Decoding the Synaptic Proteome with Long-Term Exposure to Midazolam during Early Development
Abstract
1. Introduction
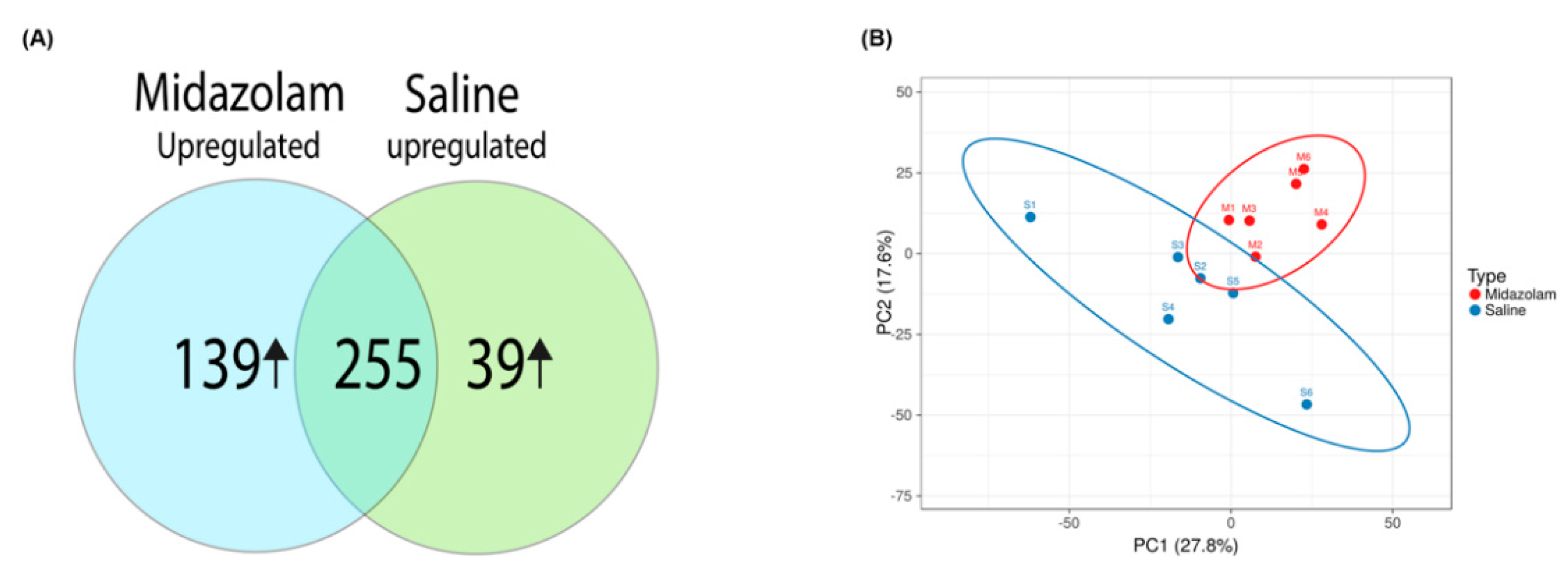
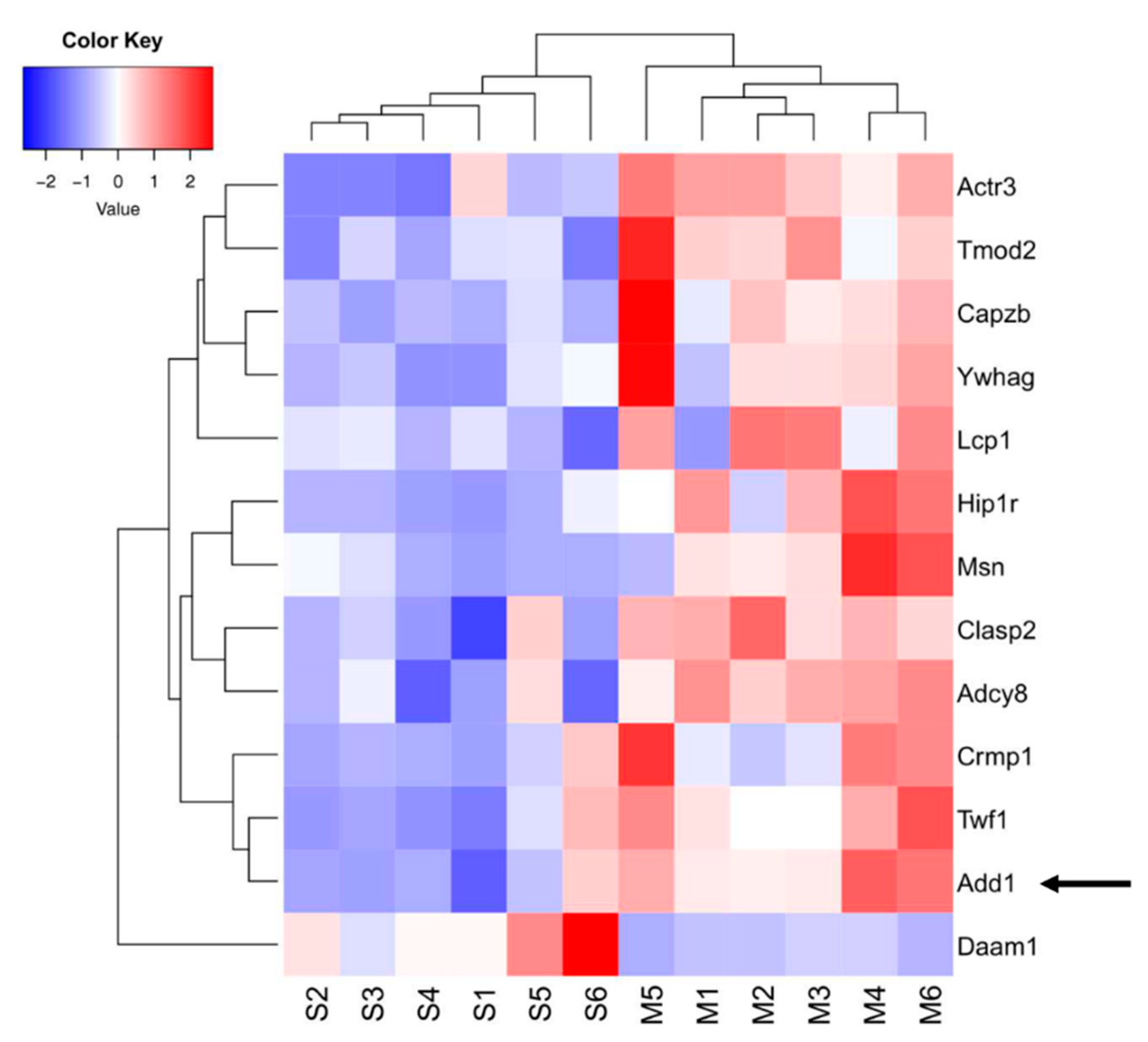
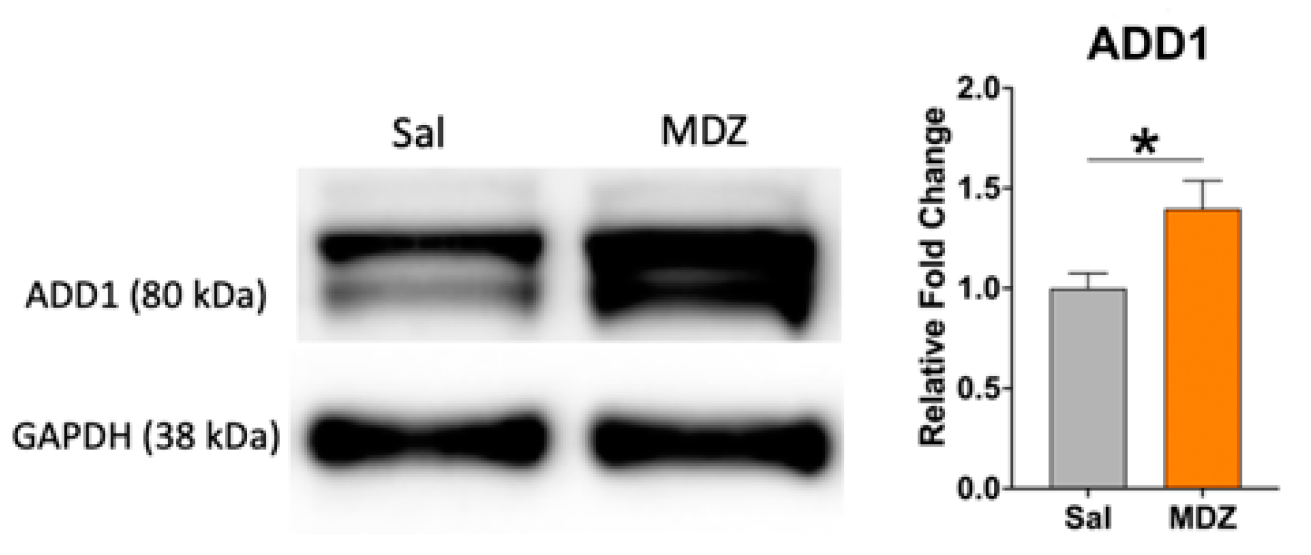
2. Results
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Midazolam Treatment
4.3. Purified Synaptosome Isolation
4.4. Mass Spectrometry Analysis
4.5. Protein Identification
4.6. Bioinformatic Analysis
4.7. Western Blot
4.8. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blencowe, H.; Cousens, S.; Oestergaard, M.Z.; Chou, D.; Moller, A.B.; Narwal, R.; Adler, A.; Vera Garcia, C.; Rohde, S.; Say, L.; et al. National, regional, and worldwide estimates of preterm birth rates in the year 2010 with time trends since 1990 for selected countries: A systematic analysis and implications. Lancet 2012, 379, 2162–2172. [Google Scholar] [CrossRef]
- Liu, L.; Oza, S.; Hogan, D.; Chu, Y.; Perin, J.; Zhu, J.; Lawn, J.E.; Cousens, S.; Mathers, C.; Black, R.E. Global, regional, and national causes of under-5 mortality in 2000-15: An updated systematic analysis with implications for the Sustainable Development Goals. Lancet 2016, 388, 3027–3035. [Google Scholar] [CrossRef]
- Ng, E.; Taddio, A.; Ohlsson, A. Intravenous midazolam infusion for sedation of infants in the neonatal intensive care unit. Cochrane Database Syst. Rev. 2017, 1, CD002052. [Google Scholar] [CrossRef] [PubMed]
- Pacifici, G.M. Clinical Pharmacology of Midazolam in Neonates and Children: Effect of Disease—A Review. Int. J. Pediatrics 2014, 2014, 309342. [Google Scholar] [CrossRef]
- Xu, J.; Mathena, R.P.; Singh, S.; Kim, J.; Long, J.J.; Li, Q.; Junn, S.; Blaize, E.; Mintz, C.D. Early Developmental Exposure to Repetitive Long Duration of Midazolam Sedation Causes Behavioral and Synaptic Alterations in a Rodent Model of Neurodevelopment. J. Neurosurg. Anesthesiol. 2019, 31, 151–162. [Google Scholar] [CrossRef]
- Iqbal O’Meara, A.M.; Miller Ferguson, N.; Zven, S.E.; Karam, O.L.; Meyer, L.C.; Bigbee, J.W.; Sato-Bigbee, C. Potential Neurodevelopmental Effects of Pediatric Intensive Care Sedation and Analgesia: Repetitive Benzodiazepine and Opioid Exposure Alters Expression of Glial and Synaptic Proteins in Juvenile Rats. Crit. Care Explor. 2020, 2, e0105. [Google Scholar] [CrossRef]
