Extracellular Vesicles Secreted by Corneal Myofibroblasts Promote Corneal Epithelial Cell Migration
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.1.1. Human Corneal Epithelial (HCE) Cells
2.1.2. Human Corneal Keratocytes (HCKs), Fibroblasts (HCFs), and Myofibroblast (HCMs)
2.2. Extracellular Vesicle (EV) Isolation
2.3. Western Blot
2.4. Nanoparticle Tracking Analysis (NTA) and Zeta (ζ) Potential Measurements
2.5. Transmission Electron Microscopy (TEM)
2.6. EV Labeling with PKH26
2.7. Tracking PKH26-Labeled EVs in HCE Cells
2.8. In Vitro HCE Cell Scratch Assay
2.9. Tandem Mass Tag (TMT) Mass Spectrometry
2.9.1. Experimental Design and Statistical Rationale
2.9.2. Sample Preparation and Proteolytic Digestion
2.9.3. TMT Labeling and Label Check
2.9.4. High-pH Reverse-Phase High-Performance Liquid Chromatography (HPLC)
2.9.5. Liquid Chromatography–Tandem Mass Spectrometry (nanoLC–MS/MS)
2.9.6. Quantitative Proteomic Analysis
2.9.7. Ingenuity Pathway Analysis (IPA) of EVs
2.10. Statistics
3. Results
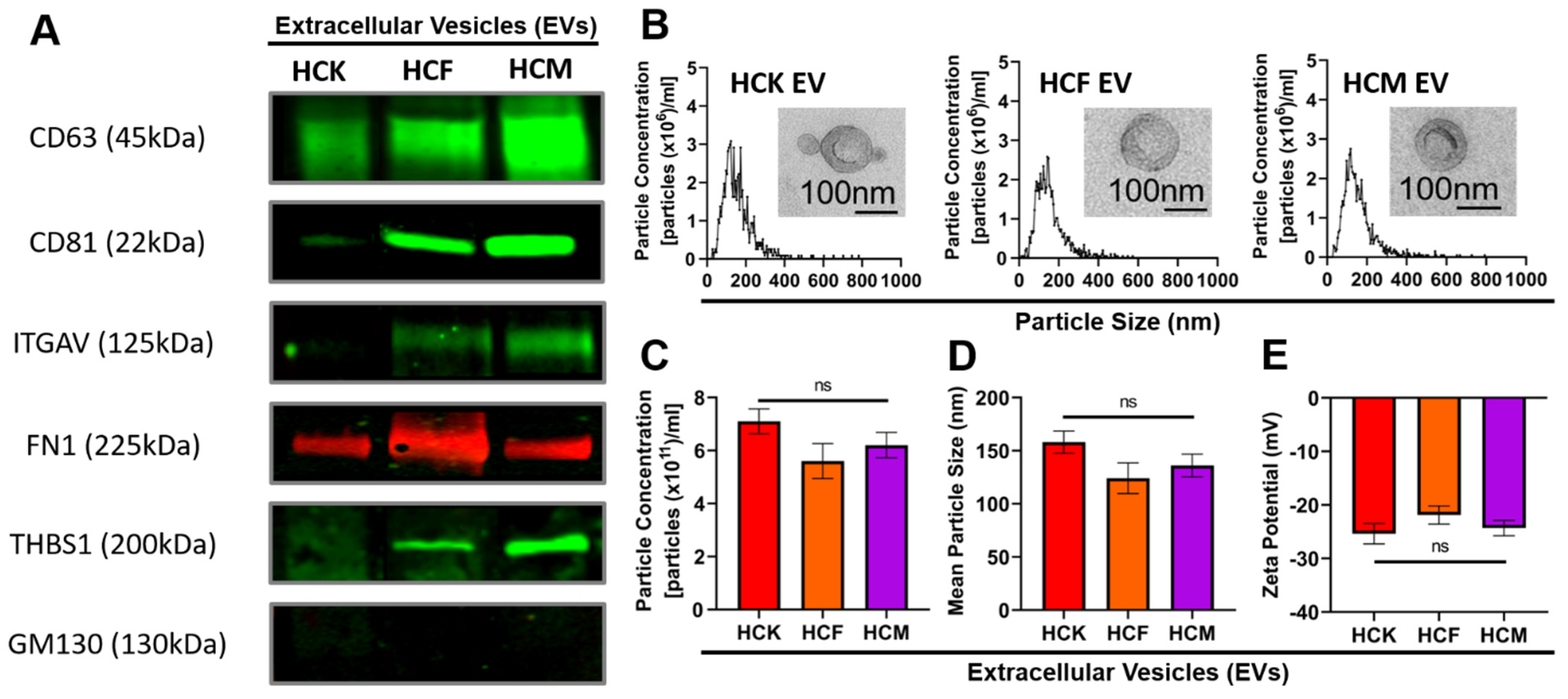
3.1. Characterization of HCK-, HCF-, and HCM-EVs
3.2. Proteomic Analysis of HCM-EV Protein Cargo
3.3. IPA of HCM-EV Proteins
3.4. HCM-EVs Promote HCE Cell Migration
3.5. HCM-EVs Increase HCE Cell Proliferation
3.6. PKH26-Labeled HCM-EVs Promote Cell Velocity
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Willoughby, C.; Ponzin, D.; Ferrari, S.; Lobo, A.; Landau, K.; Omidi, Y. Anatomy and physiology of the human eye: Effects of mucopolysaccharidoses disease on structure and function—A review. Clin. Exp. Ophthalmol. 2010, 38, 2–11. [Google Scholar] [CrossRef]
- Oie, Y.; Nishida, K. Regenerative Medicine for the Cornea. BioMed Res. Int. 2013, 2013, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Mobaraki, M.; Abbasi, R.; Omidian Vandchali, S.; Ghaffari, M.; Moztarzadeh, F.; Mozafari, M. Corneal Repair and Regeneration: Current Concepts and Future Directions. Front. Bioeng. Biotechnol. 2019, 7, 135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qazi, Y.; Wong, G.; Monson, B.; Stringham, J.; Ambati, B.K. Corneal transparency: Genesis, maintenance and dysfunction. Brain Res. Bull. 2010, 81, 198–210. [Google Scholar] [CrossRef] [Green Version]
- Meek, K.M.; Knupp, C. Corneal structure and transparency. Prog. Retin. Eye Res. 2015, 49, 1–16. [Google Scholar] [CrossRef] [Green Version]
- West-Mays, J.A.; Dwivedi, D.J. The keratocyte: Corneal stromal cell with variable repair phenotypes. Int. J. Biochem. Cell Biol. 2006, 38, 1625–1631. [Google Scholar] [CrossRef] [Green Version]
- Hay, E.D. Development of the Vertebrate Cornea. Adv. Appl. Microbiol. 1980, 63, 263–322. [Google Scholar] [CrossRef]
- Karamichos, D.; Guo, X.Q.; Hutcheon, A.E.K.; Zieske, J.D. Human Corneal Fibrosis: An In Vitro Model. Investig. Opthalmol. Vis. Sci. 2010, 51, 1382–1388. [Google Scholar] [CrossRef] [Green Version]
- Karamichos, D.; Rich, C.B.; Zareian, R.; Hutcheon, A.E.K.; Ruberti, J.W.; Trinkaus-Randall, V.; Zieske, J.D. TGF-β3 Stimulates Stromal Matrix Assembly by Human Corneal Keratocyte-Like Cells. Investig. Opthalmol. Vis. Sci. 2013, 54, 6612–6619. [Google Scholar] [CrossRef] [Green Version]
