B. GLIPR1L1 is an IZUMO-binding protein required for optimal fertilization in the mouse. BMC Biol. 2019, 17, 86. [Google Scholar] [CrossRef] [PubMed]
- He, Z.Y.; Brakebusch, C.; Fässler, R.; Kreidberg, J.A.; Primakoff, P.; Myles, D.G. None of the integrins known to be present on the mouse egg or to be ADAM receptors are essential for sperm-egg binding and fusion. Dev. Biol. 2003, 254, 226–237. [Google Scholar] [CrossRef] [Green Version]
- Yoon, S.J.; Rahman, M.S.; Kwon, W.S.; Ryu, D.Y.; Park, Y.J.; Pang, M.G. Proteomic identification of cryostress in epididymal spermatozoa. J. Anim. Sci. Biotechnol. 2016, 7, 67. [Google Scholar] [CrossRef] [Green Version]
- Yuan, J. Protein degradation and phosphorylation after freeze thawing result in spermatozoon dysfunction. Proteomics 2014, 14, 155–156. [Google Scholar] [CrossRef]
- Yoon, S.J.; Rahman, M.S.; Kwon, W.S.; Park, Y.J.; Pang, M.G. Addition of Cryoprotectant Significantly Alters the Epididymal Sperm Proteome. PLoS ONE 2016, 11, e0152690. [Google Scholar] [CrossRef] [Green Version]
- Suomalainen, L.; Dunkel, L.; Ketola, I.; Eriksson, M.; Erkkilä, K.; Oksjoki, R.; Taari, K.; Heikinheimo, M.; Pentikäinen, V. Activator protein-1 in human male germ cell apoptosis. Mol. Hum. Reprod. 2004, 10, 743–753. [Google Scholar] [CrossRef] [Green Version]
- Donnellan, E.M.; O’Brien, M.B.; Meade, K.G.; Fair, S. Comparison of the uterine inflammatory response to frozen-thawed sperm from high and low fertility bulls. Theriogenology 2021, 176, 26–34. [Google Scholar] [CrossRef]
- Fujita, Y.; Mihara, T.; Okazaki, T.; Shitanaka, M.; Kushino, R.; Ikeda, C.; Negishi, H.; Liu, Z.; Richards, J.S.; Shimada, M. Toll-like receptors (TLR) 2 and 4 on human sperm recognize bacterial endotoxins and mediate apoptosis. Hum. Reprod. 2011, 26, 2799–2806. [Google Scholar] [CrossRef] [Green Version]
- Archana, S.S.; Selvaraju, S.; Binsila, B.K.; Arangasamy, A.; Krawetz, S.A. Immune regulatory molecules as modifiers of semen and fertility: A review. Mol. Reprod. Dev. 2019, 86, 1485–1504. [Google Scholar] [CrossRef] [PubMed]
- Khan, D.R.; Ahmad, N.; Anzar, M.; Channa, A.A. Apoptosis in fresh and cryopreserved buffalo sperm. Theriogenology 2009, 71, 872–876. [Google Scholar] [CrossRef] [PubMed]
- Ran, M.-X.; Zhou, Y.-M.; Liang, K.; Wang, W.-C.; Zhang, Y.; Zhang, M.; Yang, J.-D.; Zhou, G.-B.; Wu, K.; Wang, C.-D.; et al. Comparative Analysis of MicroRNA and mRNA Profiles of Sperm with Different Freeze Tolerance Capacities in Boar (Sus scrofa) and Giant Panda (Ailuropoda melanoleuca). Biomolecules 2019, 9, 432. [Google Scholar] [CrossRef] [Green Version]
