Identification of CDPK Gene Family in Solanum habrochaites and Its Function Analysis under Stress
Abstract
:1. Introduction
2. Results
2.1. Identification of CDPK Family Members in Solanum habrochaites
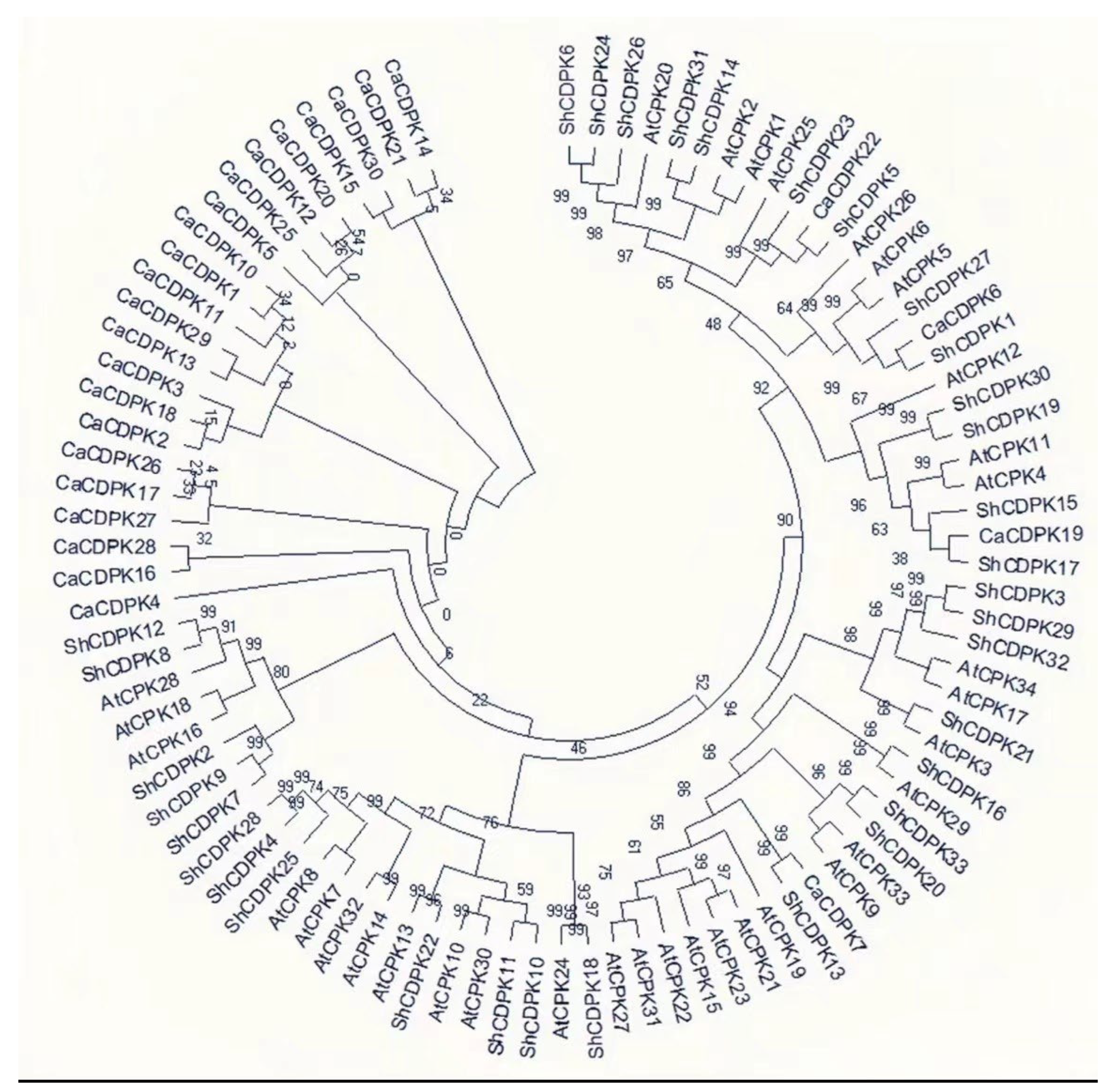
2.2. Phylogenetic Analysis of CDPK Gene in Solanum habrochaites
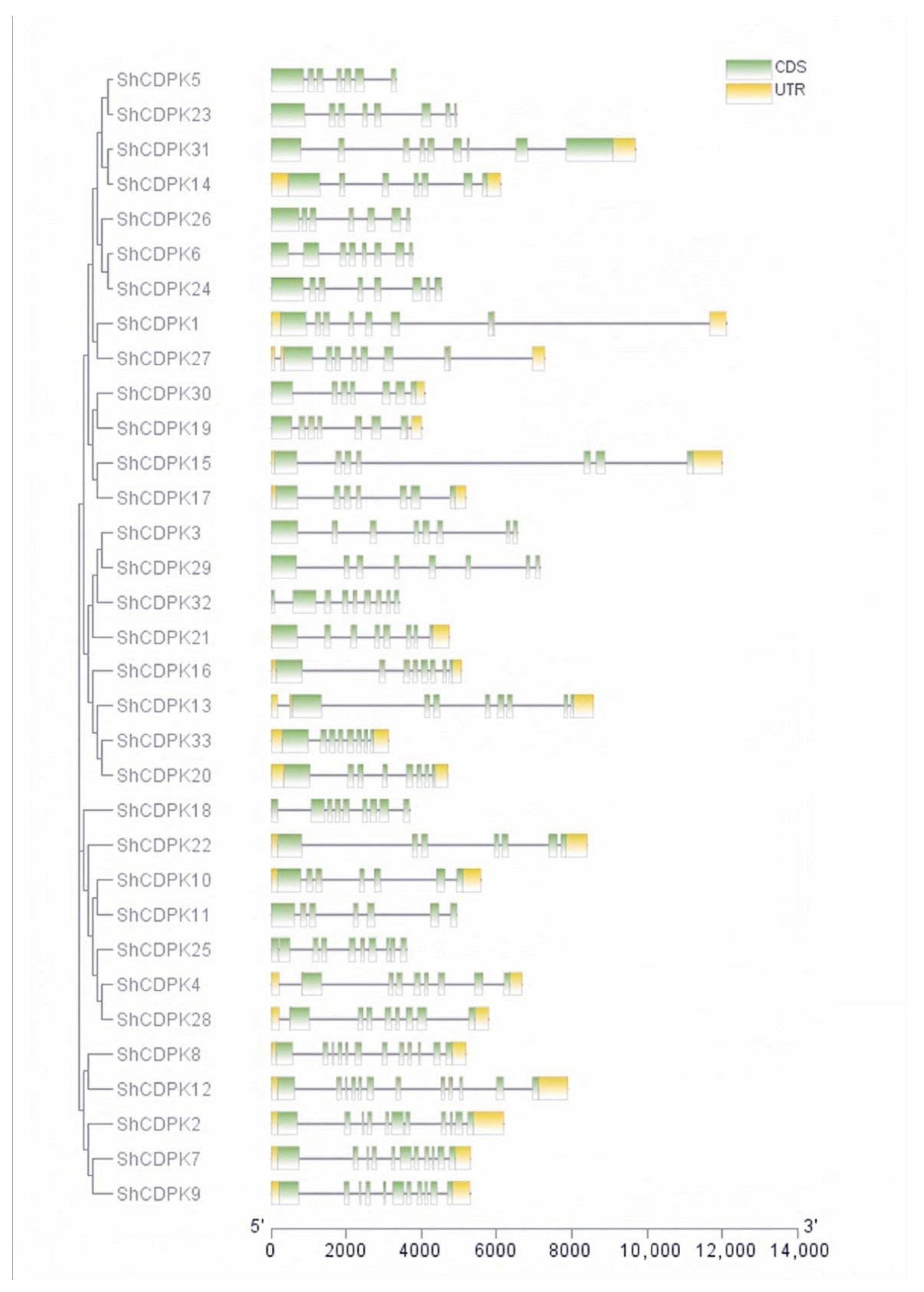
2.3. Analysis of CDPK Gene Family Structure in Solanum habrochaites
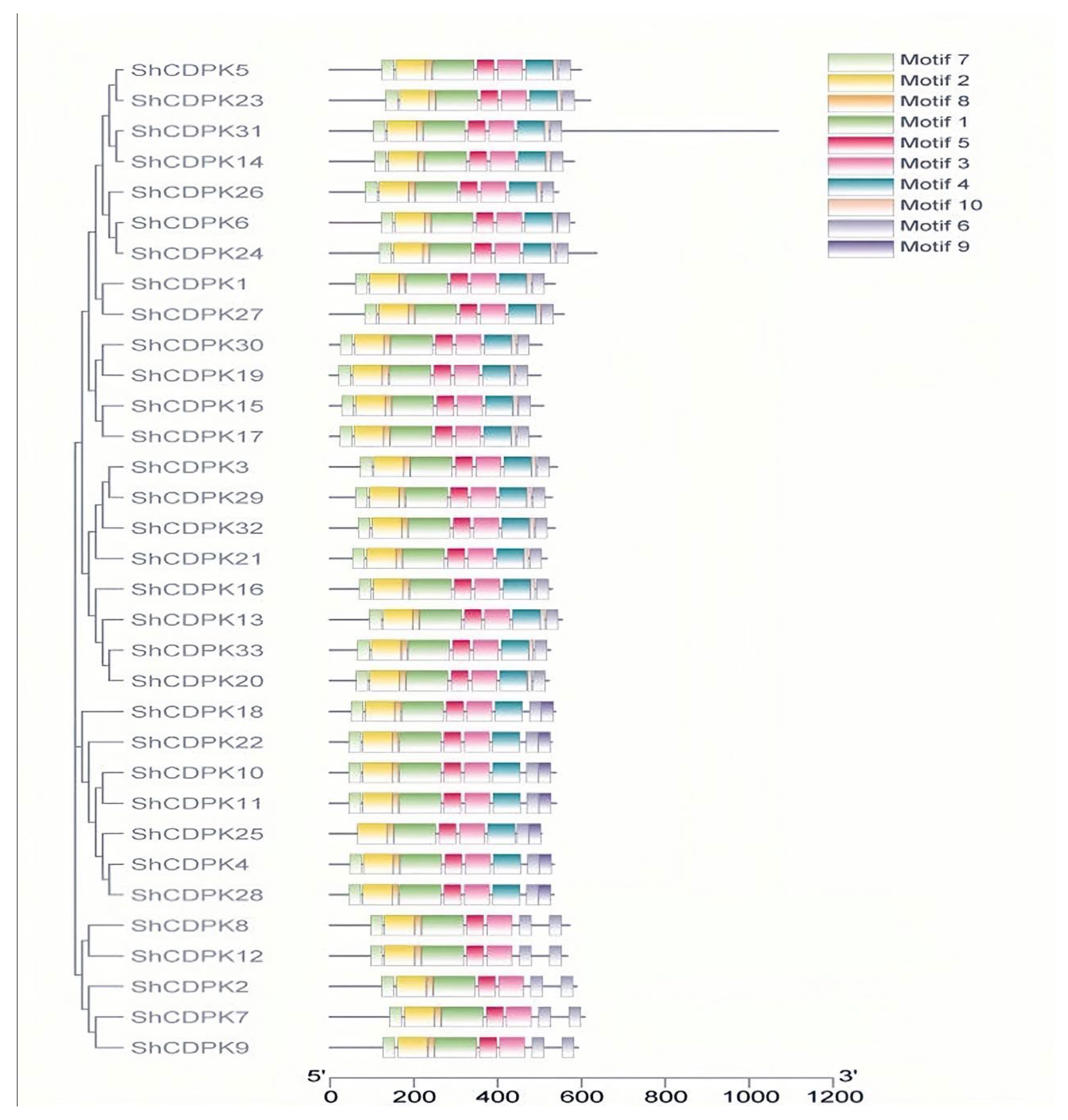
2.4. Motif Analysis of CDPK Gene Family in Solanum habrochaites
2.5. Gene Replication and Collinearity Analysis of CDPK Gene Family in Solanum habrochaites
2.6. Analysis of Cis Acting Elements of CDPK Gene Family in Solanum habrochaites
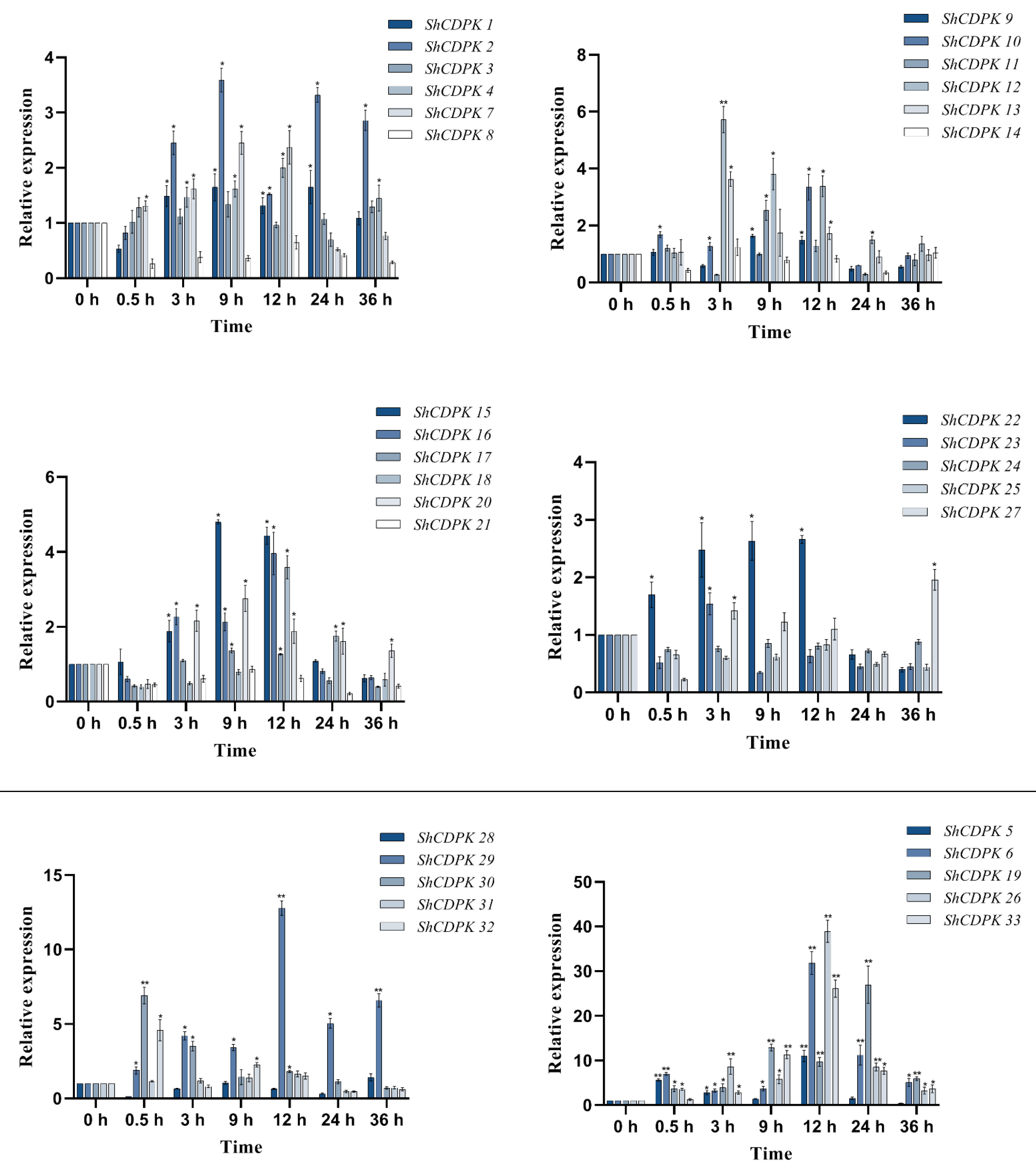
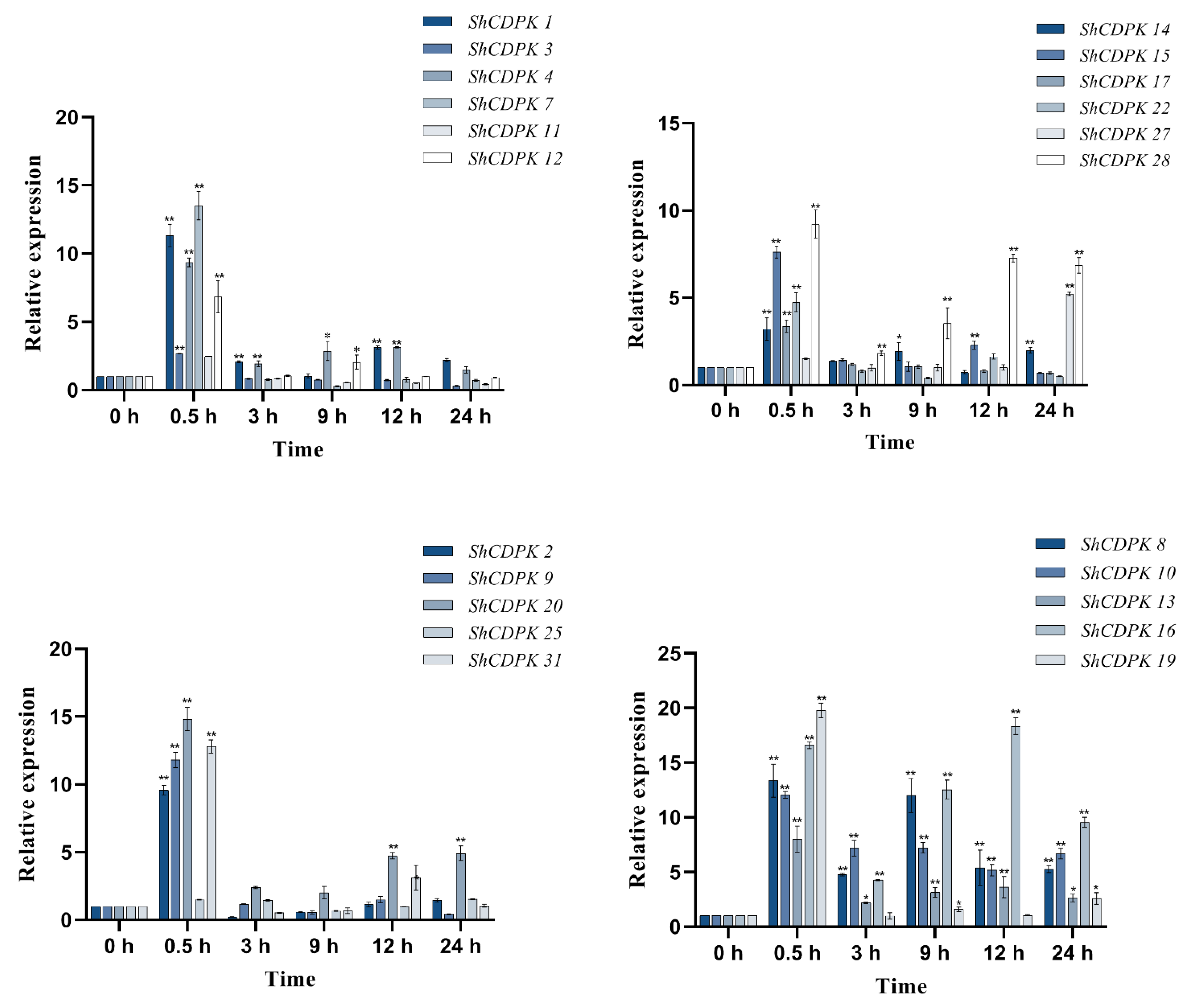
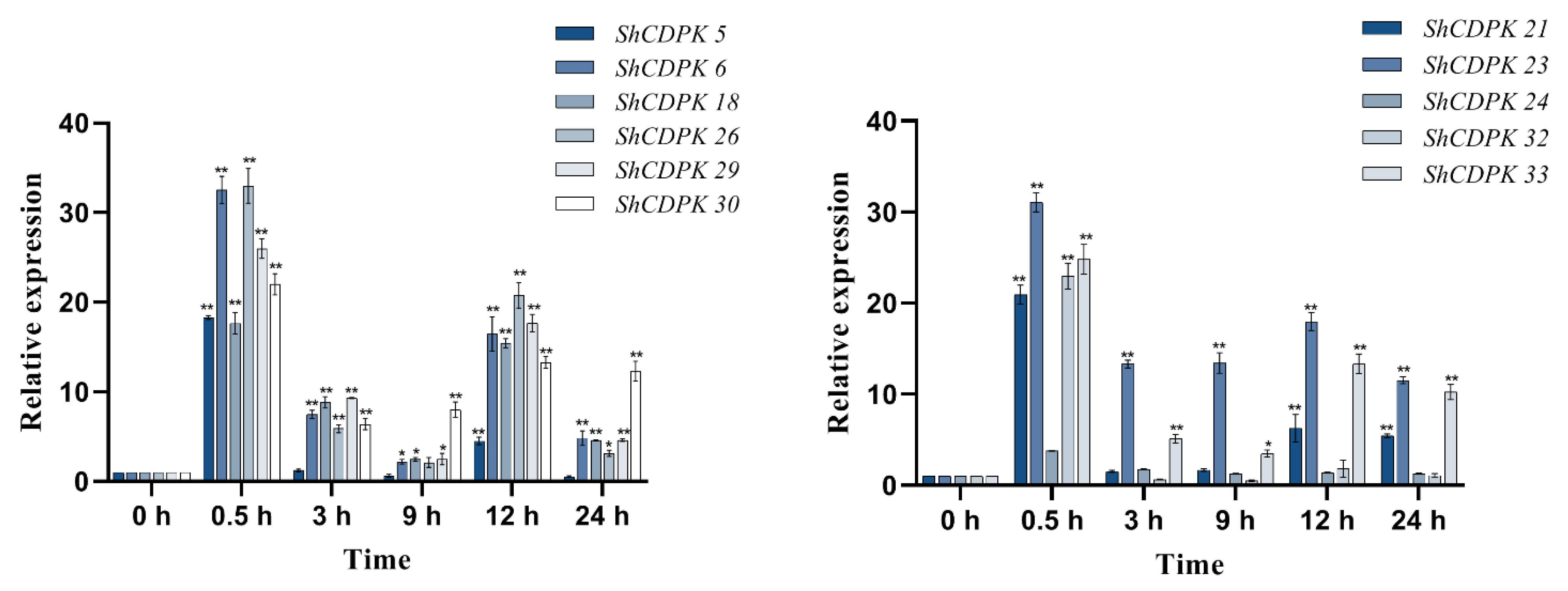
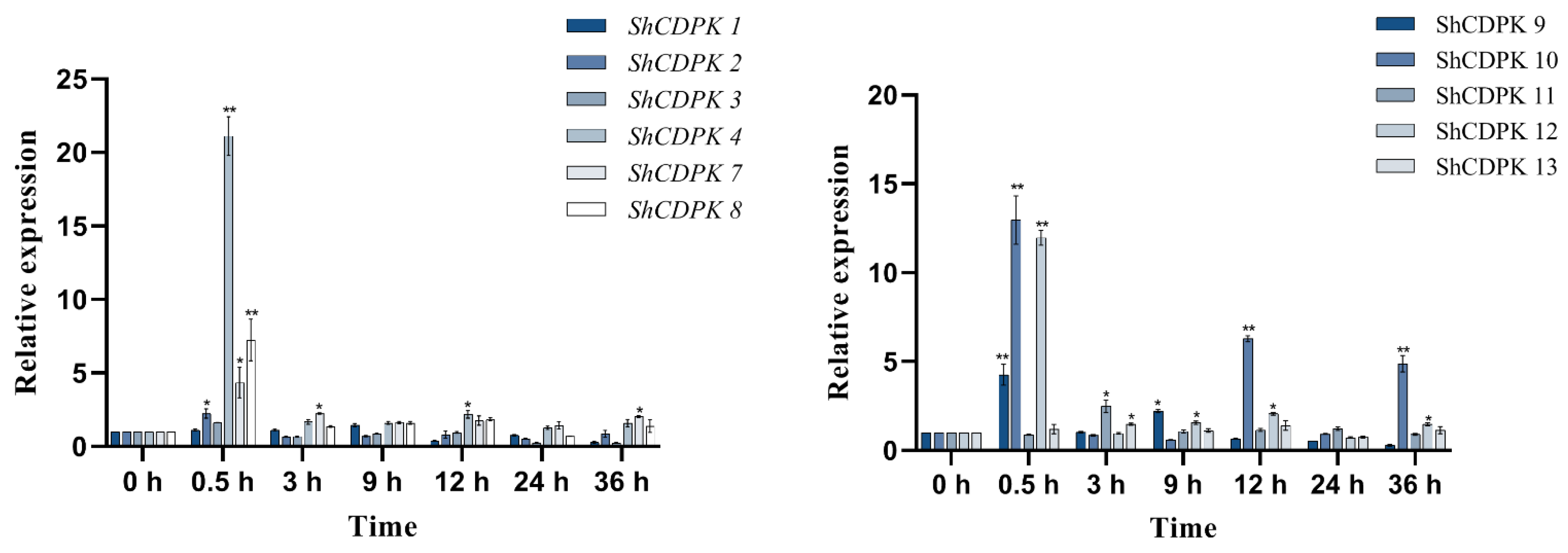
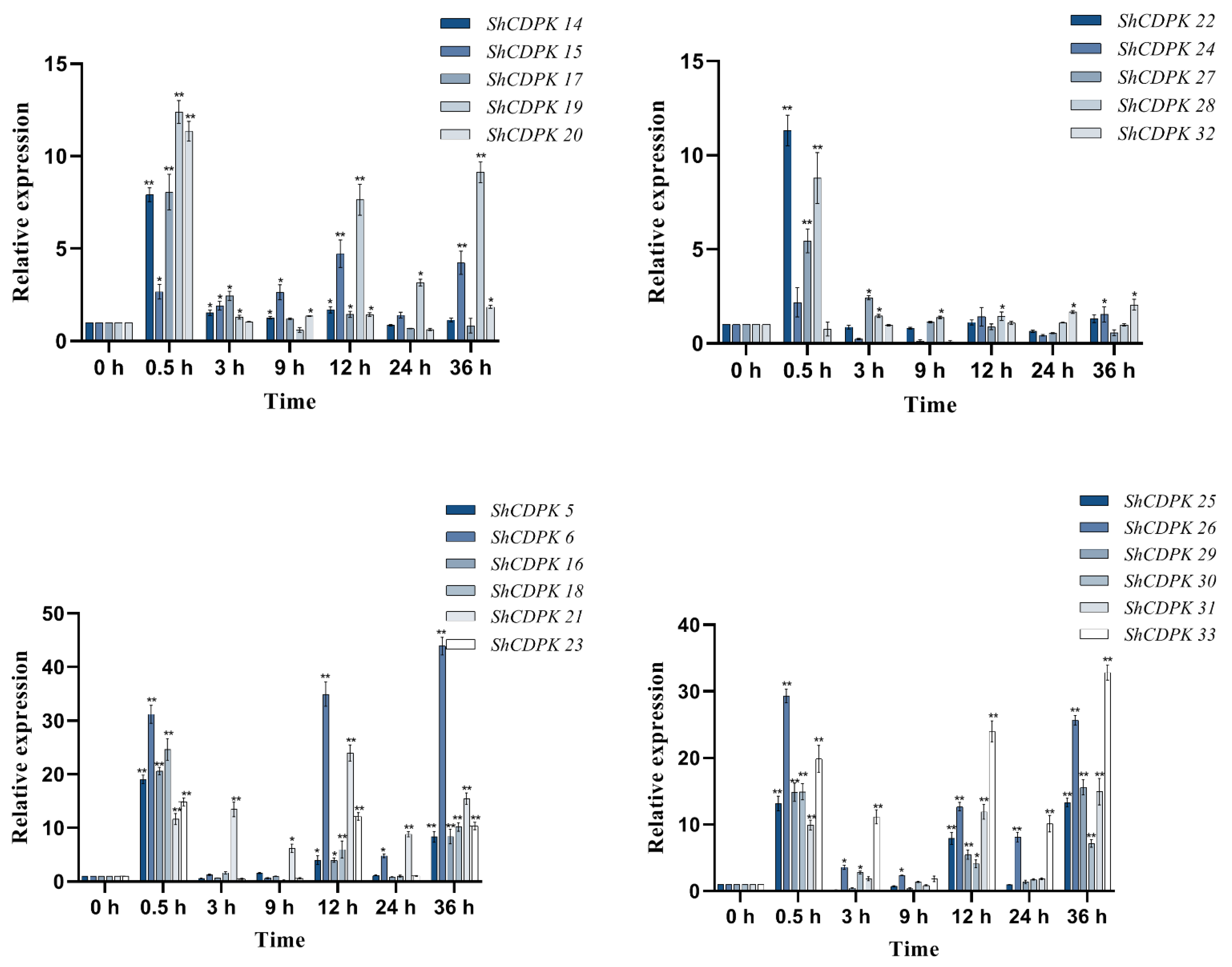
2.7. Expression Analysis of CDPK Gene Family in Solanum habrochaites under Various Stress
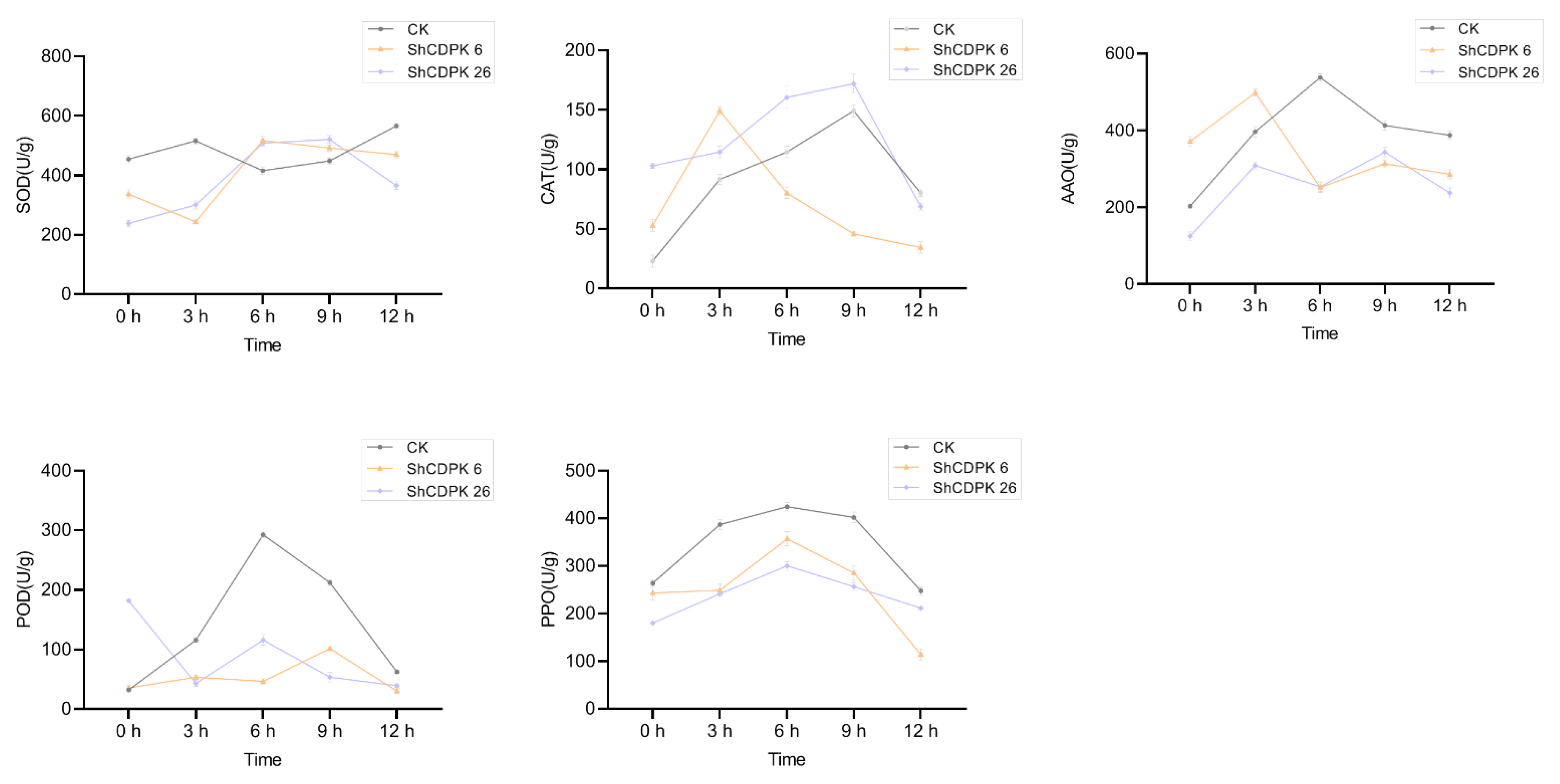
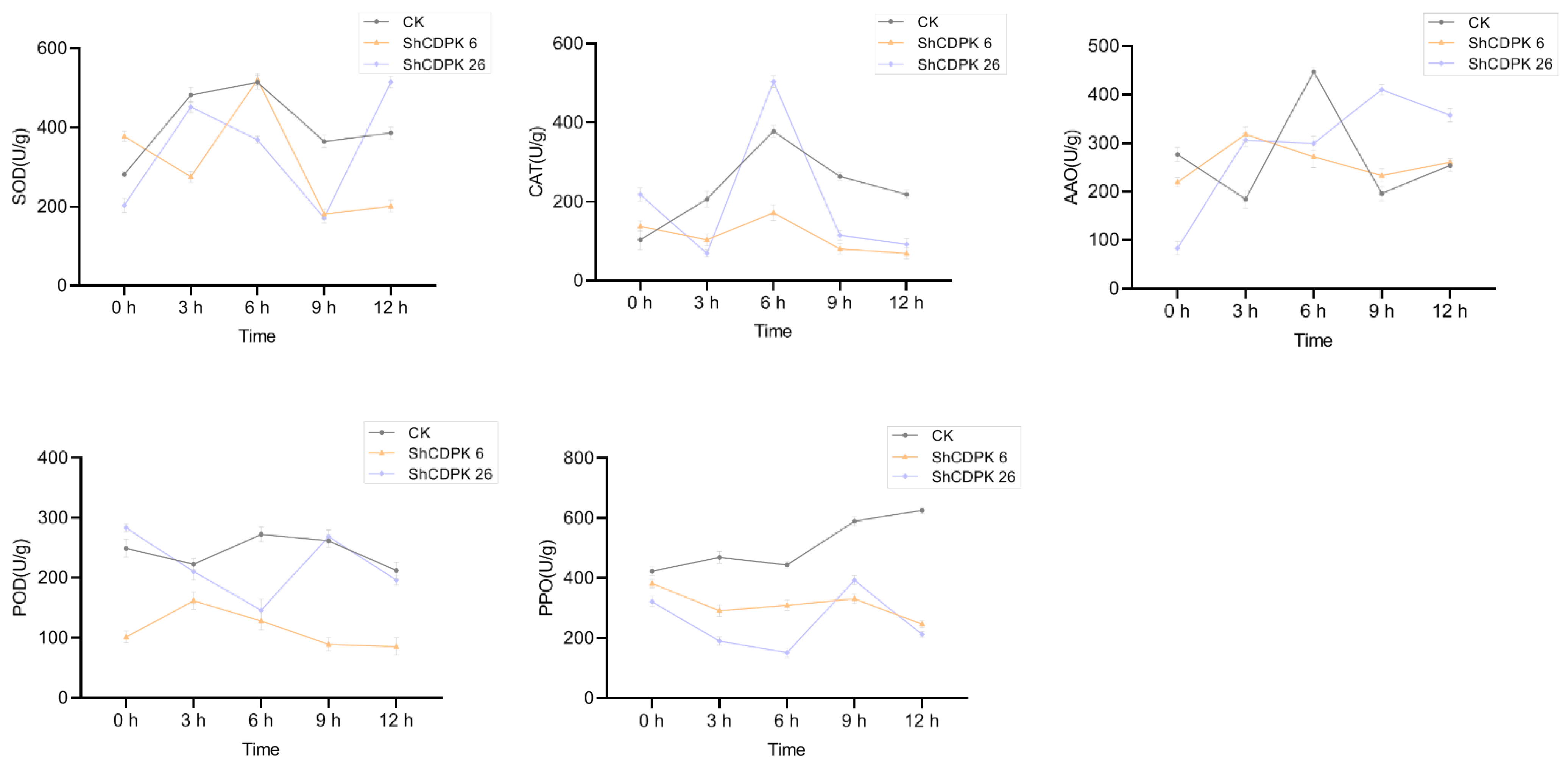
2.8. Functional Analysis of Silencing ShCDPK6 and ShCDPK26 in Solanum habrochaites
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Treatments
4.1.1. Cold Stress
4.1.2. Drought Stress
4.1.3. Botrytis Cinerea Stress
4.2. Identification of the CDPK Gene Family in Solanum habrochaites by Bioinformatic Analysis
4.2.1. Identification and Basic Information Analysis of CDPK Gene Family Members in Solanum habrochaites
4.2.2. Evolutionary Analysis of CDPK Gene Family in Solanum habrochaites
4.2.3. Analysis of Genetic Structure of CDPK Gene in Solanum habrochaites
4.2.4. Analysis of CDPK Gene Family Conservative Motif in Solanum habrochaites