- Ren, R.; Hutcheon, A.; Guo, X.; Saeidi, N.; Melotti, S.; Ruberti, J.; Zieske, J.; Trinkaus-Randall, V. Human primary corneal fibroblasts synthesize and deposit proteoglycans in long-term 3-D cultures. Dev. Dyn. 2008, 237, 2705–2715. [Google Scholar] [CrossRef] [Green Version]
- McKay, T.B.; Hutcheon, A.E.K.; Zieske, J.D.; Ciolino, J.B. Extracellular Vesicles Secreted by Corneal Epithelial Cells Promote Myofibroblast Differentiation. Cells 2020, 9, 1080. [Google Scholar] [CrossRef] [PubMed]
- Yeung, V.; Sriram, S.; Tran, J.A.; Guo, X.; Hutcheon, A.E.K.; Zieske, J.D.; Karamichos, D.; Ciolino, J.B. FAK Inhibition Attenuates Corneal Fibroblast Differentiation In Vitro. Biomolecules 2021, 11, 1682. [Google Scholar] [CrossRef]
- Fini, M.E.; Stramer, B. How the Cornea Heals. Cornea 2005, 24, S2–S11. [Google Scholar] [CrossRef] [PubMed]
- Wilson, S.E.; Liu, J.J.; Mohan, R.R. Stromal-epithelial interactions in the cornea. Prog. Retin. Eye Res. 1999, 18, 293–309. [Google Scholar] [CrossRef]
- Zieske, J.; Higashijima, S.C.; Spurr-Michaud, S.J.; Gipson, I.K. Biosynthetic responses of the rabbit cornea to a keratectomy wound. Investig. Ophthalmol. Vis. Sci. 1987, 28, 1668–1677. [Google Scholar]
- Han, K.-Y.; Tran, J.A.; Chang, J.-H.; Azar, D.T.; Zieske, J. Potential role of corneal epithelial cell-derived exosomes in corneal wound healing and neovascularization. Sci. Rep. 2017, 7, srep40548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alhamwe, B.A.; Potaczek, D.; Miethe, S.; Alhamdan, F.; Hintz, L.; Magomedov, A.; Garn, H. Extracellular Vesicles and Asthma—More Than Just a Co-Existence. Int. J. Mol. Sci. 2021, 22, 4984. [Google Scholar] [CrossRef]
- Webber, J.; Yeung, V.; Clayton, A. Extracellular vesicles as modulators of the cancer microenvironment. Semin. Cell Dev. Biol. 2015, 40, 27–34. [Google Scholar] [CrossRef]
- Shephard, A.P.; Yeung, V.; Clayton, A.; Webber, J.P. Prostate cancer exosomes as modulators of the tumor microenvironment. J. Cancer Metastasis Treat. 2017, 3, 288. [Google Scholar] [CrossRef]
- Yeung, V.; Webber, J.P.; Dunlop, E.A.; Morgan, H.; Hutton, J.; Gurney, M.; Jones, E.; Falcon-Perez, J.; Tabi, Z.; Errington, R.; et al. Rab35-dependent extracellular nanovesicles are required for induction of tumour supporting stroma. Nanoscale 2018, 10, 8547–8559. [Google Scholar] [CrossRef] [Green Version]
- Nusinow, D.P.; Szpyt, J.; Ghandi, M.; Rose, C.M.; McDonald, E.R.; Kalocsay, M.; Jané-Valbuena, J.; Gelfand, E.; Schweppe, D.K.; Jedrychowski, M.; et al. Quantitative Proteomics of the Cancer Cell Line Encyclopedia. Cell 2020, 180, 387–402.e16. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.-C.; Hsu, Y.-C.; Chiu, H.-M.; Ueda, K.; Wu, M.-S.; Kao, C.-H.; Shen, T.-L. Exploration of the Proteomic Landscape of Small Extracellular Vesicles in Serum as Biomarkers for Early Detection of Colorectal Neoplasia. Front. Oncol. 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Zanotti, S.; Gibertini, S.; Blasevich, F.; Bragato, C.; Ruggieri, A.; Saredi, S.; Fabbri, M.; Bernasconi, P.; Maggi, L.; Mantegazza, R.; et al. Exosomes and exosomal miRNAs from muscle-derived fibroblasts promote skeletal muscle fibrosis. Matrix Biol. 2018, 74, 77–100. [Google Scholar] [CrossRef] [PubMed]