- Van Spronsen, M.; Hoogenraad, C.C. Synapse pathology in psychiatric and neurologic disease. Curr. Neurol. Neurosci. Rep. 2010, 10, 207–214. [Google Scholar] [CrossRef]
- Whittaker, V.P.; Michaelson, I.A.; Kirkland, R.J. The separation of synaptic vesicles from nerve-ending particles (‘synaptosomes’). Biochem. J. 1964, 90, 293–303. [Google Scholar] [CrossRef]
- Bai, F.; Witzmann, F.A. Synaptosome proteomics. Subcell. Biochem. 2007, 43, 77–98. [Google Scholar] [CrossRef]
- Schrimpf, S.P.; Meskenaite, V.; Brunner, E.; Rutishauser, D.; Walther, P.; Eng, J.; Aebersold, R.; Sonderegger, P. Proteomic analysis of synaptosomes using isotope-coded affinity tags and mass spectrometry. Proteomics 2005, 5, 2531–2541. [Google Scholar] [CrossRef]
- Morciano, M.; Burré, J.; Corvey, C.; Karas, M.; Zimmermann, H.; Volknandt, W. Immunoisolation of two synaptic vesicle pools from synaptosomes: A proteomics analysis. J. Neurochem. 2005, 95, 1732–1745. [Google Scholar] [CrossRef] [PubMed]
- Biesemann, C.; Grønborg, M.; Luquet, E.; Wichert, S.P.; Bernard, V.; Bungers, S.R.; Cooper, B.; Varoqueaux, F.; Li, L.; Byrne, J.A.; et al. Proteomic screening of glutamatergic mouse brain synaptosomes isolated by fluorescence activated sorting. EMBO J. 2014, 33, 157–170. [Google Scholar] [CrossRef] [PubMed]
- Heo, S.; Diering, G.H.; Na, C.H.; Nirujogi, R.S.; Bachman, J.L.; Pandey, A.; Huganir, R.L. Identification of long-lived synaptic proteins by proteomic analysis of synaptosome protein turnover. Proc. Natl. Acad. Sci. USA 2018, 115, E3827–E3836. [Google Scholar] [CrossRef] [PubMed]
- Sapp, E.; Seeley, C.; Iuliano, M.; Weisman, E.; Vodicka, P.; DiFiglia, M.; Kegel-Gleason, K.B. Protein changes in synaptosomes of Huntington’s disease knock-in mice are dependent on age and brain region. Neurobiol. Dis. 2020, 141, 104950. [Google Scholar] [CrossRef]
