Pediatric Brain Tumors: Signatures from the Intact Proteome
Abstract
:1. Introduction
2. Results and Discussion
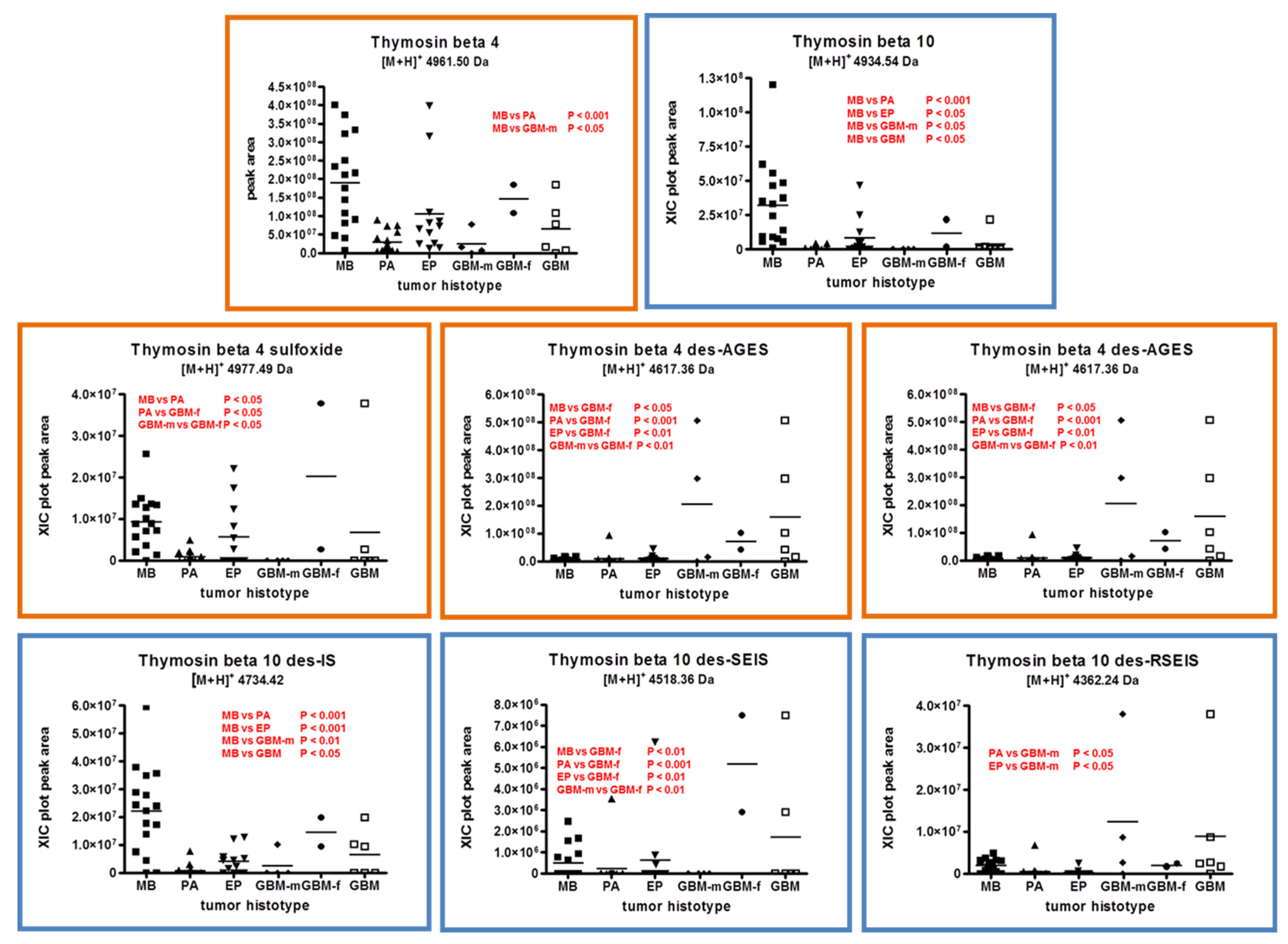
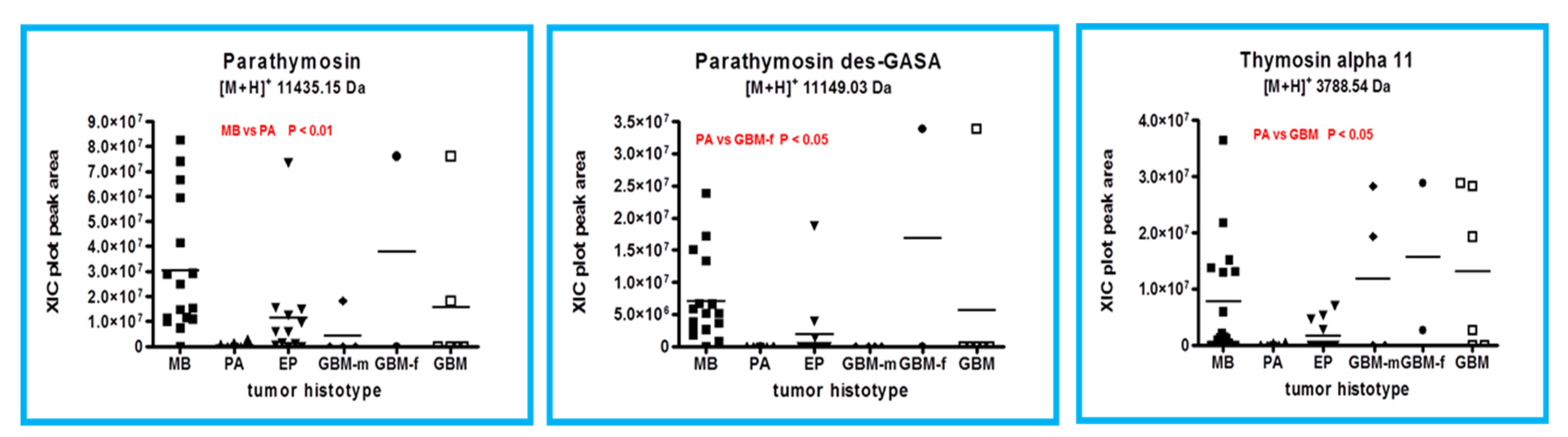
2.1. Proteins and Peptides Belonging to Thymosins’ Family
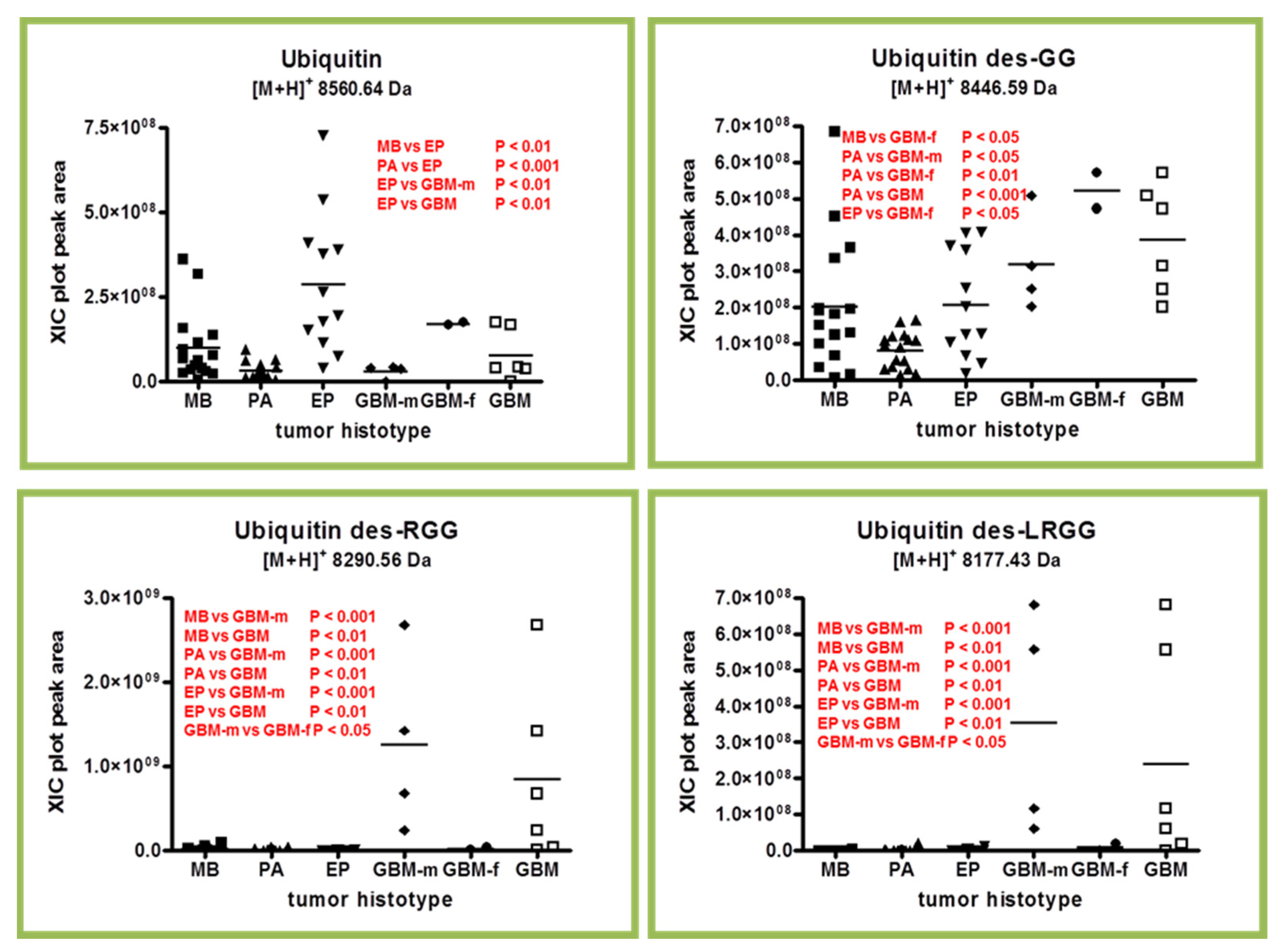
2.2. Ubiquitin and Truncated Proteoforms
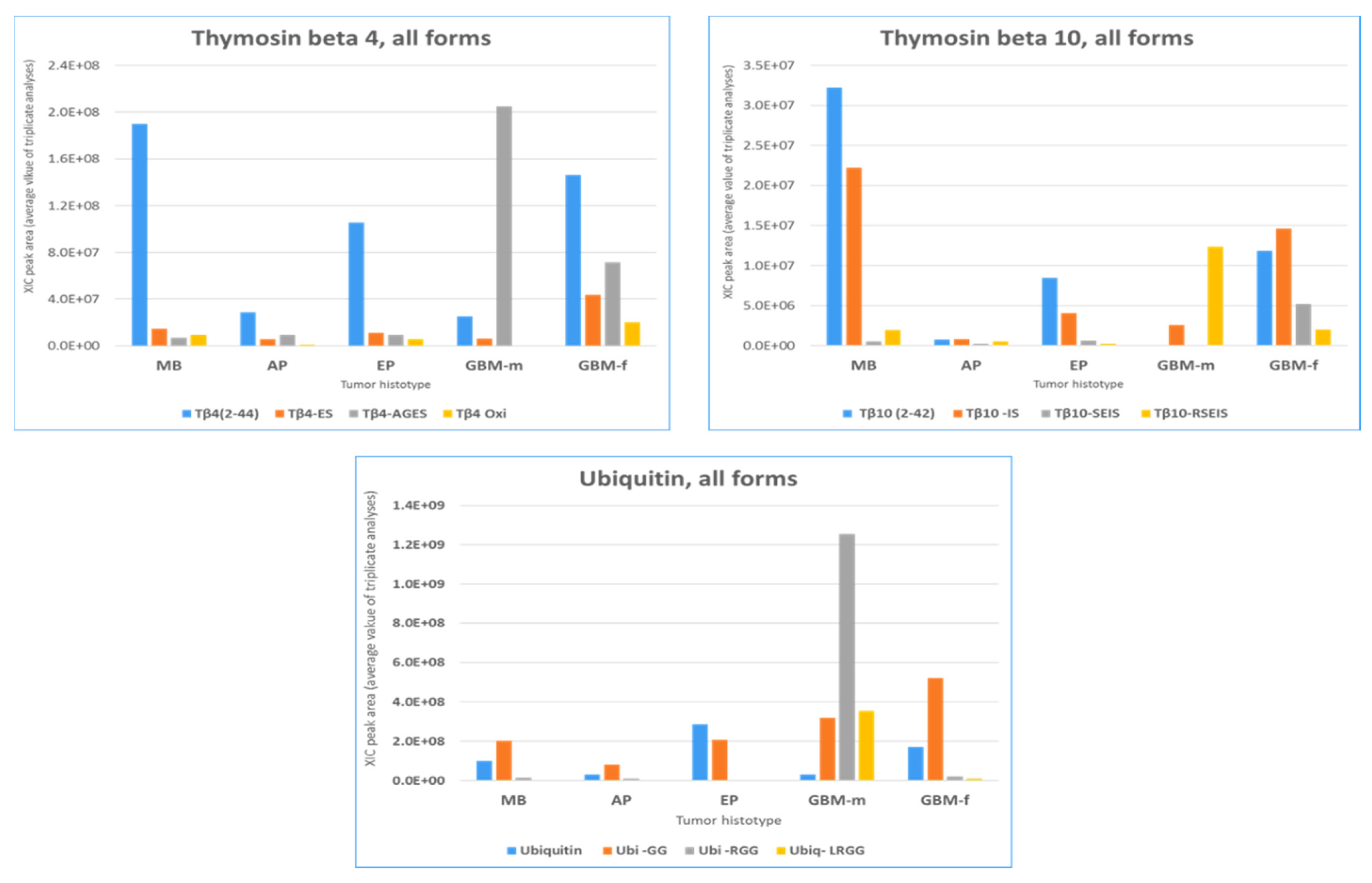
2.3. β-Thymosins and Ubiquitin Proteoforms Distribution in Tumor Histotypes
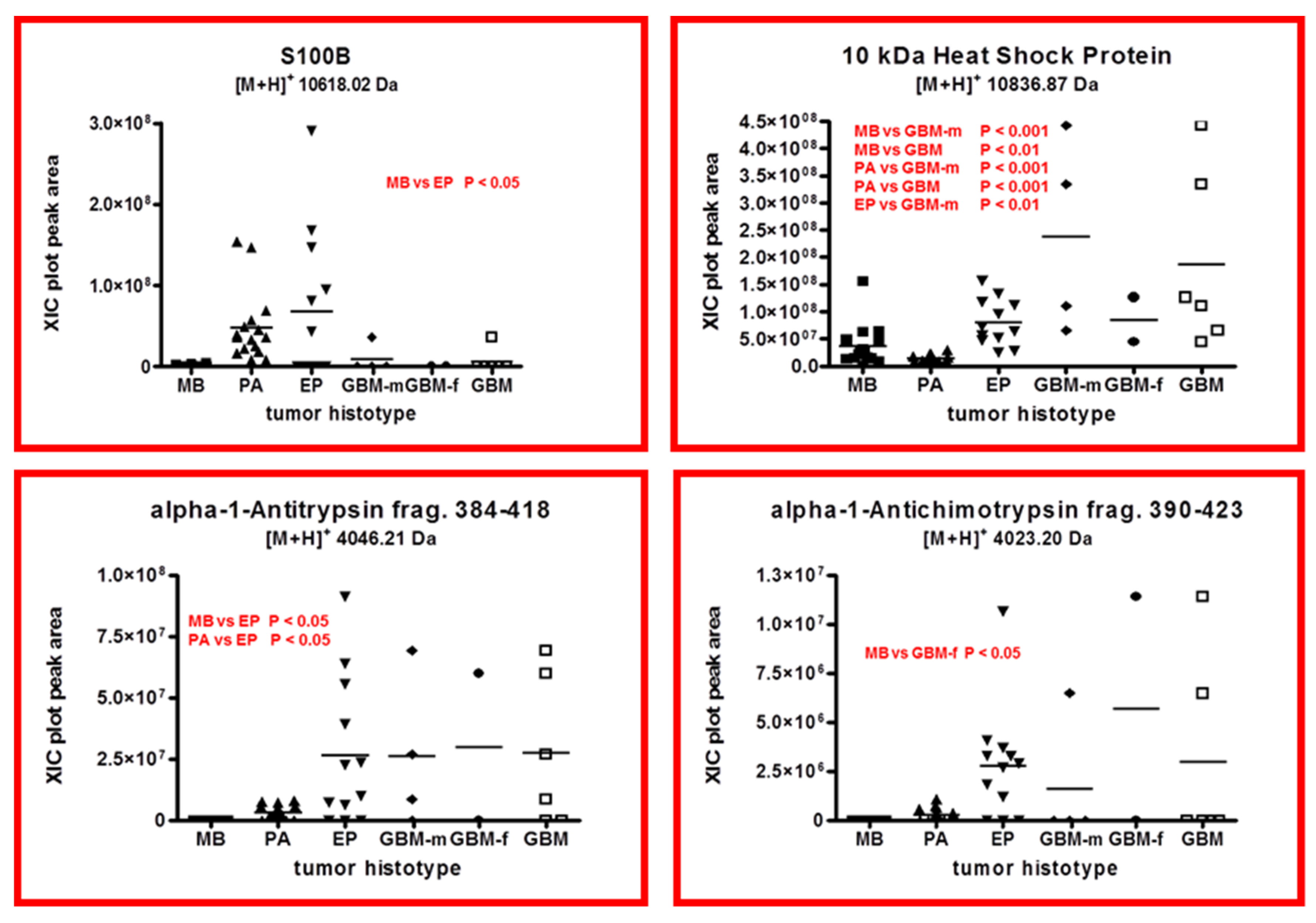
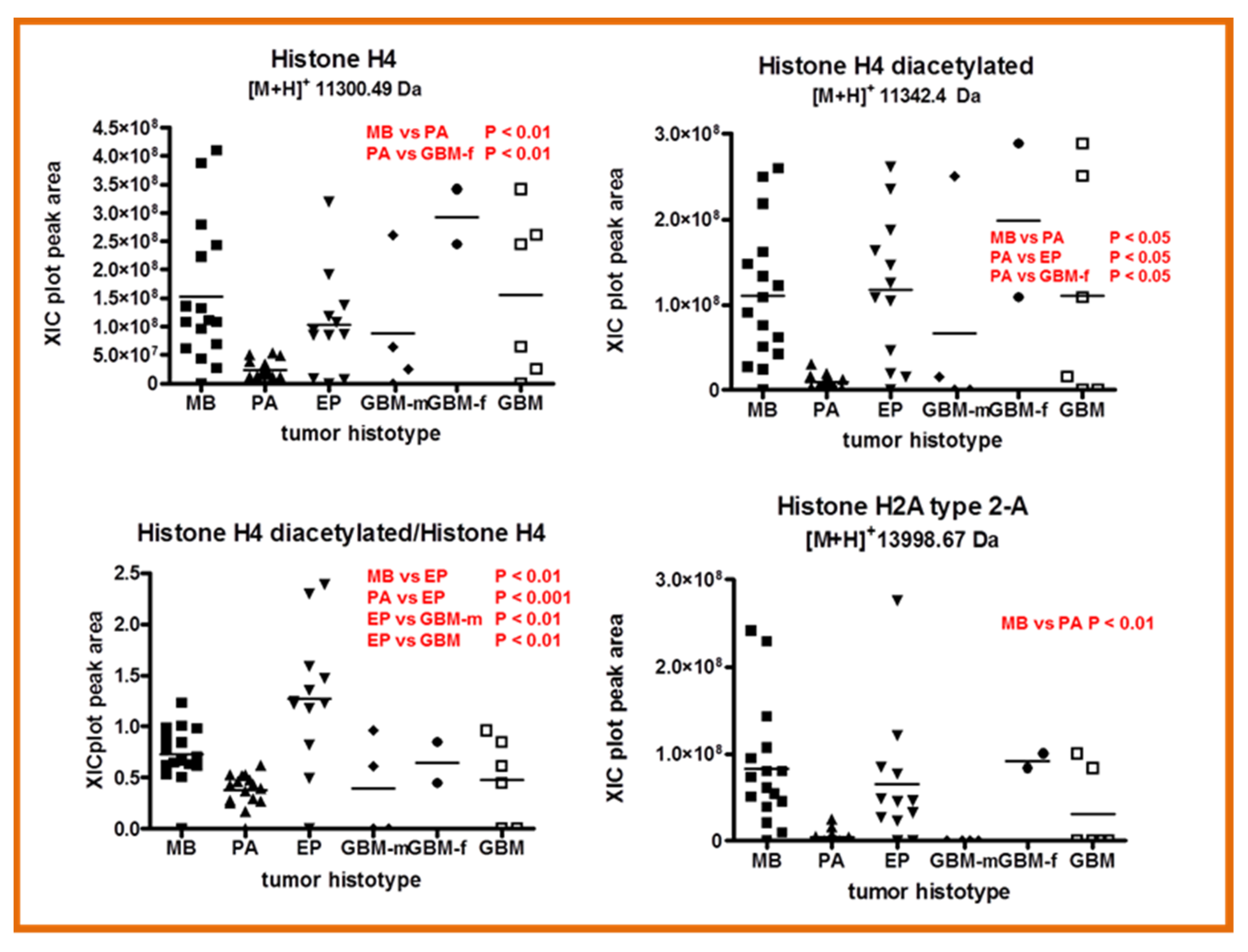
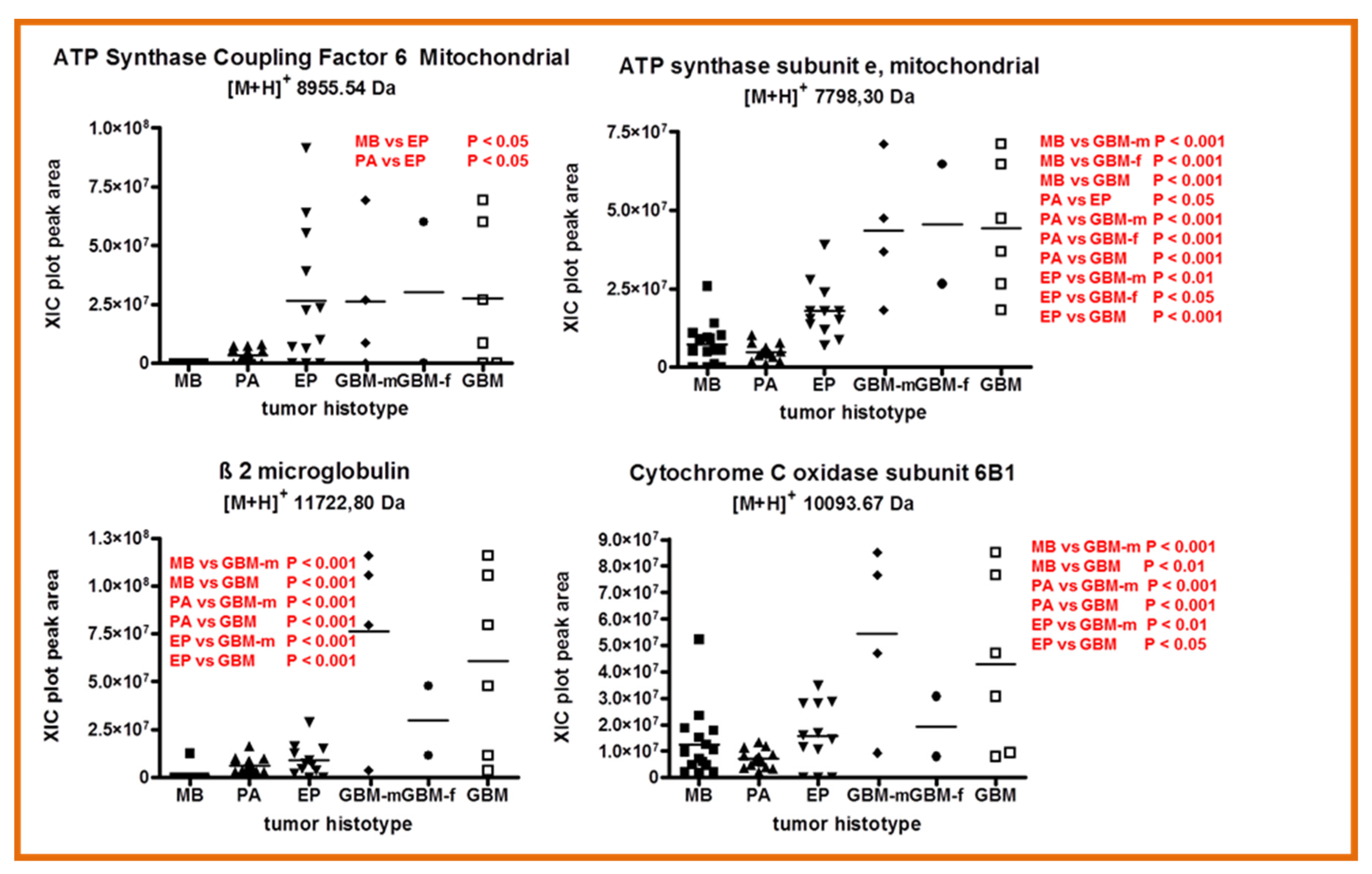
2.4. Other Protein Elements
2.5. Vimentin and Glial Fibrillary Acidic Protein Fragments: PTMs Characterization
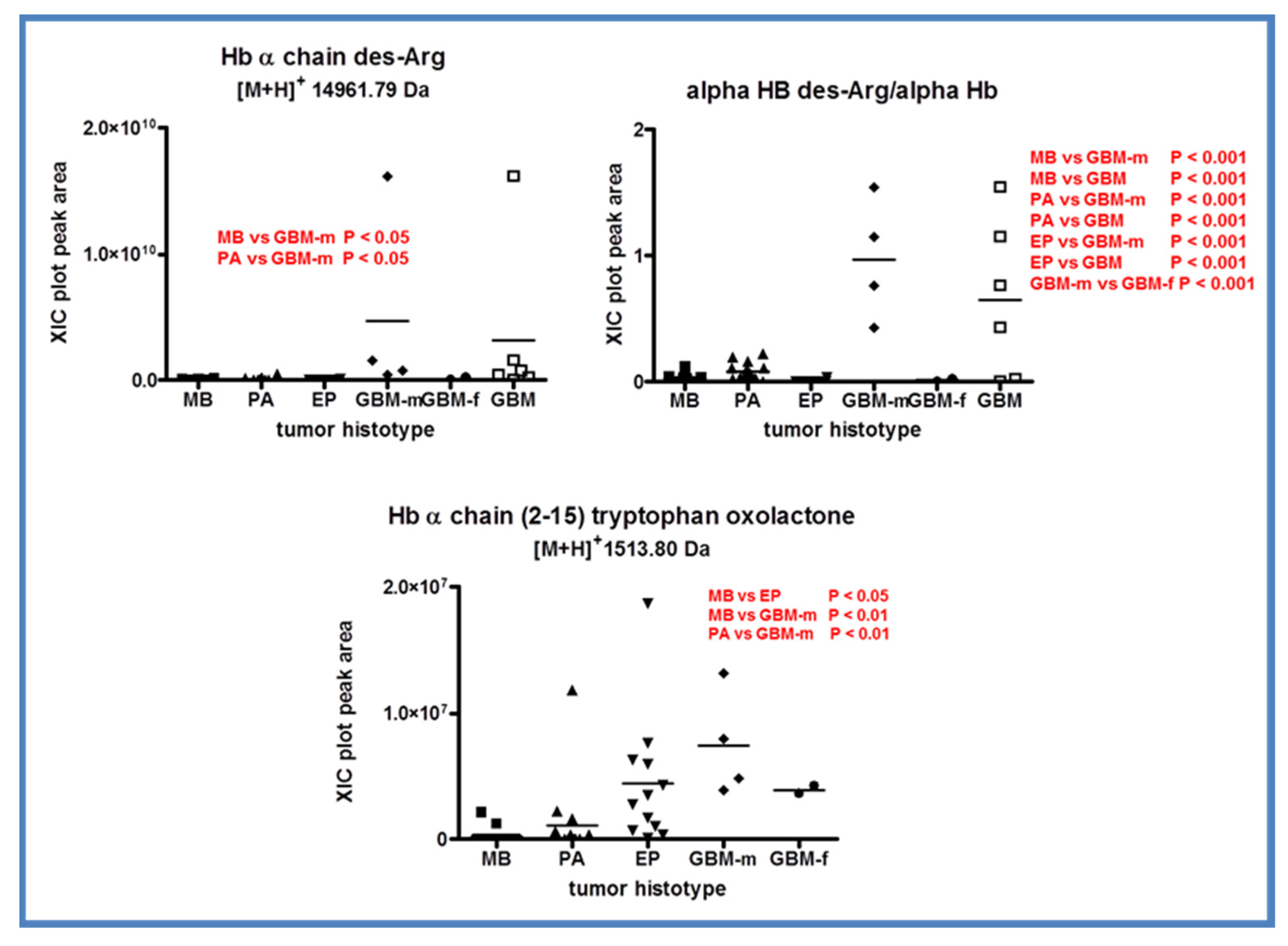
2.6. Hemoglobin
3. Materials and Methods
3.1. Chemicals
3.2. Instrumentation
3.3. Sample Collection and Treatment
3.4. LC-MS Proteomic Analysis
3.5. Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Petralia, F.; Tignor, N.; Reva, B.; Koptyra, M.; Chowdhury, S.; Rykunov, D.; Krek, A.; Ma, W.; Zhu, Y.; Ji, J.; et al. Integrated Proteogenomic Characterization across Major Histological Types of Pediatric Brain Cancer. Cell 2020, 183, 1962–1985. [Google Scholar] [CrossRef] [PubMed]
- Anagnostopoulos, A.K.; Tsangaris, G.T. The proteomics of pediatric brain tumors. Expert Rev. Proteom. 2014, 11, 641–648. [Google Scholar] [CrossRef] [PubMed]
- Tsangaris, G.T.; Anagnostopoulos, A.K. Pediatric brain tumors: Update of proteome-based studies. J. Proteom. 2018, 188, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Samuel, N.; Remke, M.; Rutka, J.T.; Raught, B.; Malkin, D. Proteomic analyses of CSF aimed at biomarker development for pediatric brain tumors. J. Neurooncol. 2014, 118, 225–238. [Google Scholar] [CrossRef] [PubMed]
- Tsangaris, G.T.; Anastasoviti, M.C.; Anagnostopoulos, A.K. Proteomics of pediatric ependymomas: A review. Childs Nerv. Syst. 2021, 37, 767–770. [Google Scholar] [CrossRef] [PubMed]
- Desiderio, C.; Rossetti, D.V.; Castagnola, M.; Massimi, L.; Tamburrini, G. Adamantinomatous craniopharyngioma: Advances in proteomic research. Childs Nerv. Syst. 2021, 37, 789–797. [Google Scholar] [CrossRef] [PubMed]
- Iavarone, F.; Desiderio, C.; Vitali, A.; Messana, I.; Martelli, C.; Castagnola, M.; Cabras, T. Cryptides: Latent peptides everywhere. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 246–263. [Google Scholar] [CrossRef] [PubMed]
- Martelli, C.; Iavarone, F.; D’Angelo, L.; Arba, M.; Vincenzoni, F.; Inserra, I.; Delfino, D.; Rossetti, D.V.; Caretto, M.; Massimi, L.; et al. Integrated proteomic platforms for the comparative characterization of medulloblastoma and pilocytic astrocytoma pediatric brain tumors: A preliminary study. Mol. Biosyst. 2015, 11, 1668–1683. [Google Scholar] [CrossRef] [PubMed]
- Rossetti, D.V.; Massimi, L.; Martelli, C.; Vincenzoni, F.; Di Silvestre, S.; Scorpio, G.; Tamburrini, G.; Caldarelli, M.; Urbani, A.; Desiderio, C. Ependymoma Pediatric Brain Tumor Protein Fingerprinting by Integrated Mass Spectrometry Platforms: A Pilot Investigation. Cancers 2020, 12, 674. [Google Scholar] [CrossRef] [Green Version]
- Delfino, D.; Rossetti, D.V.; Martelli, C.; Inserra, I.; Vincenzoni, F.; Castagnola, M.; Urbani, A.; Scarpa, S.; Fuso, A.; Cavallaro, R.A.; et al. Exploring the brain tissue proteome of TgCRND8 Alzheimer’s Disease model mice under B vitamin deficient diet induced hyperhomocysteinemia by LC-MS top-down platform. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2019, 1124, 165–172. [Google Scholar] [CrossRef]