- Martin-Medina, A.; Lehmann, M.; Burgy, O.; Hermann, S.; Baarsma, H.; Wagner, D.E.; De Santis, M.M.; Ciolek, F.; Hofer, T.P.; Frankenberger, M.; et al. Increased Extracellular Vesicles Mediate WNT5A Signaling in Idiopathic Pulmonary Fibrosis. Am. J. Respir. Crit. Care Med. 2018, 198, 1527–1538. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Miao, J.; Wang, C.; Zhou, S.; Chen, S.; Ren, Q.; Hong, X.; Wang, Y.; Hou, F.F.; Zhou, L.; et al. Tubule-derived exosomes play a central role in fibroblast activation and kidney fibrosis. Kidney Int. 2020, 97, 1181–1195. [Google Scholar] [CrossRef] [PubMed]
- Ailawadi, S.; Wang, X.; Gu, H.; Fan, G.-C. Pathologic function and therapeutic potential of exosomes in cardiovascular disease. Biochim. Biophys. Acta Mol. Basis Dis. 2015, 1852, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Connolly, K.; Guschina, I.A.; Yeung, V.; Clayton, A.; Draman, M.S.; Von Ruhland, C.; Ludgate, M.; James, P.E.; Rees, D.A. Characterisation of adipocyte-derived extracellular vesicles released pre- and post-adipogenesis. J. Extracell. Vesicles 2015, 4, 29159. [Google Scholar] [CrossRef]
- Gartz, M.; Strande, J.L. Examining the Paracrine Effects of Exosomes in Cardiovascular Disease and Repair. J. Am. Hear. Assoc. 2018, 7, e007954. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Qiu, L.; Hu, W.; Deng, X.; Xu, H.; Cao, Y.; Xiao, Z.; Peng, L.; Johnson, S.; Alexey, L.; et al. Genetically-modified bone mesenchymal stem cells with TGF-β 3 improve wound healing and reduce scar tissue formation in a rabbit model. Exp. Cell Res. 2018, 367, 24–29. [Google Scholar] [CrossRef]
- Yeung, V.; Willis, G.R.; Taglauer, E.; Mitsialis, S.A.; Kourembanas, S. Paving the Road for Mesenchymal Stem Cell-Derived Exosome Therapy in Bronchopulmonary Dysplasia and Pulmonary Hypertension. In Stem Cell-Based Therapy for Lung Disease; Springer: Berlin/Heidelberg, Germany, 2019; pp. 131–152. [Google Scholar] [CrossRef]
- Jiang, T.; Wang, Z.; Sun, J. Human bone marrow mesenchymal stem cell-derived exosomes stimulate cutaneous wound healing mediates through TGF-β/Smad signaling pathway. Stem Cell Res. Ther. 2020, 11, 1–10. [Google Scholar] [CrossRef]
- Willis, G.R.; Fernandez-Gonzalez, A.; Reis, M.; Yeung, V.; Liu, X.; Ericsson, M.; Andrews, N.A.; Mitsialis, S.A.; Kourembanas, S. Mesenchymal stromal cell-derived small extracellular vesicles restore lung architecture and improve exercise capacity in a model of neonatal hyperoxia-induced lung injury. J. Extracell. Vesicles 2020, 9, 1790874. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Gonzalez, A.; Willis, G.R.; Yeung, V.; Reis, M.; Liu, X.; Mitsialis, S.A.; Kourembanas, S. Therapeutic Effects of Mesenchymal Stromal Cell-Derived Small Extracellular Vesicles in Oxygen-Induced Multi-Organ Disease: A Developmental Perspective. Front. Cell Dev. Biol. 2021, 9, 647025. [Google Scholar] [CrossRef] [PubMed]
- Reis, M.; Willis, G.R.; Fernandez-Gonzalez, A.; Yeung, V.; Taglauer, E.; Magaletta, M.; Parsons, T.; Derr, A.; Liu, X.; Maehr, R.; et al. Mesenchymal Stromal Cell-Derived Extracellular Vesicles Restore Thymic Architecture and T Cell Function Disrupted by Neonatal Hyperoxia. Front. Immunol. 2021, 12, 1203. [Google Scholar] [CrossRef]