4.2.5. Gene Replication of CDPK Gene Family in Solanum habrochaites
4.3. Real-Time Quantitative PCR (qRT-PCR) Analysis
4.4. Virus-Induced Gene Silencing (VIGS)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tuteja, N.; Mahajan, S. Calcium Signaling Network in Plants. Plant Signal. Behav. 2007, 2, 79–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liese, A.; Romeis, T. Biochemical regulation of in vivo function of plant calcium-dependent protein kinases (CDPK). BBA Mol. Cell Res. 2013, 1833, 1582–1589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simeunovic, A.; Mair, A.; Wurzinger, B.; Teige, M. Know where your clients are: Subcellular localization and targets of calcium-dependent protein kinases. J. Exp. Bot. 2016, 67, 3855–3872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almonte, G.R.; Arthur, K.; Higgins, C.M.; MacDiarmid, R.M. Calcium-dependent protein kinases in plants: Evolution, expression and function. Plant Cell Physiol. 2014, 55, 551–569. [Google Scholar] [CrossRef] [Green Version]
- Knight, H. Calcium signaling during abiotic stress in plants. Int. Rev. Cytol. 1999, 195, 269–324. [Google Scholar]
- Sanders, D.; Brownlee, C.; Harper, J.F. Communicating with calcium. Plant Cell 1999, 11, 691–706. [Google Scholar] [CrossRef] [Green Version]
- Kudla, J.; Batistič, O.; Hashimoto, K. Calcium signals: The lead currency of plant information processing. Plant Cell 2010, 22, 541–563. [Google Scholar] [CrossRef]
- Dodd, A.N.; Kudla, J.; Sanders, D. The language of calcium signaling. Annu. Rev. Plant Biol. 2010, 61, 593–620. [Google Scholar] [CrossRef]
- Knight, H.; Knight, M.R. Abiotic stress signalling pathways: Specifificity and cross-talk. Trends Plant Sci. 2001, 6, 262–267. [Google Scholar] [CrossRef]