- Martelli, C.; Serra, R.; Inserra, I.; Rossetti, D.V.; Iavarone, F.; Vincenzoni, F.; Castagnola, M.; Urbani, A.; Tamburrini, G.; Caldarelli, M.; et al. Investigating the Protein Signature of Adamantinomatous Craniopharyngioma Pediatric Brain Tumor Tissue: Towards the Comprehension of Its Aggressive Behavior. Dis. Markers. 2019, 2019, 3609789. [Google Scholar] [CrossRef] [PubMed]
- Holla, F.K.; Postma, T.J.; Blankenstein, M.A.; van Mierlo, T.J.M.; Vos, M.J.; Sizoo, E.M.; de Groot, M.; Uitdehaag, B.M.J.; Buter, J.; Klein, M.; et al. Prognostic value of the S100B protein in newly diagnosed and recurrent glioma patients: A serial analysis. J. Neurooncol. 2016, 129, 525–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czarnecka, A.M.; Campanella, C.; Zummo, G.; Cappello, F. Heat shock protein 10 and signal transduction: A “capsula eburnea” of carcinogenesis? Cell Stress Chaperones. 2006, 11, 287–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hannappel, E.; Huff, T. The thymosins. Prothymosin alpha, parathymosin, and beta-thymosins: Structure and function. Vitam Horm. 2003, 66, 257–296. [Google Scholar] [CrossRef] [PubMed]
- Kuzan, A. Thymosin β as an Actin-binding Protein with a Variety of Functions. Adv. Clin. Exp. Med. 2016, 25, 1331–1336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sribenja, S.; Wongkham, S.; Wongkham, C.; Yao, Q.; Chen, C. Roles and mechanisms of β-thymosins in cell migration and cancer metastasis: An update. Cancer Investig. 2013, 31, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Sribenja, S.; Li, M.; Wongkham, S.; Wongkham, C.; Yao, Q.; Chen, C. Advances in thymosin beta10 research: Differential expression, molecular mechanisms, and clinical implications in cancer and other conditions. Cancer Investig. 2009, 27, 1016–1022. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Li, M.; Yang, H.; Chai, H.; Fisher, W.; Yao, Q. Roles of thymosins in cancers and other organ systems. World J. Surg. 2005, 29, 264–270. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.Z. Altered histone modifications in gliomas. Brain Tumor Res. Treat. 2014, 2, 7–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Wang, D.; Han, X.; Tang, F.; Gao, D. Mechanism of methylation and acetylation of high GDNF transcription in glioma cells: A review. Heliyon 2019, 5, e01951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, M.J.; Singleton, W.G.; Lowis, S.P.; Malik, K.; Kurian, K.M. Therapeutic Targeting of Histone Modifications in Adult and Pediatric High-Grade Glioma. Front Oncol. 2017, 7, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, W.; Song, Y.; Ren, Z.; Cheng, X.; Li, P.; Song, H.; Jia, L. Glioma cells are resistant to inflammation-induced alterations of mitochondrial dynamics. Int. J. Oncol. 2020, 57, 1293–1306. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Rubin, J.B.; Lathia, J.D.; Berens, M.E.; Barnholtz-Sloan, J.S. Females have the survival advantage in glioblastoma. Neuro Oncol. 2018, 20, 576–577. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Warrington, N.M.; Taylor, S.J.; Whitmire, P.; Carrasco, E.; Singleton, K.W.; Wu, N.; Lathia, J.D.; Berens, M.E.; Kim, A.H.; et al. Sex differences in GBM revealed by analysis of patient imaging, transcriptome, and survival data. Sci. Transl. Med. 2019, 11, eaao5253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCrea, H.J.; Bander, E.D.; Venn, R.A.; Reiner, A.S.; Iorgulescu, J.B.; Puchi, L.A.; Schaefer, P.M.; Cederquist, G.; Greenfield, J.P. Sex, Age, Anatomic Location, and Extent of Resection Influence Outcomes in Children With High-grade Glioma. Neurosurgery 2015, 77, 443–452, discussion 452–453. [Google Scholar] [CrossRef] [PubMed]