- Taglauer, E.S.; Fernandez-Gonzalez, A.; Willis, G.R.; Reis, M.; Yeung, V.; Liu, X.; Prince, L.S.; Mitsialis, S.A.; Kourembanas, S. Antenatal Mesenchymal Stromal Cell Extracellular Vesicle Therapy Prevents Preeclamptic Lung Injury in Mice. Am. J. Respir. Cell Mol. Biol. 2022, 66, 86–95. [Google Scholar] [CrossRef] [PubMed]
- Taglauer, E.S.; Fernandez-Gonzalez, A.; Willis, G.R.; Reis, M.; Yeung, V.; Liu, X.; Mitsialis, S.A.; Kourembanas, S. Mesenchymal stromal cell-derived extracellular vesicle therapy prevents preeclamptic physiology through intrauterine immunomodulation. Biol. Reprod. 2021, 104, 457–467. [Google Scholar] [CrossRef]
- Willis, G.R.; Reis, M.; Gheinani, A.H.; Fernandez-Gonzalez, A.; Taglauer, E.S.; Yeung, V.; Liu, X.; Ericsson, M.; Haas, E.; Mitsialis, S.A.; et al. Extracellular Vesicles Protect the Neonatal Lung from Hyperoxic Injury through the Epigenetic and Transcriptomic Reprogramming of Myeloid Cells. Am. J. Respir. Crit. Care Med. 2021, 204, 1418–1432. [Google Scholar] [CrossRef]
- Samaeekia, R.; Rabiee, B.; Putra, I.; Shen, X.; Park, Y.J.; Hematti, P.; Eslani, M.; Djalilian, A.R. Effect of Human Corneal Mesenchymal Stromal Cell-derived Exosomes on Corneal Epithelial Wound Healing. Investig. Ophthalmol. Vis. Sci. 2018, 59, 5194–5200. [Google Scholar] [CrossRef] [Green Version]
- Shojaati, G.; Khandaker, I.; Funderburgh, M.L.; Mann, M.M.; Basu, R.; Stolz, D.B.; Geary, M.L.; Dos Santos, A.; Deng, S.X.; Funderburgh, J.L. Mesenchymal Stem Cells Reduce Corneal Fibrosis and Inflammation via Extracellular Vesicle-Mediated Delivery of miRNA. Stem Cells Transl. Med. 2019, 8, 1192–1201. [Google Scholar] [CrossRef] [Green Version]
- Yu, C.; Chen, P.; Xu, J.; Liu, Y.; Li, H.; Wang, L.; Di, G. hADSCs derived extracellular vesicles inhibit NLRP3inflammasome activation and dry eye. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- McKay, T.B.; Yeung, V.; Hutcheon, A.E.K.; Guo, X.; Zieske, J.D.; Ciolino, J.B. Extracellular Vesicles in the Cornea: Insights from Other Tissues. Anal. Cell. Pathol. 2021, 2021, 1–12. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keerthikumar, S.; Chisanga, D.; Ariyaratne, D.; Al Saffar, H.; Anand, S.; Zhao, K.; Samuel, M.; Pathan, M.; Jois, M.; Chilamkurti, N.; et al. ExoCarta: A Web-Based Compendium of Exosomal Cargo. J. Mol. Biol. 2016, 428, 688–692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, R.C.; Tan, S.S.; Yeo, R.W.Y.; Choo, A.B.H.; Reiner, A.; Su, Y.; Shen, Y.; Fu, Z.; Alexander, L.; Sze, S.K.; et al. MSC secretes at least 3 EV types each with a unique permutation of membrane lipid, protein and RNA. J. Extracell. Vesicles 2016, 5, 29828. [Google Scholar] [CrossRef] [PubMed]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pathan, M.; Fonseka, P.; Chitti, S.V.; Kang, T.; Sanwlani, R.; Van Deun, J.; Hendrix, A.; Mathivanan, S. Vesiclepedia 2019: A compendium of RNA, proteins, lipids and metabolites in extracellular vesicles. Nucleic Acids Res. 2019, 47, D516–D519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Araki-Sasaki, K.; Ohashi, Y.; Sasabe, T.; Hayashi, K.; Watanabe, H.; Tano, Y.; Handa, H. An SV40-immortalized human corneal epithelial cell line and its characterization. Investig. Ophthalmol. Vis. Sci. 1995, 36, 614–621. [Google Scholar]