- Marcec, M.J.; Gilroy, S.; Poovaiah, B.W.; Tanaka, K. Mutual interplay of Ca2+ and ROS signaling in plant immune response. Plant Sci. 2019, 283, 343–354. [Google Scholar] [CrossRef]
- Tian, W.; Wang, C.; Gao, Q.; Li, L.; Luan, S. Calcium spikes, waves and oscillations in plant development and biotic interactions. Nat. Plants 2020, 6, 750–759. [Google Scholar] [CrossRef] [PubMed]
- White, P.; Broadley, M. Calcium in plants. Ann. Bot. 2003, 92, 487–511. [Google Scholar] [CrossRef] [PubMed]
- Sanders, D.; Pelloux, J.; Brownlee, C.; Harper, J.F. Calcium at the crossroads of signaling. Plant Cell 2002, 14, S401–S417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Batistic, O.; Kudla, J. Analysis of calcium signaling pathways in plants. Biochim. Biophys. Acta 2012, 1820, 1283–1293. [Google Scholar] [CrossRef] [PubMed]
- Schulz, P.; Herde, M.; Romeis, T. Calcium-dependent protein kinases: Hubs in plant stress signaling and development. Plant Physiol. 2013, 163, 523–530. [Google Scholar] [CrossRef] [Green Version]
- Harper, J.F.; Breton, G.; Harmon, A. Decoding Ca2+ signals through plant protein kinases. Annu. Rev. Plant Biol. 2004, 55, 263–288. [Google Scholar] [CrossRef]
- Monaghan, J.; Matschi, S.; Shorinola, O.; Rovenich, H.; Matei, A.; Segonzac, C.; Malinovsky, F.G.; Rathjen, J.P.; MacLean, D.; Romeis, T.; et al. The calcium-dependent protein kinase CPK28 buffers plant immunity and regulates BIK1 turn over. Cell Host Microbe 2014, 16, 605–615. [Google Scholar] [CrossRef] [Green Version]
- Hong, Y.; Takano, M.; Liu, C.; Gasch, A.; Chye, M.; Chua, N. Expression of three members of the calcium-dependent protein kinase gene family in Arabidopsis thaliana. Plant Mol. Biol. 1996, 30, 1259–1275. [Google Scholar] [CrossRef] [Green Version]