- György, B.; Tóth, E.; Tarcsa, E.; Falus, A.; Buzás, E.I. Citrullination: A posttranslational modification in health and disease. Int. J. Biochem. Cell Biol. 2006, 38, 1662–1677. [Google Scholar] [CrossRef] [PubMed]
- Yuzhalin, A.E. Citrullination in Cancer. Cancer Res. 2019, 79, 1274–1284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Z.; Cui, Y.; Wang, L.; Zhao, Y.; Yan, S.; Chang, X. Investigating citrullinated proteins in tumour cell lines. World J. Surg. Oncol. 2013, 11, 260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willumsen, N.; Bager, C.L.; Leeming, D.J.; Smith, V.; Christiansen, C.; Karsdal, M.A.; Dornan, D.; Bay-Jensen, A.C. Serum biomarkers reflecting specific tumor tissue remodeling processes are valuable diagnostic tools for lung cancer. Cancer Med. 2014, 3, 1136–1145. [Google Scholar] [CrossRef] [PubMed]
- Ishigami, A.; Ohsawa, T.; Hiratsuka, M.; Taguchi, H.; Kobayashi, S.; Saito, Y.; Murayama, S.; Asaga, H.; Toda, T.; Kimura, N.; et al. Abnormal accumulation of citrullinated proteins catalyzed by peptidylarginine deiminase in hippocampal extracts from patients with Alzheimer’s disease. J. Neurosci. Res. 2005, 80, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Jang, B.; Jin, J.K.; Jeon, Y.C.; Cho, H.J.; Ishigami, A.; Choi, K.C.; Carp, R.I.; Maruyama, N.; Kim, Y.S.; Choi, E.K. Involvement of peptidylarginine deiminase-mediated post-translational citrullination in pathogenesis of sporadic Creutzfeldt-Jakob disease. Acta Neuropathol. 2010, 119, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.C.; Liao, Y.F.; Lin, C.L.; Lin, W.H.; Liu, G.Y.; Hung, H.C. Vimentin is involved in peptidylarginine deiminase 2-induced apoptosis of activated Jurkat cells. Mol. Cells 2014, 37, 426–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, X.; Han, J.; Pang, L.; Zhao, Y.; Yang, Y.; Shen, Z. Increased PADI4 expression in blood and tissues of patients with malignant tumors. BMC Cancer 2009, 9, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, X.; Hou, X.; Pan, J.; Fang, K.; Wang, L.; Han, J. Investigating the pathogenic role of PADI4 in oesophageal cancer. Int. J. Biol. Sci. 2011, 7, 769–781. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, R.; Gan, Y.; Ying, S. The roles of PAD2- and PAD4-mediated protein citrullination catalysis in cancers. Int. J. Cancer 2021, 148, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Jang, B.; Kim, M.J.; Lee, Y.J.; Ishigami, A.; Kim, Y.S.; Choi, E.K. Vimentin citrullination probed by a novel monoclonal antibody serves as a specific indicator for reactive astrocytes in neurodegeneration. Neuropathol. Appl. Neurobiol. 2020, 46, 751–769. [Google Scholar] [CrossRef] [PubMed]
- Damgaard, D.; Senolt, L.; Nielsen, M.F.; Pruijn, G.J.; Nielsen, C.H. Demonstration of extracellular peptidylarginine deiminase (PAD) activity in synovial fluid of patients with rheumatoid arthritis using a novel assay for citrullination of fibrinogen. Arthritis Res Ther. 2014, 16, 498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vossenaar, E.R.; Radstake, T.R.; van der Heijden, A.; van Mansum, M.A.; Dieteren, C.; de Rooij, D.J.; Barrera, P.; Zendman, A.J.; van Venrooij, W.J. Expression and activity of citrullinating peptidylarginine deiminase enzymes in monocytes and macrophages. Ann. Rheum. Dis. 2004, 63, 373–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brentville, V.A.; Vankemmelbeke, M.; Metheringham, R.L.; Durrant, L.G. Post-translational modifications such as citrullination are excellent targets for cancer therapy. Semin. Immunol. 2020, 47, 101393. [Google Scholar] [CrossRef] [PubMed]