- Xu, Y.; Song, X.; Wang, D.; Wang, Y.; Li, P.; Li, J. Proteomic insights into synaptic signaling in the brain: The past, present and future. Mol. Brain 2021, 14, 37. [Google Scholar] [CrossRef]
- Ivannikov, M.V.; Sugimori, M.; Llinás, R.R. Synaptic vesicle exocytosis in hippocampal synaptosomes correlates directly with total mitochondrial volume. J. Mol. Neurosci. MN 2013, 49, 223–230. [Google Scholar] [CrossRef]
- Weiss, B. Vulnerability of children and the developing brain to neurotoxic hazards. Environ. Health Perspect. 2000, 108 (Suppl. 3), 375–381. [Google Scholar] [CrossRef]
- Grandjean, P.; Landrigan, P.J. Neurobehavioural effects of developmental toxicity. Lancet Neurol. 2014, 13, 330–338. [Google Scholar] [CrossRef]
- Pratt, J.A.; Brett, R.R.; Laurie, D.J. Benzodiazepine dependence: From neural circuits to gene expression. Pharmacol. Biochem. Behav. 1998, 59, 925–934. [Google Scholar] [CrossRef]
- Wirak, G.S.; Gabel, C.V.; Connor, C.W. Isoflurane Exposure in Juvenile Caenorhabditis elegans Causes Persistent Changes in Neuron Dynamics. Anesthesiology 2020, 133, 569–582. [Google Scholar] [CrossRef]
- Griessner, J.; Pasieka, M.; Böhm, V.; Grössl, F.; Kaczanowska, J.; Pliota, P.; Kargl, D.; Werner, B.; Kaouane, N.; Strobelt, S.; et al. Central amygdala circuit dynamics underlying the benzodiazepine anxiolytic effect. Mol. Psychiatry 2021, 26, 534–544. [Google Scholar] [CrossRef]
- Tau, G.Z.; Peterson, B.S. Normal development of brain circuits. Neuropsychopharmacology 2010, 35, 147–168. [Google Scholar] [CrossRef]
- De Roo, M.; Klauser, P.; Briner, A.; Nikonenko, I.; Mendez, P.; Dayer, A.; Kiss, J.Z.; Muller, D.; Vutskits, L. Anesthetics rapidly promote synaptogenesis during a critical period of brain development. PLoS ONE 2009, 4, e7043. [Google Scholar] [CrossRef]
- Citri, A.; Malenka, R.C. Synaptic Plasticity: Multiple Forms, Functions, and Mechanisms. Neuropsychopharmacology 2008, 33, 18–41. [Google Scholar] [CrossRef]
- Abraham, W.C.; Jones, O.D.; Glanzman, D.L. Is plasticity of synapses the mechanism of long-term memory storage? NPJ Sci. Learn. 2019, 4, 9. [Google Scholar] [CrossRef]