- Ye, S.; Wang, L.; Xie, W.; Wan, B.; Li, X.; Lin, Y. Expression profifile of calcium-dependent protein kinase (CDPKs) genes during the whole lifespan and under phytohormone treatment conditions in rice (Oryza sativa L. ssp. indica). Plant Mol. Biol. 2009, 70, 311–325. [Google Scholar] [CrossRef]
- Boudsocq, M.; Sheen, J. CDPKs in immune and stress signaling. Trends Plant Sci. 2013, 18, 30–40. [Google Scholar] [CrossRef] [Green Version]
- Harmon, A.C.; Gribskov, M.; Gubrium, E.; Harper, J.F. The CDPK superfamily of protein kinases. New Phytol. 2001, 151, 175–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ho, S.L.; Huang, L.F.; Lu, C.A.; He, S.L.; Wang, C.C.; Yu, S.P.; Chen, J.; Yu, S.-M. Sugar starvation-and GA-inducible calcium dependent protein kinase 1 feedback regulates GA biosynthesis and activates a 14-3-3 protein to confer drought tolerance in rice seedlings. Plant Mol. Biol. 2013, 81, 347–361. [Google Scholar] [CrossRef] [PubMed]
- Edel, K.H.; Kudla, J. Integration of calcium and ABA signaling. Curr. Opin. Plant Biol. 2016, 33, 83–91. [Google Scholar] [PubMed]
- Zhang, H.F.; Liu, D.Y.; Yang, B.; Liu, W.Z.; Mu, B.B.; Song, H.X.; Chen, B.Y.; Li, Y.; Ren, D.T.; Deng, H.Q.; et al. Arabidopsis CPK6 positively regulates ABA signaling and drought tolerance through phosphorylating ABA-responsive element-binding factors. J. Exp. Bot. 2020, 71, 188–203. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Wu, C.; Luo, C.; Wei, M.; Qu, S.; Wang, S. Overexpression of MdCPK1a gene, a calcium dependent protein kinase in apple, increase tobacco cold tolerance via scavenging ROS accumulation. PLoS ONE 2020, 15, e0242139. [Google Scholar] [CrossRef]