- Guo, X.; Hutcheon, A.E.K.; Melotti, S.A.; Zieske, J.D.; Trinkaus-Randall, V.; Ruberti, J.W. Morphologic characterization of organized extracellular matrix deposition by ascorbic acid-stimulated human corneal fibroblasts. Investig. Opthalmol. Vis. Sci. 2007, 48, 4050–4060. [Google Scholar] [CrossRef]
- Long, C.J.; Roth, M.R.; Tasheva, E.S.; Funderburgh, M.; Smit, R.; Conrad, G.W.; Funderburgh, J.L. Fibroblast Growth Factor-2 Promotes Keratan Sulfate Proteoglycan Expression by Keratocytes in Vitro. J. Biol. Chem. 2000, 275, 13918–13923. [Google Scholar] [CrossRef] [Green Version]
- Foster, J.; Gouveia, R.M.; Connon, C.J. Low-glucose enhances keratocyte-characteristic phenotype from corneal stromal cells in serum-free conditions. Sci. Rep. 2015, 5, 10839. [Google Scholar] [CrossRef] [Green Version]
- Yusoff, N.Z.B.M.; Riau, A.K.; Yam, G.H.F.; Halim, N.S.H.B.; Mehta, J.S. Isolation and Propagation of Human Corneal Stromal Keratocytes for Tissue Engineering and Cell Therapy. Cells 2022, 11, 178. [Google Scholar] [CrossRef]
- Beales, M.P.; Funderburgh, J.L.; Jester, J.; Hassell, J.R. Proteoglycan synthesis by bovine keratocytes and corneal fibroblasts: Maintenance of the keratocyte phenotype in culture. Investig. Ophthalmol. Vis. Sci. 1999, 40, 1658–1663. [Google Scholar]
- Gardiner, C.; Ferreira, Y.J.; Dragovic, R.A.; Redman, C.W.G.; Sargent, I.L. Extracellular vesicle sizing and enumeration by nanoparticle tracking analysis. J. Extracell. Vesicles 2013, 2, 19671. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006, 30, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Cheung, C.H.Y.; Juan, H.-F. Quantitative proteomics in lung cancer. J. Biomed. Sci. 2017, 24, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Marchione, D.M.; Ilieva, I.; Devins, K.; Sharpe, D.; Pappin, D.J.; Garcia, B.A.; Wilson, J.P.; Wojcik, J.B. HYPERsol: High-Quality Data from Archival FFPE Tissue for Clinical Proteomics. J. Proteome Res. 2020, 19, 973–983. [Google Scholar] [CrossRef] [PubMed]
- Krasny, L.; Huang, P.H. Data-independent acquisition mass spectrometry (DIA-MS) for proteomic applications in oncology. Mol. Omics 2021, 17, 29–42. [Google Scholar] [CrossRef]
- Giansanti, P.; Tsiatsiani, L.; Low, T.Y.; Heck, A.J.R. Six alternative proteases for mass spectrometry–based proteomics beyond trypsin. Nat. Protoc. 2016, 11, 993–1006. [Google Scholar] [CrossRef]
- Rose, C.; Isasa, M.; Ordureau, A.; Prado, M.; Beausoleil, S.A.; Jedrychowski, M.P.; Finley, D.J.; Harper, J.; Gygi, S.P. Highly Multiplexed Quantitative Mass Spectrometry Analysis of Ubiquitylomes. Cell Syst. 2016, 3, 395–403.e4. [Google Scholar] [CrossRef] [Green Version]
- Mulholland, B.; Tuft, S.J.; Khaw, P.T. Matrix metalloproteinase distribution during early corneal wound healing. Eye 2004, 19, 584–588. [Google Scholar] [CrossRef] [PubMed]
- Ljubimov, A.V.; Saghizadeh, M. Progress in corneal wound healing. Prog. Retin. Eye Res. 2015, 49, 17–45. [Google Scholar] [CrossRef] [Green Version]