- Jevtovic-Todorovic, V.; Hartman, R.E.; Izumi, Y.; Benshoff, N.D.; Dikranian, K.; Zorumski, C.F.; Olney, J.W.; Wozniak, D.F. Early exposure to common anesthetic agents causes widespread neurodegeneration in the developing rat brain and persistent learning deficits. J. Neurosci. Off. J. Soc. Neurosci. 2003, 23, 876–882. [Google Scholar] [CrossRef]
- Puia-Dumitrescu, M.; Comstock, B.A.; Li, S.; Heagerty, P.J.; Perez, K.M.; Law, J.B.; Wood, T.R.; Gogcu, S.; Mayock, D.E.; Juul, S.E. Assessment of 2-Year Neurodevelopmental Outcomes in Extremely Preterm Infants Receiving Opioids and Benzodiazepines. JAMA Netw. Open 2021, 4, e2115998. [Google Scholar] [CrossRef]
- Spence, E.F.; Soderling, S.H. Actin Out: Regulation of the Synaptic Cytoskeleton. J. Biol. Chem. 2015, 290, 28613–28622. [Google Scholar] [CrossRef]
- Dutta, P.; Bharti, P.; Kumar, J.; Maiti, S. Role of actin cytoskeleton in the organization and function of ionotropic glutamate receptors. Curr. Res. Struct. Biol. 2021, 3, 277–289. [Google Scholar] [CrossRef]
- Nelson, J.C.; Stavoe, A.K.H.; Colón-Ramos, D.A. The actin cytoskeleton in presynaptic assembly. Cell Adhes. Migr. 2013, 7, 379–387. [Google Scholar] [CrossRef]
- Milligan, R.A.; Whittaker, M.; Safer, D. Molecular structure of F-actin and location of surface binding sites. Nature 1990, 348, 217–221. [Google Scholar] [CrossRef]
- Matus, A. Actin-based plasticity in dendritic spines. Science 2000, 290, 754–758. [Google Scholar] [CrossRef]
- Cingolani, L.A.; Goda, Y. Actin in action: The interplay between the actin cytoskeleton and synaptic efficacy. Nat. Rev. Neurosci. 2008, 9, 344–356. [Google Scholar] [CrossRef]
- Pielage, J.; Bulat, V.; Zuchero, J.B.; Fetter, R.D.; Davis, G.W. Hts/Adducin Controls Synaptic Elaboration and Elimination. Neuron 2011, 69, 1114–1131. [Google Scholar] [CrossRef]
- Stevens, R.J.; Littleton, J.T. Synaptic Growth: Dancing with Adducin. Curr. Biol. 2011, 21, R402–R405. [Google Scholar] [CrossRef][Green Version]
- Manunta, P.; Barlassina, C.; Bianchi, G. Adducin in essential hypertension. FEBS Lett. 1998, 430, 41–44. [Google Scholar] [CrossRef]
- Hughes, C.A.; Bennett, V. Adducin: A physical model with implications for function in assembly of spectrin-actin complexes. J. Biol. Chem. 1995, 270, 18990–18996. [Google Scholar] [CrossRef]