- Weckwerth, P.; Ehlert, B.; Romeis, T. ZmCPK1, a calcium-independent kinase member of the Zea mays CDPK gene family, functions as a negative regulator in cold stress signalling. Plant Cell Environ. 2015, 38, 544–558. [Google Scholar] [CrossRef]
- Ma, S.; Wu, W. AtCPK23 functions in Arabidopsis responses to drought and salt stresses. Plant Mol. Biol. 2007, 65, 511–518. [Google Scholar] [CrossRef]
- Romeis, T.; Ludwig, A.A.; Martin, R.; Jones, J.D. Calcium-dependent protein kinases play an essential role in a plant defence response. EMBO J. 2001, 20, 5556–5567. [Google Scholar] [CrossRef] [Green Version]
- Murillo, I.; Jaeck, E.; Cordero, M.J.; San Segundo, B. Transcriptional activation of a maize calcium-dependent protein kinase gene in response to fungal elicitors and infection. Plant Mol. Biol. 2001, 45, 145–158. [Google Scholar] [CrossRef]
- Cao, X.; Jiang, F.; Wang, X.; Zang, Y.; Wu, Z. Comprehensive evaluation and screening for chilling-tolerance in tomato lines at the seedling stage. Euphytica 2015, 205, 569–584. [Google Scholar] [CrossRef]
- Foolad, M.R.; Lin, G.Y. Relationship between cold tolerance during seed germination and vegetative growth in tomato: Germplasm evaluation. J. Am. Soc. Hortic. Sci. 2000, 125, 679–683. [Google Scholar] [CrossRef]
- Venema, J.H.; Linger, P.; Van Heusden, A.W.; Van Hasselt, P.R.; Bruggemann, W. The inheritance of chilling tolerance in tomato (Lycopersicon spp.). Plant Biol. 2005, 7, 118–130. [Google Scholar] [CrossRef] [PubMed]
- Egashira, H.; Kuwashima, A.; Ishiguro, H.; Fukushima, K.; Kaya, T.; Imanishi, S. Screening of wild accessions resistance to gray mold (Botrytis cinerea Pers.) in Lycopersicon. Acta Physiol. Plant. 2000, 22, 324–326. [Google Scholar] [CrossRef]