- Podor, T.J.; Singh, D.; Chindemi, P.; Foulon, D.M.; McKelvie, R.; Weitz, J.I.; Austin, R.; Boudreau, G.; Davies, R. Vimentin exposed on activated platelets and platelet microparticles localizes vitronectin and plasminogen activator inhibitor complexes on their surface. J. Biol. Chem. 2002, 277, 7529–7539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moisan, E.; Girard, D. Cell surface expression of intermediate filament proteins vimentin and lamin B1 in human neutrophil spontaneous apoptosis. J. Leukoc. Biol. 2006, 79, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.A.; Southwood, S.; Sette, A.; Jevnikar, A.M.; Bell, D.A.; Cairns, E. Cutting edge: The conversion of arginine to citrulline allows for a high-affinity peptide interaction with the rheumatoid arthritis-associated HLA-DRB1*0401 MHC class II molecule. J. Immunol. 2003, 171, 538–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ireland, J.; Herzog, J.; Unanue, E.R. Cutting edge: Unique T cells that recognize citrullinated peptides are a feature of protein immunization. J. Immunol. 2006, 177, 1421–1425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brentville, V.A.; Metheringham, R.L.; Gunn, B.; Symonds, P.; Daniels, I.; Gijon, M.; Cook, K.; Xue, W.; Durrant, L.G. Citrullinated Vimentin Presented on MHC-II in Tumor Cells Is a Target for CD4+ T-Cell-Mediated Antitumor Immunity. Cancer Res. 2016, 76, 548–560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brentville, V.A.; Metheringham, R.L.; Daniels, I.; Atabani, S.; Symonds, P.; Cook, K.W.; Vankemmelbeke, M.; Choudhury, R.; Vaghela, P.; Gijon, M.; et al. Combination vaccine based on citrullinated vimentin and enolase peptides induces potent CD4-mediated anti-tumor responses. J. Immunother. Cancer 2020, 8, e000560. [Google Scholar] [CrossRef] [PubMed]
- Benham, H.; Nel, H.J.; Law, S.C.; Mehdi, A.M.; Street, S.; Ramnoruth, N.; Pahau, H.; Lee, B.T.; Ng, J.; Brunck, M.E.; et al. Citrullinated peptide dendritic cell immunotherapy in HLA risk genotype-positive rheumatoid arthritis patients. Sci. Transl. Med. 2015, 7, 290ra87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desiderio, C.; D’Angelo, L.; Rossetti, D.V.; Iavarone, F.; Giardina, B.; Castagnola, M.; Massimi, L.; Tamburrini, G.; Di Rocco, C. Cerebrospinal fluid top-down proteomics evidenced the potential biomarker role of LVV- and VV-hemorphin-7 in posterior cranial fossa pediatric brain tumors. Proteomics 2012, 12, 2158–2166. [Google Scholar] [CrossRef] [PubMed]
- Altinoz, M.A.; Elmaci, I.; Ince, B.; Ozpinar, A.; Sav, A.M. Hemoglobins, Hemorphins, and 11p15.5 Chromosomal Region in Cancer Biology and İmmunity with Special Emphasis for Brain Tumors. J. Neurol. Surg. A Cent. Eur. Neurosurg. 2016, 77, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Marti, H.R.; Beale, D.; Lehmann, H. Haemoglobin Koelliker: A new acquired haemoglobin appearing after severe haemolysis: Alpha-2 minus 141 Arg beta-2. Acta Haematol. 1967, 37, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Schilirò, G.; Russo, A.; Azzia, N. Hemoglobin Koelliker (alpha 2 minus 141 arg beta 2) in favism. Acta Haematol. 1982, 67, 229. [Google Scholar] [CrossRef] [PubMed]