- Vukojevic, V.; Gschwind, L.; Vogler, C.; Demougin, P.; de Quervain, D.J.; Papassotiropoulos, A.; Stetak, A. A role for α-adducin (ADD-1) in nematode and human memory. EMBO J. 2012, 31, 1453–1466. [Google Scholar] [CrossRef]
- Leite, S.C.; Sampaio, P.; Sousa, V.F.; Nogueira-Rodrigues, J.; Pinto-Costa, R.; Peters, L.L.; Brites, P.; Sousa, M.M. The Actin-Binding Protein α-Adducin Is Required for Maintaining Axon Diameter. Cell Rep. 2016, 15, 490–498. [Google Scholar] [CrossRef]
- Kadenbach, B.; Jarausch, J.; Hartmann, R.; Merle, P. Separation of mammalian cytochrome c oxidase into 13 polypeptides by a sodium dodecyl sulfate-gel electrophoretic procedure. Anal. Biochem. 1983, 129, 517–521. [Google Scholar] [CrossRef]
- Dhar, S.S.; Johar, K.; Wong-Riley, M.T.T. Bigenomic transcriptional regulation of all thirteen cytochrome c oxidase subunit genes by specificity protein 1. Open Biol. 2013, 3, 120176. [Google Scholar] [CrossRef] [PubMed]
- Mjaatvedt, A.E.; Wong-Riley, M.T. Relationship between synaptogenesis and cytochrome oxidase activity in Purkinje cells of the developing rat cerebellum. J. Comp. Neurol. 1988, 277, 155–182. [Google Scholar] [CrossRef] [PubMed]
- Wong-Riley, M.T.T. Bigenomic regulation of cytochrome c oxidase in neurons and the tight coupling between neuronal activity and energy metabolism. Adv. Exp. Med. Biol. 2012, 748, 283–304. [Google Scholar] [CrossRef] [PubMed]
- Gunter, T.E.; Gunter, K.K.; Sheu, S.S.; Gavin, C.E. Mitochondrial calcium transport: Physiological and pathological relevance. Am. J. Physiol. 1994, 267, C313–C339. [Google Scholar] [CrossRef]
- Schwarz, T.L. Mitochondrial trafficking in neurons. Cold Spring Harb. Perspect. Biol. 2013, 5, a011304. [Google Scholar] [CrossRef]
- Tang, J.; Oliveros, A.; Jang, M.H. Dysfunctional Mitochondrial Bioenergetics and Synaptic Degeneration in Alzheimer Disease. Int. Neurourol. J. 2019, 23, S5–S10. [Google Scholar] [CrossRef]