- Karabulut, S.; Demiroğlu-Zergeroğlu, A.; Yılmaz, E.; Kutlu, P.; Keskin, İ. Effects of human sperm cryopreservation on apoptotic markers in normozoospermic and non-normozoospermic patients. Zygote 2018, 26, 308–313. [Google Scholar] [CrossRef] [PubMed]
- Becker, J.; Craig, E.A. Heat-shock proteins as molecular chaperones. Eur. J. Biochem. 1994, 219, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Nollen, E.A.; Morimoto, R.I. Chaperoning signaling pathways: Molecular chaperones as stress-sensing ‘heat shock’ proteins. J. Cell Sci. 2002, 115, 2809–2816. [Google Scholar] [CrossRef]
- Wang, P.; Wang, Y.F.; Wang, H.; Wang, C.W.; Zan, L.S.; Hu, J.H.; Li, Q.W.; Jia, Y.H.; Ma, G.J. HSP90 expression correlation with the freezing resistance of bull sperm. Zygote 2014, 22, 239–245. [Google Scholar] [CrossRef]
- Chen, X.; Zhu, H.; Hu, C.; Hao, H.; Zhang, J.; Li, K.; Zhao, X.; Qin, T.; Zhao, K.; Zhu, H.; et al. Identification of differentially expressed proteins in fresh and frozen-thawed boar spermatozoa by iTRAQ-coupled 2D LC-MS/MS. Reproduction 2014, 147, 321–330. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.Y.; Kuo, Y.H.; Lee, W.C.; Tsou, H.L.; Lee, Y.P.; Chang, H.L.; Wu, J.J.; Yang, P.C. Substantial decrease of heat-shock protein 90 precedes the decline of sperm motility during cooling of boar spermatozoa. Theriogenology 1999, 51, 1007–1016. [Google Scholar] [CrossRef]
- Cao, W.L.; Wang, Y.X.; Xiang, Z.Q.; Li, Z. Cryopreservation-induced decrease in heat-shock protein 90 in human spermatozoa and its mechanism. Asian J. Androl. 2003, 5, 43–46. [Google Scholar]
- Zhang, X.G.; Hu, S.; Han, C.; Zhu, Q.C.; Yan, G.J.; Hu, J.H. Association of heat shock protein 90 with motility of post-thawed sperm in bulls. Cryobiology 2015, 70, 164–169. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Shu, Z.; He, L.; Cui, X.; Wang, Y.; Gao, D. The pertinence of expression of heat shock proteins (HSPs) to the efficacy of cryopreservation in HELAs. Cryo Lett. 2005, 26, 7–16. [Google Scholar]
- Boulanger, J.; Faulds, D.; Eddy, E.M.; Lingwood, C.A. Members of the 70 kDa heat shock protein family specifically recognize sulfoglycolipids: Role in gamete recognition and mycoplasma-related infertility. J. Cell. Physiol. 1995, 165, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Miller, D.; Brough, S.; al-Harbi, O. Characterization and cellular distribution of human spermatozoal heat shock proteins. Hum. Reprod. 1992, 7, 637–645. [Google Scholar] [CrossRef]