- Fasan, G.; Grandgeorge, M.; Vigneron, C.; Dellacherie, E. Preparation of unaltered hemoglobin from human placentas for possible use in blood substitutes. J. Biochem. Biophys. Methods 1991, 23, 53–66. [Google Scholar] [CrossRef]
- Michel, B.; Igić, R.; Leray, V.; Deddish, P.A.; Erdös, E.G. Removal of Arg141 from the alpha chain of human hemoglobin by carboxypeptidases N and M. Circ. Res. 1996, 78, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Gittleman, H.; Ostrom, Q.T.; Stetson, L.C.; Waite, K.; Hodges, T.R.; Wright, C.H.; Wright, J.; Rubin, J.B.; Berens, M.E.; Lathia, J.; et al. Sex is an important prognostic factor for glioblastoma but not for nonglioblastoma. Neurooncol. Pract. 2019, 6, 451–462. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Buehler, P.W.; Boykins, R.A.; Venable, R.M.; Alayash, A.I. Structural basis of peroxide-mediated changes in human hemoglobin: A novel oxidative pathway. J. Biol. Chem. 2007, 282, 4894–4907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fedorova, M.; Todorovsky, T.; Kuleva, N.; Hoffmann, R. Quantitative evaluation of tryptophan oxidation in actin and troponin I from skeletal muscles using a rat model of acute oxidative stress. Proteomics 2010, 10, 2692–2700. [Google Scholar] [CrossRef] [PubMed]
- Schöneich, C. Novel chemical degradation pathways of proteins mediated by tryptophan oxidation: Tryptophan side chain fragmentation. J. Pharm. Pharmacol. 2018, 70, 655–665. [Google Scholar] [CrossRef] [Green Version]
- Barelli, S.; Canellini, G.; Thadikkaran, L.; Crettaz, D.; Quadroni, M.; Rossier, J.S.; Tissot, J.D.; Lion, N. Oxidation of proteins: Basic principles and perspectives for blood proteomics. Proteom. Clin. Appl. 2008, 2, 142–157. [Google Scholar] [CrossRef] [Green Version]
- Fellers, R.T.; Greer, J.B.; Early, B.P.; Yu, X.; LeDuc, R.D.; Kelleher, N.L.; Thomas, P.M. ProSight Lite: Graphical software to analyze top-down mass spectrometry data. Proteomics 2015, 15, 1235–1238. [Google Scholar] [CrossRef]
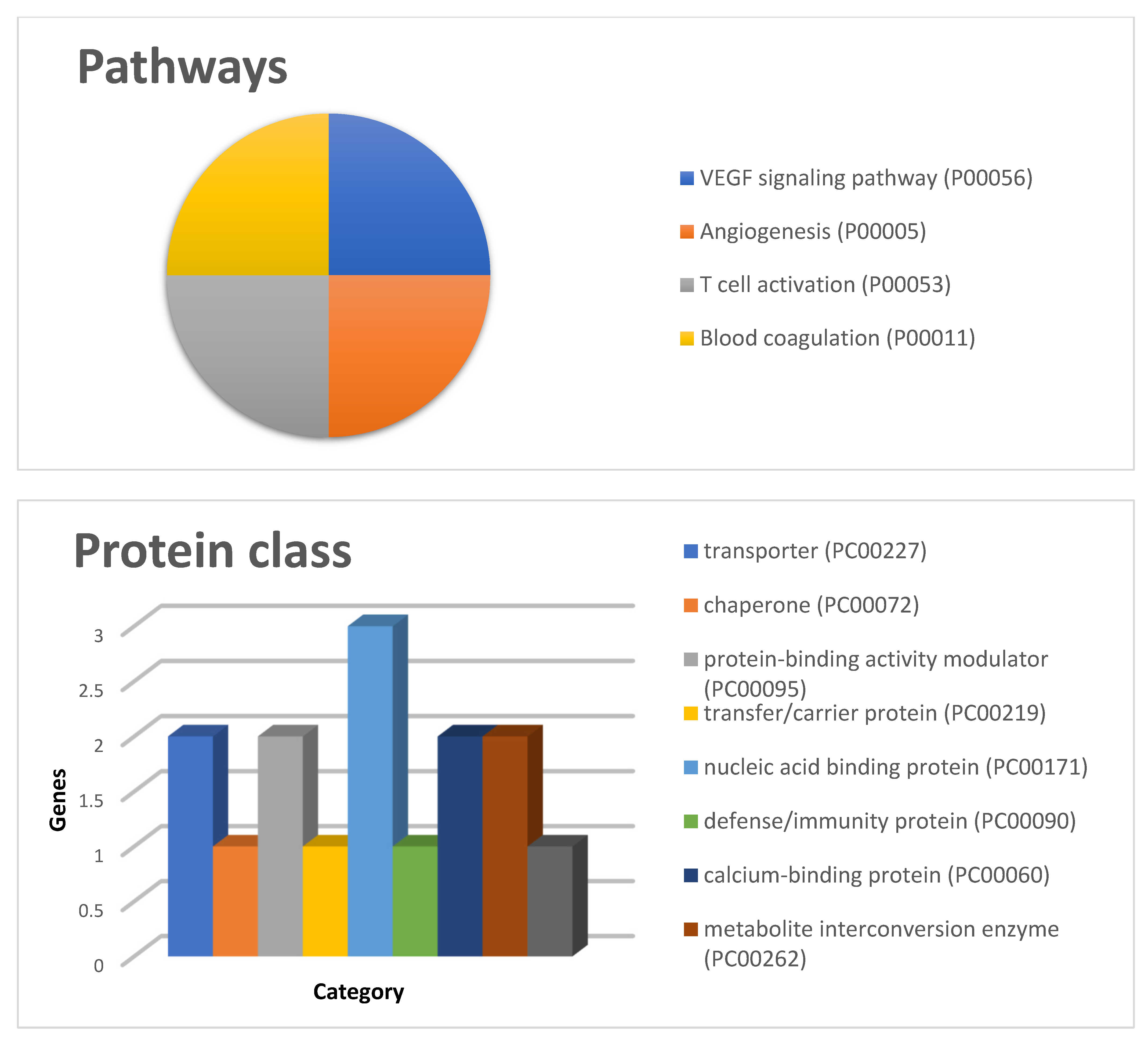
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–Protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Mi, H.; Huang, X.; Muruganujan, A.; Tang, H.; Mills, C.; Kang, D.; Thomas, P.D. PANTHER version 11: Expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017, 45, D183–D189. [Google Scholar] [CrossRef] [Green Version]


| Uniprot Accession | Protein Name | Amino Acid Position * | [M+H]+ Theoretical Monoisotopic | [M+H]+ Experimental Monoisotopic | PTMs |
|---|---|---|---|---|---|
| P69905 | Hemoglobin α-Chain | 2-142 (Chain) | 15,117.89 | 15,117.90 | - |
| Hemoglobin α-Chain des-R (C-terminal) | 2-141 | 14,961.79 | - | ||
| Hemoglobin α-Chain frag. | 2-32 | 3195.65 | 3195.66 | - | |
| Hemoglobin α-Chain frag. | 2-15 | 1513.80 | 1513.80 | Oxidation to oxolactone W15 | |
| P68871 | Hemoglobin β-Chain | 2-147 (Chain) | 15,858.26 | 15,858.25 | - |
| P63313 | Thymosin β10 | 2-44 (Chain) | 4934.53 | 4934.54 | Acetylation N-terminal |
| Thymosin β10 des-IS [10,11] | 2-42 | 4734.41 | 4734.42 | Acetylation N-terminal | |
| Thymosin β10 des-SEIS | 2-40 | 4518.34 | 4518.36 | Acetylation N-terminal | |
| Thymosin β10 des-RSEIS | 2-39 | 4362.24 | 4362.24 | Acetylation N-terminal | |
| Thymosin β10 frag. | 2-27 | 2964.50 | 2964.51 | Acetylation N-terminal | |
| Thymosin β10 frag. | 28-41 | 1687.89 | 1687.89 | - | |
| P62328 | Thymosin β4 | 2-44 (Chain) | 4961.50 | 4961.50 | Acetylation N-terminal |
| Thymosin β4 sulfoxide | 2-44 (Chain) | 4977.49 | 4977.49 | Acetylation N-terminal Oxidation M7 | |