- Bi, R.; Zhang, W.; Zhang, D.-F.; Xu, M.; Fan, Y.; Hu, Q.-X.; Jiang, H.-Y.; Tan, L.; Li, T.; Fang, Y.; et al. Genetic association of the cytochrome c oxidase-related genes with Alzheimer’s disease in Han Chinese. Neuropsychopharmacology 2018, 43, 2264–2276. [Google Scholar] [CrossRef]
- Kissin, I.; Brown, P.T.; Bradley, E.L., Jr. Sedative and Hypnotic Midazolam-Morphine Interactions in Rats. Anesth. Analg. 1990, 71, 137–143. [Google Scholar] [CrossRef]
- Luo, J.; Guo, J.; Han, D.; Li, H. Comparison of dexmedetomidine and midazolam on neurotoxicity in neonatal mice. Sheng Wu Yi Xue Gong Cheng Xue Za Zhi 2013, 30, 607–610. [Google Scholar]
- Pendyala, G.; Buescher, J.L.; Fox, H.S. Isolation of Synaptosomes from Archived Brain Tissues. In Current Laboratory Methods in Neuroscience Research; Xiong, H., Gendelman, H.E., Eds.; Springer: New York, NY, USA, 2014; pp. 145–152. [Google Scholar]
- Chand, S.; Jo, A.; Vellichirammal, N.N.; Gowen, A.; Guda, C.; Schaal, V.; Odegaard, K.; Lee, H.; Pendyala, G.; Yelamanchili, S.V. Comprehensive Characterization of Nanosized Extracellular Vesicles from Central and Peripheral Organs: Implications for Preclinical and Clinical Applications. ACS Appl. Nano Mater. 2020, 3, 8906–8919. [Google Scholar] [CrossRef]
- Odegaard, K.E.; Schaal, V.L.; Clark, A.R.; Koul, S.; Gowen, A.; Sankarasubramani, J.; Xiao, P.; Guda, C.; Lisco, S.J.; Yelamanchili, S.V.; et al. Characterization of the intergenerational impact of in utero and postnatal oxycodone exposure. Transl. Psychiatry 2020, 10, 329. [Google Scholar] [CrossRef] [PubMed]
- Odegaard, K.E.; Schaal, V.L.; Clark, A.R.; Koul, S.; Sankarasubramanian, J.; Xia, Z.; Mellon, M.; Uberti, M.; Liu, Y.; Stothert, A.; et al. A Holistic Systems Approach to Characterize the Impact of Pre- and Post-natal Oxycodone Exposure on Neurodevelopment and Behavior. Front. Cell Dev. Biol. 2020, 8, 619199. [Google Scholar] [CrossRef] [PubMed]
- Pendyala, G.; Buescher, J.L.; Fox, H.S. Methamphetamine and inflammatory cytokines increase neuronal Na+/K+-ATPase isoform 3: Relevance for HIV associated neurocognitive disorders. PLoS ONE 2012, 7, e37604. [Google Scholar] [CrossRef]