- Zhao, R.; Sun, H.L.; Mei, C.; Wang, X.J.; Yan, L.; Liu, R.; Zhang, X.F.; Wang, X.F.; Zhang, D.P. The Arabidopsis Ca2+-dependent protein kinase CPK12 negatively regulates abscisic acid signaling in seed germination and post-germination growth. New Phytol. 2011, 192, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.Y.; Yu, X.C.; Wang, X.J.; Zhao, R.; Li, Y.; Fan, R.C.; Shang, Y.; Du, S.Y.; Wang, X.F.; Wu, F.Q.; et al. Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell 2007, 19, 3019–3036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, L.; Lan, W.; Jiang, Y.; Fang, W.; Luan, S. A calcium-dependent protein kinase interacts with and activates a calcium channel to regulate pollen tube growth. Mol. Plant 2014, 7, 369–376. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.W.; Ji, J.; Lu, L.; Qi, X.H.; Chen, X.H. Cloning and expression analysis of Cucumis sativus calcium-dependent protein kinase 5 gene (CsCDPK5) under waterlogging stress. Acta Hortic. Sin. 2016, 43, 704–714. [Google Scholar]
- Klimecka, M.; Muszyńska, G. Structure and functions of plant calcium-dependent protein kinases. Acta Biochim. Pol. 2007, 54, 219–233. [Google Scholar] [CrossRef] [Green Version]
- Zou, J.J.; Wei, F.J.; Wang, C.; Wu, J.J.; Ratnasekera, D.; Liu, W.X.; Wu, W.H. Arabidopsis calcium-dependent protein kinase CPK10 functions in abscisic acid- and Ca2 +-mediated stomatal regulation in response to drought stress. Plant Physiol. 2010, 154, 1232–1243. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Liu, M.; Lu, L.; Qu, W.Q. Genome-wide analysis and expression of the calcium-dependent protein kinase gene family in cucumber. Mol. Genet. Genom. 2015, 290, 1403–1414. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, Y.; He, Q.; Chai, M.; Huang, Y.; Chen, F.; Wang, X.; Liu, Y.; Cai, H.; Qin, Y. Genome-wide investigation of calcium-dependent protein kinase gene family in pineapple: Evolution and expression profiles during development and stress. BMC Genom. 