| Thymosin β4 des-ES (C-terminal) | 2-42 | 4745.42 | 4745.43 | Acetylation N-terminal | |
| Thymosin β4 des-AGES (C-terminal) | 2-40 | 4617.36 | 4617.36 | Acetylation N-terminal | |
| Thymosin β4 frag. | 2-19 | 2151.10 | 2151.10 | Acetylation N-terminal | |
| Thymosin β4 frag. | 2-14 | 1566.70 | 1566.70 | Acetylation N-terminal | |
| Thymosin β4 frag. | 2-12 | 1304.60 | 1304.61 | Acetylation N-terminal | |
| Thymosin β4 frag. | 2-15 | 1694.79 | 1694.80 | Acetylation N-terminal | |
| Thymosin β4 frag. | 2-16 | 1781.83 | 1781.83 | Acetylation N-terminal | |
| P20962 | Parathymosin (2-102) | 2-102 (Chain) | 11,435.17 | 11,435.15 | Acetylation N-terminal |
| Parathymosin frag. Des-GASA | 2-98 | 11,149.04 | 11,149.03 | Acetylation N-terminal | |
| P06454 | Prothymosin α1 frag. | 2-15 | 1466.69 | 1466.69 | Acetylation N-terminal |
| Thymosin α1 | 2-29 | 3107.51 | 3107.52 | Acetylation N-terminal | |
| Thymosin α11 | 2-36 | 3788.83 | 3788.54 | Acetylation N-terminal | |
| P61604 | 10 kDa Heat Shock Protein | 2-102 (Chain) | 10,836.85 | 10,836.87 | Acetylation N-terminal |
| P59665 | α-Defensin 1 [12,13] | 65-94 | 3440.52 | 3440.53 | Disulfide bonds (66→94, 68→83, 73→93) |
| P59665/6 | α-Defensin 2 [12,13] | 66-94 | 3369.48 | 3369.49 | Disulfide bonds (66→94, 68→83, 73→93) |
| P59666 | α-Defensin 3 [12,13] | 65-94 | 3484.51 | 3484.51 | Disulfide bonds (66→94, 68→83, 73→93) |
| P56385 | ATP Synthase Subunit e, Mitochondrial | 2-69 (Chain) | 7798.29 | 7798.30 | - |
| P18859 | ATP Synthase Coupling Factor 6, Mitochondrial | 33-108 (Chain) | 8955.55 | 8955.48 | - |
| P14854 | Cytochrome C Oxidase Subunit 6B1 | 2-86 (Chain) | 10,093.67 | 10,093.66 | Acetylation N-terminal Disulfide bonds (30→65, 40→54) |
| P14136 | Glial Fibrillary Acidic Protein | 388-432 | 5206.73 | 5206.74 | - |
| 388-432 | 5207.72 | 5207.74 | Deamidation Q388 or Citrullination R390 | ||
| 388-432 | 5208.70 | 5208.72 | Deamidation Q388 or Citrullination R390 Citrullination R416 | ||
| 398-432 | 5208.70 | 5208.72 | Citrullination R406, 416 | ||
| 388-432 | 5209.68 | 5209.69 | Deamidation Q388 or Citrullination R390 Citrullinations R406, R416 | ||
| 388-431 | 5075.69 | 5075.70 | - | ||
| 388-431 | 5076.67 | 5076.69 | Citrullination R390 or 406 or 416 | ||
| 388-431 | 5077.66 | 5077.67 | Deamidation Q388 or Citrullination R390 Citrullination R416 | ||
| 15-36 | 2185.13 | 2185.14 | - | ||
| 15-38 | 2385.25 | 2385.26 | - | ||
| 15-38 | 2386.23 | 2386.24 | Citrullination | ||
| 15-38 | 2387.22 | 2387.22 | Citrullinations R29, R36 | ||
| 41-59 | 2052.11 | 2052,11 | - | ||
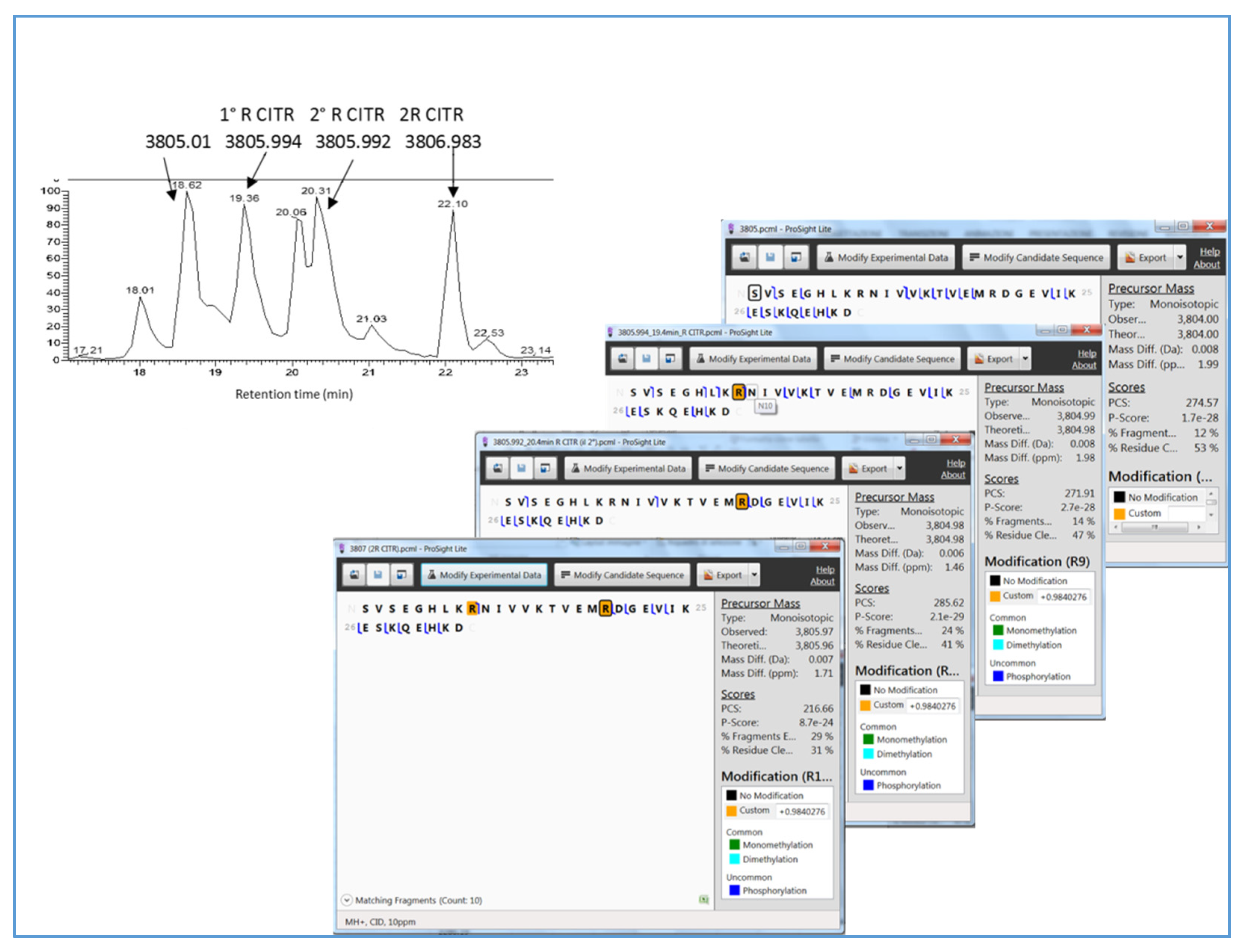
| 398-430 | 3805.00 | 3805.01 | - | ||
| 398-430 | 3805.99 | 3805.99 | Citrullination R406 | ||
| 398-430 | 3805.99 | 3805.99 | Deamidation N407 or Citrullination R416 | ||
| 398-430 | 3805.99 | 3805.99 | Citrullination R416 | ||
| 398-430 | 3806.97 | 3806.98 | Citrullination R406, R416 | ||
| 388-430 | 4976.62 | 4976.61 | - | ||
| 388-430 | 4977.61 | 4977.63 | Deamidation Q388 or Citrullination R390 | ||
| 388-430 | 4978.59 | 4978.60 | Deamidation Q388 or Citrullination R390 Citrullination R416 | ||
| 388-430 | 4979.57 | 4979.59 | Deamidation Q388 or Citrullination R390 Citrullinations R406, R416 | ||
| 398-432 | 4035.11 | 4035.12 | - | ||
| 398-432 | 4036.10 | 4036.11 | Citrullinations R406 or Deamidation N407 | ||
| 398-432 | 4036.10 | 4036.10 | Citrullinations R416 | ||
| 398-432 | 4037.08 | 4037.08 | Citrullinations R406, R416 | ||
| 388-405 | 2028.07 | 2028.08 | - | ||