- Pastar, I.; Stojadinovic, O.; Yin, N.C.; Ramirez, H.; Nusbaum, A.G.; Sawaya, A.; Patel, S.B.; Khalid, L.; Isseroff, R.R.; Tomic-Canic, M. Epithelialization in Wound Healing: A Comprehensive Review. Adv. Wound Care 2014, 3, 445–464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bomb, R.; Heckle, M.R.; Sun, Y.; Mancarella, S.; Guntaka, R.V.; Gerling, I.C.; Weber, K.T. Myofibroblast secretome and its auto-/paracrine signaling. Expert Rev. Cardiovasc. Ther. 2016, 14, 591–598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jalili Angourani, K.; Mazhari, S.; Farivar, S.; Salman Mahini, D.; Rouintan, A.; Baghaei, K. Fibroblast-myofibroblast crosstalk after exposure to mesenchymal stem cells secretome. Gastroenterol. Hepatol. Bed Bench 2018, 11, S73–S79. [Google Scholar] [PubMed]
- Kalimuthu, S.; Gangadaran, P.; Rajendran, R.L.; Zhu, L.; Oh, J.M.; Lee, H.W.; Gopal, A.; Baek, S.H.; Jeong, S.Y.; Lee, S.-W.; et al. A New Approach for Loading Anticancer Drugs into Mesenchymal Stem Cell-Derived Exosome Mimetics for Cancer Therapy. Front. Pharmacol. 2018, 9, 1116. [Google Scholar] [CrossRef] [PubMed]
- Sung, B.H.; von Lersner, A.; Guerrero, J.; Krystofiak, E.S.; Inman, D.; Pelletier, R.; Zijlstra, A.; Ponik, S.M.; Weaver, A.M. A live cell reporter of exosome secretion and uptake reveals pathfinding behavior of migrating cells. Nat. Commun. 2020, 11, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.M.; Johanesen, P.A.; Wendt, M.K.; Binion, D.G.; Dwinell, M.B. CXCL12 activation of CXCR4 regulates mucosal host defense through stimulation of epithelial cell migration and promotion of intestinal barrier integrity. Am. J. Physiol. Liver Physiol. 2005, 288, G316–G326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shieh, J.-M.; Tsai, Y.-J.; Tsou, C.-J.; Wu, W.-B. CXCL1 Regulation in Human Pulmonary Epithelial Cells by Tumor Necrosis Factor. Cell. Physiol. Biochem. 2014, 34, 1373–1384. [Google Scholar] [CrossRef]
- Park, G.-Y.; Pathak, H.B.; Godwin, A.K.; Kwon, Y. Epithelial-stromal communication via CXCL1-CXCR2 interaction stimulates growth of ovarian cancer cells through p38 activation. Cell. Oncol. 2021, 44, 77–92. [Google Scholar] [CrossRef] [PubMed]
- Kuo, P.-L.; Shen, K.-H.; Hung, S.-H.; Hsu, Y.-L. CXCL1/GROα increases cell migration and invasion of prostate cancer by decreasing fibulin-1 expression through NF-κB/HDAC1 epigenetic regulation. Carcinogenesis 2012, 33, 2477–2487. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.; Ma, Q.; Li, L.; Wang, H. The CXCL12–CXCR4 axis promotes migration, invasiveness, and EMT in human papillary thyroid carcinoma B-CPAP cells via NF-κB signaling. Biochem. Cell Biol. 2018, 96, 619–626. [Google Scholar] [CrossRef]
- Chen, Q.; Liu, D.; Hu, Z.; Luo, C.; Zheng, S.L. miRNA-101-5p inhibits the growth and aggressiveness of NSCLC cells through targeting CXCL6. OncoTargets Ther. 2019, 12, 835–848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, G.; An, L.; Zhang, H.; Du, P.; Sheng, Y. Activation of CXCL6/CXCR1/2 Axis Promotes the Growth and Metastasis of Osteosarcoma Cells in vitro and in vivo. Front. Pharmacol. 