2020, 21, 72. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Han, Y.T.; Zhao, F.L.; Hu, Y.; Gao, Y.R.; Ma, Y.F.; Zheng, Y.; Wang, Y.J.; Wen, Y.Q. Genome-wide identification and expression analysis of the CDPK gene family in grape, Vitis spp. BMC Plant Biol. 2015, 15, 164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolfe, K.H.; Gouy, M.; Yang, Y.W.; Sharp, P.M.; Li, W.H. Date of the monocot-dicot divergence estimated from chloroplast DNA sequence data. Proc. Natl. Acad. Sci. USA 1989, 86, 6201–6205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mcainsh, M.R.; Pinttman, J.K. Shaping the calcium signature: Tansley review. New Phytol. 2009, 181, 275–294. [Google Scholar] [CrossRef]
- Jiang, S.S.; Zhang, D.; Kong, X.P.; Zhou, Y.; Li, D.Q. Research progress of structural characteristics and functions of calcium-dependent protein kinases in plants. Biotechnol. Bull. 2013, 29, 12–19. [Google Scholar]
- Geiger, D.; Scherzer, S.; Mumm, P.; Marten, I.; Ache, P.; Matschi, S.; Liese, A.; Wellmann, C.; Al-Rasheid, K.A.; Grill, E.; et al. Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affifinities. Proc. Natl. Acad. Sci. USA 2010, 107, 8023–8028. [Google Scholar] [CrossRef] [Green Version]
- Liu, E.; Page, J.E. Optimized cDNA libraries for virus-induced gene silencing (VIGS) using tobacco rattle virus. Plant Methods 2008, 4, 5. [Google Scholar] [CrossRef] [Green Version]
- Cui, Y.N.; Jiang, J.B.; Yang, H.H.; Zhao, T.T.; Xu, X.Y.; Li, J.F. Virus-induced gene silencing (VIGS) of the NBS-LRR gene SlNLC1 compromises Sm-mediated disease resistance to Stemphylium lycopersici in tomato. Biochem. Bioph. Res. Commun. 2018, 503, 1524–1529. [Google Scholar] [CrossRef]