- Tsujimoto, Y.; Shimizu, S. Bcl-2 family: Life-or-death switch. FEBS Lett. 2000, 466, 6–10. [Google Scholar] [CrossRef] [Green Version]
- de Oliveira, M.R.; Nabavi, S.F.; Manayi, A.; Daglia, M.; Hajheydari, Z.; Nabavi, S.M. Resveratrol and the mitochondria: From triggering the intrinsic apoptotic pathway to inducing mitochondrial biogenesis, a mechanistic view. Biochim. Biophys. Acta 2016, 1860, 727–745. [Google Scholar] [CrossRef]
- Nasimi, P.; Roohi, S. Cell Death in Animals and Plants; Apoptosis, Necrosis & Autophagy; Islamic Azad University of Masjed Soleyman Press: Khuzestan, Iran, 2012. [Google Scholar]
- Belka, C.; Budach, W. Anti-apoptotic Bcl-2 proteins: Structure, function and relevance for radiation biology. Int. J. Radiat. Biol. 2002, 78, 643–658. [Google Scholar] [CrossRef]
- Llambi, F.; Moldoveanu, T.; Tait, S.W.; Bouchier-Hayes, L.; Temirov, J.; McCormick, L.L.; Dillon, C.P.; Green, D.R. A unified model of mammalian BCL-2 protein family interactions at the mitochondria. Mol. Cell 2011, 44, 517–531. [Google Scholar] [CrossRef] [Green Version]
- Kontos, C.K.; Christodoulou, M.I.; Scorilas, A. Apoptosis-related BCL2-family members: Key players in chemotherapy. Anti-Cancer Agents Med. Chem. 2014, 14, 353–374. [Google Scholar] [CrossRef]
- Dalal, J.; Kumar, A.; Honparkhe, M.; Deka, D.; Singh, N. Minimization of apoptosis-like changes in cryopreserved buffalo bull sperm by supplementing extender with Bcl-2 protein. Vet. World 2016, 9, 432–436. [Google Scholar] [CrossRef] [Green Version]
- He, W.-H.; Zhai, X.-H.; Duan, X.-J.; Di, H.-S. Effect of resveratrol treatment on apoptosis and apoptotic pathways during boar semen freezing. J. Zhejiang Univ. Sci. B 2020, 21, 485–494. [Google Scholar] [CrossRef] [PubMed]
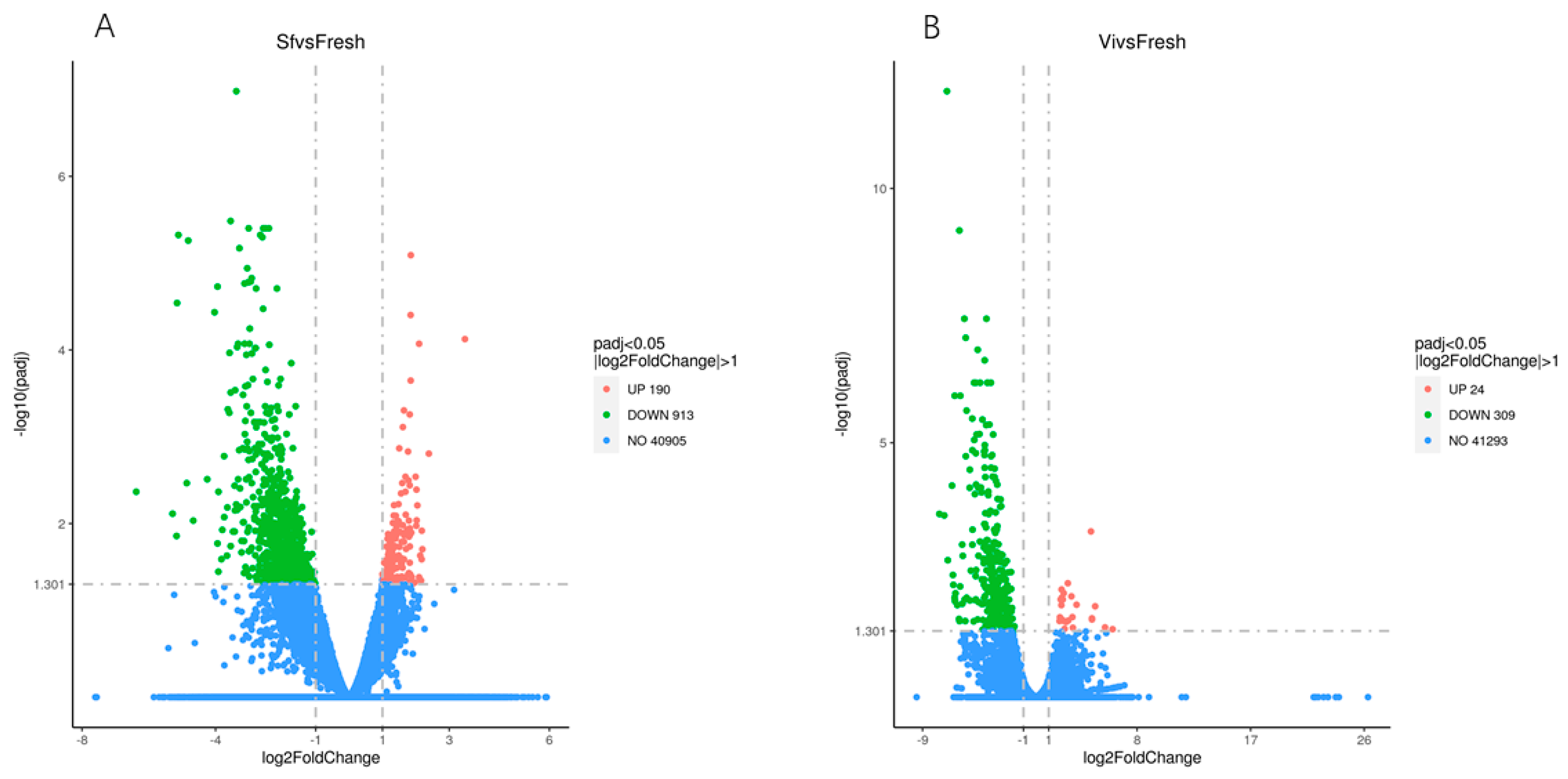

| Group | Raw Reads | Raw Bases | Clean Reads | Clean Bases | GC Content % | %Q30 | %Q20 | Total cczzMapped Reads | Unique cczzMapped Reads | Positive Mapped Reads | Negative Mapped Reads |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Fresh | 192,180,478 | 28.83G | 183,898,966 | 27.58G | 48.35 | 88.87 | 94.27 | 136,571,453 (74.26%) | 128,873,987 (70.07%) | 64,747,047 (35.2%) | 64,126,940 (34.87%) |