2019, 10, 307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, C.; Yu, H.; Chen, R.; Tao, K.; Jian, L.; Peng, M.; Li, X.; Liu, M.; Liu, S. CXCL1 stimulates migration and invasion in ER-negative breast cancer cells via activation of the ERK/MMP2/9 signaling axis. Int. J. Oncol. 2019, 55, 684–696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ojeda, A.F.; Munjaal, R.P.; Lwigale, P.Y. Expression of CXCL12 and CXCL14 during eye development in chick and mouse. Gene Expr. Patterns 2013, 13, 303–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pulukuri, S.M.K.; Rao, J.S. Matrix metalloproteinase-1 promotes prostate tumor growth and metastasis. Int. J. Oncol. 2008, 32, 757–765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, Y.; Liang, F.; Zhang, Z.-Y. PRL1 Promotes Cell Migration and Invasion by Increasing MMP2 and MMP9 Expression through Src and ERK1/2 Pathways. Biochemistry 2009, 48, 1838–1846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Webb, A.H.; Gao, B.T.; Goldsmith, Z.K.; Irvine, A.S.; Saleh, N.; Lee, R.P.; Lendermon, J.B.; Bheemreddy, R.; Zhang, Q.; Brennan, R.C.; et al. Inhibition of MMP-2 and MMP-9 decreases cellular migration, and angiogenesis in in vitro models of retinoblastoma. BMC Cancer 2017, 17, 1–11. [Google Scholar] [CrossRef]
- Wang, X.; Yang, B.; She, Y.; Ye, Y. The lncRNA TP73-AS1 promotes ovarian cancer cell proliferation and metastasis via modulation of MMP2 and MMP9. J. Cell. Biochem. 2018, 119, 7790–7799. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Lv, L.; Tang, Y.; Zhang, L.; Wang, L. MMP-1 is overexpressed in triple-negative breast cancer tissues and the knockdown of MMP-1 expression inhibits tumor cell malignant behaviors in vitro. Oncol. Lett. 2018, 17, 1732–1740. [Google Scholar] [CrossRef] [Green Version]
- Stevens, L.J.; Page-McCaw, A. A secreted MMP is required for reepithelialization during wound healing. Mol. Biol. Cell 2012, 23, 1068–1079. [Google Scholar] [CrossRef]
- Caley, M.P.; Martins, V.; O’Toole, E. Metalloproteinases and Wound Healing. Adv. Wound Care 2015, 4, 225–234. [Google Scholar] [CrossRef] [Green Version]
- Rohani, M.G.; Parks, W.C. Matrix remodeling by MMPs during wound repair. Matrix Biol. 2015, 44, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Daniels, J.T.; Limb, G.A.; Saarialho-Kere, U.; Murphy, G.; Khaw, P.T. Human corneal epithelial cells require MMP-1 for HGF-mediated migration on collagen I. Investig. Opthalmol. Vis. Sci. 2003, 44, 1048–1055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Couture, C.; Zaniolo, K.; Carrier, P.; Lake, J.; Patenaude, J.; Germain, L.; Guérin, S.L. The tissue-engineered human cornea as a model to study expression of matrix metalloproteinases during corneal wound healing. Biomaterials 2016, 78, 86–101. [Google Scholar] [CrossRef] [PubMed]
- Movahedan, A.; Majdi, M.; Afsharkhamseh, N.; Sagha, H.M.; Saadat, N.S.; Shalileh, K.; Milani, B.Y.; Ying, H.; Djalilian, A.R. Notch Inhibition during Corneal Epithelial Wound Healing Promotes Migration. Investig. Opthalmol. Vis. Sci. 2012, 53, 7476–7483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arranz-Valsero, I.; Romaní, L.S.; García-Posadas, L.; López-García, A.; Diebold, Y. IL-6 as a corneal wound healing mediator in an in vitro scratch assay. Exp. Eye Res. 2014, 125, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Kowtharapu, B.S.; Murín, R.; Jünemann, A.G.M.; Stachs, O. Role of Corneal Stromal Cells on Epithelial Cell Function during Wound Healing. Int. J. Mol. Sci. 2018, 19, 464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zehra, M.; Mushtaq, S.; Musharraf, S.G.; Ghani, R.; Ahmed, N. Association of Cyclin Dependent Kinase 10 and Transcription Factor 2 during Human Corneal Epithelial Wound Healing in vitro model. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hocine, H.R.; Brunel, S.; Chen, Q.; Giustiniani, J.; San Roman, M.J.; Ferrat, Y.J.; Palacios, I.; De La Rosa, O.; Lombardo, E.; Bensussan, A.; et al. Extracellular Vesicles Released by Allogeneic Human Cardiac Stem/Progenitor Cells as Part of their Therapeutic Benefit. Stem Cells Transl. Med. 2019, 8, 911–924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hettich, B.F.; Greenwald, M.B.; Werner, S.; Leroux, J. Exosomes for Wound Healing: Purification Optimization and Identification of Bioactive Components. Adv. Sci. 2020, 7, 2002596. [Google Scholar] [CrossRef]
- Merjaneh, M.; Langlois, A.; LaRochelle, S.; Cloutier, C.B.; Ricard-Blum, S.; Moulin, V.J. Pro-angiogenic capacities of microvesicles produced by skin wound myofibroblasts. Angiogenesis 2017, 20, 385–398. [Google Scholar] [CrossRef] [PubMed]
- Arif, S.; LaRochelle, S.; Moulin, V.J. PLGF-1 contained in normal wound myofibroblast-derived microvesicles stimulated collagen production by dermal fibroblasts. J. Cell Commun. Signal. 2020, 14, 427–438. [Google Scholar] [CrossRef] [PubMed]
- Holmberg, C.; Ghesquière, B.; Impens, F.; Gevaert, K.; Kumar, J.D.; Cash, N.; Kandola, S.; Hegyi, P.; Wang, T.C.; Dockray, G.J.; et al. Mapping Proteolytic Processing in the Secretome of Gastric Cancer-Associated Myofibroblasts Reveals Activation of MMP-1, MMP-2, and MMP-3. J. Proteome Res. 2013, 12, 3413–3422. [Google Scholar] [CrossRef] [PubMed]
- McKaig, B.C.; Makh, S.S.; Hawkey, C.J.; Podolsky, D.K.; Mahida, Y.R. Normal human colonic subepithelial myofibroblasts enhance epithelial migration (restitution) via TGF-β3. Am. J. Physiol. Liver Physiol. 1999, 276, G1087–G1093. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yeung, V.; Zhang, T.C.; Yuan, L.; Parekh, M.; Cortinas, J.A.; Delavogia, E.; Hutcheon, A.E.K.; Guo, X.; Ciolino, J.B. Extracellular Vesicles Secreted by Corneal Myofibroblasts Promote Corneal Epithelial Cell Migration. Int. J. Mol. Sci. 2022, 23, 3136. https://doi.org/10.3390/ijms23063136
Yeung V, Zhang TC, Yuan L, Parekh M, Cortinas JA, Delavogia E, Hutcheon AEK, Guo X, Ciolino JB. Extracellular Vesicles Secreted by Corneal Myofibroblasts Promote Corneal Epithelial Cell Migration. International Journal of Molecular Sciences. 2022; 23(6):3136. https://doi.org/10.3390/ijms23063136
Chicago/Turabian StyleYeung, Vincent, Tancy C. Zhang, Ling Yuan, Mohit Parekh, John A. Cortinas, Eleni Delavogia, Audrey E. K. Hutcheon, Xiaoqing Guo, and Joseph B. Ciolino. 2022. "Extracellular Vesicles Secreted by Corneal Myofibroblasts Promote Corneal Epithelial Cell Migration" International Journal of Molecular Sciences 23, no. 6: 3136. https://doi.org/10.3390/ijms23063136