- Odegaard, K.E. In Utero and Postnatal Oxycodone Exposure: Implications for Intergenerational Effects. Ph.D. Thesis, University of Nebraska Medical Center, Omaha, NE, USA, 2021. DigitalCommons@UNMC. [Google Scholar]
- Koul, S.; Schaal, V.L.; Chand, S.; Pittenger, S.T.; Nanoth Vellichirammal, N.; Kumar, V.; Guda, C.; Bevins, R.A.; Yelamanchili, S.V.; Pendyala, G. Role of Brain Derived Extracellular Vesicles in Decoding Sex Differences Associated with Nicotine Self-Administration. Cells 2020, 9, 1883. [Google Scholar] [CrossRef]
- Guda, R.S.; Odegaard, K.E.; Tan, C.; Schaal, V.L.; Yelamanchili, S.V.; Pendyala, G. Integrated Systems Analysis of Mixed Neuroglial Cultures Proteome Post Oxycodone Exposure. Int. J. Mol. Sci. 2021, 22, 6421. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef]
- Chand, S.; Gowen, A.; Savine, M.; Moore, D.; Clark, A.; Huynh, W.; Wu, N.; Odegaard, K.; Weyrich, L.; Bevins, R.A.; et al. A comprehensive study to delineate the role of an extracellular vesicle-associated microRNA-29a in chronic methamphetamine use disorder. J. Extracell. Vesicles 2021, 10, e12177. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, N.M.; Vellichirammal, N.N.; Guda, C.; Pendyala, G. Decoding the Synaptic Proteome with Long-Term Exposure to Midazolam during Early Development. Int. J. Mol. Sci. 2022, 23, 4137. https://doi.org/10.3390/ijms23084137
Nguyen NM, Vellichirammal NN, Guda C, Pendyala G. Decoding the Synaptic Proteome with Long-Term Exposure to Midazolam during Early Development. International Journal of Molecular Sciences. 2022; 23(8):4137. https://doi.org/10.3390/ijms23084137
Chicago/Turabian StyleNguyen, Nghi M., Neetha N. Vellichirammal, Chittibabu Guda, and Gurudutt Pendyala. 2022. "Decoding the Synaptic Proteome with Long-Term Exposure to Midazolam during Early Development" International Journal of Molecular Sciences 23, no. 8: 4137. https://doi.org/10.3390/ijms23084137
APA StyleNguyen, N. M., Vellichirammal, N. N., Guda, C., & Pendyala, G. (2022). Decoding the Synaptic Proteome with Long-Term Exposure to Midazolam during Early Development. International Journal of Molecular Sciences, 23(8), 4137. https://doi.org/10.3390/ijms23084137






_Guda.png)