| Slow freezing | 171,745,512 | 25.76G | 162,929,510 | 24.44G | 50.1 | 88.24 | 93.67 | 112,130,004 (68.82%) | 104,616,702 (64.2%) | 52,606,155 (32.28%) | 52,010,547 (31.92%) |
| Vitrification | 183,160,670 | 27.48G | 173,830,938 | 26.07G | 47.71 | 89.22 | 94.3 | 121,859,357 (70.1%) | 115,152,448 (66.24%) | 57,780,422 (33.23%) | 57,372,026 (33.0%) |
| GO ID | Term | p-Value | padj |
|---|---|---|---|
| GO:0043312 | neutrophil degranulation | 2.69 × 10−20 | 7.53 × 10−17 |
| GO:0002283 | neutrophil activation involved in immune response | 3.95 × 10−20 | 7.53 × 10−17 |
| GO:0002446 | neutrophil mediated immunity | 4.58 × 10−20 | 7.53 × 10−17 |
| GO:0036230 | granulocyte activation | 6.67 × 10−20 | 8.21 × 10−17 |
| GO:0042119 | neutrophil activation | 1.22 × 10−19 | 1.21 × 10−16 |
| GO:0009566 | fertilization | 6.64 × 10−9 | 5.45 × 10−6 |
| GO:0098542 | defense response to other organism | 4.52 × 10−8 | 3.04 × 10−5 |
| GO:0050663 | cytokine secretion | 5.39 × 10−8 | 3.04 × 10−5 |
| GO:0007338 | single fertilization | 5.55 × 10−8 | 3.04 × 10−5 |
| GO:0046916 | cellular transition metal ion homostasis | 1.11 × 10−7 | 5.45 × 10−5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, M.; Todorov, P.; Wang, W.; Isachenko, E.; Rahimi, G.; Mallmann, P.; Isachenko, V. Cryoprotectants-Free Vitrification and Conventional Freezing of Human Spermatozoa: A Comparative Transcript Profiling. Int. J. Mol. Sci. 2022, 23, 3047. https://doi.org/10.3390/ijms23063047
Wang M, Todorov P, Wang W, Isachenko E, Rahimi G, Mallmann P, Isachenko V. Cryoprotectants-Free Vitrification and Conventional Freezing of Human Spermatozoa: A Comparative Transcript Profiling. International Journal of Molecular Sciences. 2022; 23(6):3047. https://doi.org/10.3390/ijms23063047
Chicago/Turabian StyleWang, Mengying, Plamen Todorov, Wanxue Wang, Evgenia Isachenko, Gohar Rahimi, Peter Mallmann, and Vladimir Isachenko. 2022. "Cryoprotectants-Free Vitrification and Conventional Freezing of Human Spermatozoa: A Comparative Transcript Profiling" International Journal of Molecular Sciences 23, no. 6: 3047. https://doi.org/10.3390/ijms23063047
APA StyleWang, M., Todorov, P., Wang, W., Isachenko, E., Rahimi, G., Mallmann, P., & Isachenko, V. (2022). Cryoprotectants-Free Vitrification and Conventional Freezing of Human Spermatozoa: A Comparative Transcript Profiling. International Journal of Molecular Sciences, 23(6), 3047. https://doi.org/10.3390/ijms23063047