| 388-405 | 2029.06 | 2029.06 | Deamidation Q388 or Citrullination R390 | ||
| 416-432 | 2028.02 | 2028.02 | - | ||
| 22-36 | 1463.86 | 1463.86 | - | ||
| 388-415 | 3197.73 | 3197.74 | - | ||
| 388-415 | 3198.72 | 3198.72 | Deamidation Q388 or Citrullination R390 | ||
| 406-432 | 3197.68 | 3197.68 | - | ||
| 406-432 | 3198.66 | 3198.67 | Citrullination R416 | ||
| 406-430 | 2967.57 | 2967.57 | - | ||
| P08670 | Vimentin | 424-466 | 4953.53 | 4953.54 | - |
| 424-466 | 4954.51 | 4954.53 | Citrullination R440 or R450 or Deamidation Q453 | ||
| 422-466 | 5180.66 | 5180.67 | - | ||
| 422-466 | 5181.64 | 5181.65 | Deamidation N422 or Citrullination R424 | ||
| 422-466 | 5182.63 | 5182.64 | Deamidation N422 or 427 or Citrullination R424 Citrullination R424 or 450 | ||
| 430-466 | 4225.18 | 4225.19 | - | ||
| 430-466 (Rt 23.2 min) | 4226.17 | 4226.18 | Citrullination R440 | ||
| 430-466 (Rt 24.0 min) | 4226.17 | 4226.18 | Citrullination R450 | ||
| 434-466 | 3814.93 | 3814.94 | - | ||
| 443-466 | 2777.37 | 2777.37 | - | ||
| 443-466 | 2778.35 | 2778.37 | Deamidation N456 | ||
| 443-466 | 2778.35 | 2778.36 | Citrullination R450 or Deamidation Q453 | ||
| 444-466 | 2664.29 | 2664.29 | - | ||
| 444-466 | 2665.27 | 2665.28 | Deamidation Q460 | ||
| 444-466 | 2665.27 | 2665.27 | Citrullination R450 | ||
| 447-466 | 2322.06 | 2322.06 | - | ||
| 447-466 | 2323.04 | 2323.05 | Citrullination R450 or Deamidation Q453 | ||
| 54-69 | 1565.80 | 1565.81 | - | ||
| P07108 | AcylCoA Binding Protein | 2-87 (Chain) | 9950.00 | 9950.03 | Acetylation N-terminal |
| AcylCoA Binding Protein natural variant M71→V | 2-87 (Chain) | 9918.03 | 9918.04 | Acetylation N-terminal | |
| P04271 | S100B | 2-92 (Chain) | 10,618.03 | 10,618.02 | Acetylation N-terminal |
| P06703 | S100A6 | 2-90 (Chain) | 10,085.32 | 10,085.32 | Acetylation N-terminal |
| 2-90 (Chain) | 10,204.32 | 10,204.40 | Acetylation N-terminal Cysteinylation C3 | ||
| 2-90 (Chain) | 10,390.39 | 10,390.38 | Acetylation N-terminal Gluthathionylation C3 | ||
| P01011 | α-1-Antichimotrypsin | 390-423 | 4023.18 | 4023.20 | - |
| 387-423 | 4352.34 | 4352.36 | - | ||
| P01009 | α-1-Antitrypsin | 384-418 | 4046.20 | 4046.21 | - |
| Q16555-2 | Dihydropyrimidinase-related protein | 521-570 | 5305.80 | 5305.81 | - |
| 521-572 | 5475.91 | 5475.92 | - | ||
| P0CG48 | Ubiquitin | 1-76 (Chain) | 8560.63 | 8560.64 | - |
| Ubiquitin des-GG (C-terminal) | 1-74 | 8446.58 | 8446.59 | - | |
| Ubiquitin des-RGG (C-terminal) | 1-73 | 8290.48 | 8290.56 | - | |
| Ubiquitin des-LRGG (C-terminal) | 1-72 | 8177.40 | 8177.43 | - | |
| P05204 | Non-histone chromosomal protein HMG-17 | 2-90 (Chain) | 9258.01 | 9258.02 | Deamidation N72 |
| P62805 | Histone H4 | 2-103 (Chain) | 11,300.39 | 11,300.49 | Acetylation N-terminal Dimethylation K21 |
| 2-103 (Chain) | 11,342.40 | 11,342.36 | Acetylation N-terminal Acetylation K17 or K32 Dimethylation K21 | ||
| Q6FI13 | Histone H2A type 2-A | 2-130 (Chain) | 13,998.87 | 13,998.91 | Acetylation N-terminal |
| Q5QNW6 | Histone H2B Type 2-F | 2-126 (Chain) | 13,781.54 | 13,781.57 | - |
| P61769 | β2-microglobulin | 21-119 (Chain) | 11,722.78 | 11,722.78 | Disulfide bond 45 ↔ 100 |
| P14174 | Macrophage migration inhibitory factor | 2-115 (Chain) | 12,338.19 | 12,338.20 | - |
| P00441 | Superoxide dismutase [Cu-Zn] | 2-154 (Chain) | 15,835.87 | 15,836.00 | Acetylation N-terminal, Disulfide bond 58 ↔ 147 |
| P02511 | α-crystallin B | 1-175 (Chain) | 20,189.44 | 20,189.44 | Acetylation N-terminal |
| P80723 | BASP1 | 2-227 (Chain) | 22,760.18 | 22,759.15 | Myristoylation N-terminal |
| Specimen ID | Patient Age (y = Years, m = Months) | Patient Sex | Tumor Grade | Diagnosis |
|---|---|---|---|---|
| MB1 | 7 y | M | WHO IV | Medulloblastoma |
| MB2 | 5 y 6 m | F | WHO IV | Medulloblastoma |
| MB3 | 6 y | M | WHO IV | Medulloblastoma |
| MB4 | 16 y | M | WHO IV | Medulloblastoma |
| MB5 | 8 y | F | WHO IV | Medulloblastoma |
| MB6 | 9 y | F | WHO IV | Medulloblastoma |
| MB7 | 20 y | M | WHO IV | Medulloblastoma |
| MB8 | 3 y | M | WHO IV | Medulloblastoma |
| MB9 | 14 y | M | WHO IV | Medulloblastoma |
| MB10 | 10 y | M | WHO IV | Medulloblastoma |
| MB11 | 16 y | F | WHO IV | Medulloblastoma |
| MB12 | 6 y | F | WHO IV | Medulloblastoma |
| MB13 | 0 y 5 m | M | WHO IV | Medulloblastoma |
| MB14 | 8 y | F | WHO IV | Medulloblastoma |
| MB15 | 8 y | M | WHO IV | Medulloblastoma |
| MB16 | 6 y | F | WHO IV | Medulloblastoma |
| PA1 | 3 y 6 m | M | WHO I | Pilocytic Astrocytoma |
| PA2 | 17 y | M | WHO I | Pilocytic Astrocytoma |
| PA3 | 9 y | M | WHO I | Pilocytic Astrocytoma |
| PA4 | 4 y | F | WHO I | Pilocytic Astrocytoma |
| PA5 | 21y | M | WHO I | Pilocytic Astrocytoma |
| PA6 | 12 y | M | WHO I | Pilocytic Astrocytoma |
| PA7 | 6 y | M | WHO I | Pilocytic Astrocytoma |
| PA8 | 4 y | F | WHO I | Pilocytic Astrocytoma |
| PA9 | 0 y 8 m | M | WHO I | Pilocytic Astrocytoma |
| PA10 | 11 y | F | WHO I | Pilocytic Astrocytoma |
| PA11 | 8 y | M | WHO I | Pilocytic Astrocytoma |
| PA12 | 12 y | M | WHO I | Pilocytic Astrocytoma |
| PA13 | 8 y | F | WHO I | Pilocytic Astrocytoma |
| PA14 | 4 y | M | WHO I | Pilocytic Astrocytoma |
| PA15 | 3 y | F | WHO I | Pilocytic Astrocytoma |
| PA16 | 18y | F | WHO I | Pilocytic Astrocytoma |
| EP1 | 15 y | M | WHO II | Ependymoma |
| EP2 | 2 y | M | WHO II | Ependymoma |
| EP3 | 8 y | M | WHO II | Ependymoma |
| EP4 | 12 y | M | WHO II | Ependymoma |
| EP5 | 16 y | F | WHO II | Ependymoma |
| EP6 | 8 y | F | WHO III | Ependymoma |
| EP7 | 12 y | M | WHO III | Ependymoma |
| EP8 | 13 y | M | WHO III | Ependymoma |
| EP9 | 0 y 7 m | M | WHO III | Ependymoma |
| EP10 | 6 y | M | WHO III | Ependymoma |
| EP11 | 1 y | F | WHO III | Ependymoma |
| EP12 | 1 y | M | WHO III | Ependymoma |
| GBM1 | 7 y | M | WHO IV | Glioblastoma |
| GBM2 | 8 y | M | WHO IV | Glioblastoma |
| GBM3 | 8 y | M | WHO IV | Glioblastoma |
| GBM4 | 11 y | F | WHO IV | Glioblastoma |
| GBM5 | 11 y | M | WHO IV | Glioblastoma |
| GBM6 | 3 y | F | WHO IV | Glioblastoma |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rossetti, D.V.; Inserra, I.; Nesticò, A.; Vincenzoni, F.; Iavarone, F.; Messana, I.; Castagnola, M.; Massimi, L.; Tamburrini, G.; Caldarelli, M.; et al. Pediatric Brain Tumors: Signatures from the Intact Proteome. Int. J. Mol. Sci. 2022, 23, 3196. https://doi.org/10.3390/ijms23063196
Rossetti DV, Inserra I, Nesticò A, Vincenzoni F, Iavarone F, Messana I, Castagnola M, Massimi L, Tamburrini G, Caldarelli M, et al. Pediatric Brain Tumors: Signatures from the Intact Proteome. International Journal of Molecular Sciences. 2022; 23(6):3196. https://doi.org/10.3390/ijms23063196
Chicago/Turabian StyleRossetti, Diana Valeria, Ilaria Inserra, Alessia Nesticò, Federica Vincenzoni, Federica Iavarone, Irene Messana, Massimo Castagnola, Luca Massimi, Gianpiero Tamburrini, Massimo Caldarelli, and et al. 2022. "Pediatric Brain Tumors: Signatures from the Intact Proteome" International Journal of Molecular Sciences 23, no. 6: 3196. https://doi.org/10.3390/ijms23063196