- Virk, N.; Liu, B.; Zhang, H.J.; Li, X.H.; Zhang, Y.F.; Li, D.Y.; Song, F.M. Tomato SlMPK4 is required for resistance against Botrytis cinerea and tolerance to drought stress. Acta Physiol. Plant. 2013, 35, 1211–1221. [Google Scholar] [CrossRef]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Liang, Z.; Aranda, M.A.; Hong, N.; Liu, L.; Kang, B.; Gu, Q. A cucumber green mottle mosaic virus vector for virus-induced gene silencing in cucurbit plants. Plant Methods 2020, 16, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Gene Name | Sequence Accession | Gene Length (bp) | Protein Length (aa) | Mv/Da (ku) | PI | Chromosome | Chromosome Starting Position | Chromosome Termination Position | Subcellula Localization |
|---|---|---|---|---|---|---|---|---|---|
| ShCDPK1 | Solhab01g001000 | 1608 | 535 | 60,015.8 | 5.8 | Chr1 | 153266 | 165357 | 1 |
| ShCDPK2 | Solhab01g038800 | 1767 | 588 | 65,836 | 9.25 | Chr1 | 2898264 | 2904451 | 1 |
| ShCDPK3 | Solhab01g408500 | 1626 | 541 | 61,077.6 | 5.52 | Chr1 | 101718341 | 101724893 | 1 |
| ShCDPK4 | Solhab01g412600 | 1602 | 533 | 60,048.3 | 7.28 | Chr1 | 102121744 | 102128426 | 1 |
| ShCDPK5 | Solhab01g424200 | 1797 | 598 | 67,570.6 | 5.22 | Chr1 | 103438229 | 103441551 | 1 |
| ShCDPK6 | Solhab01g425400 | 1749 | 582 | 64,677 | 5.93 | Chr1 | 103522036 | 103525799 | 1 |
| ShCDPK7 | Solhab02g041900 | 1824 | 607 | 67,931.8 | 9.51 | Chr2 | 3447317 | 3452629 | 1 |
| ShCDPK8 | Solhab02g106200 | 1713 | 570 | 64,271.5 | 9.77 | Chr2 | 8674949 | 8680139 | 1.2 |
| ShCDPK9 | Solhab02g234000 | 1776 | 591 | 66,246.1 | 8.72 | Chr2 | 19980037 | 19985317 | 1 |
| ShCDPK10 | Solhab03g087000 | 1617 | 538 | 60,922.5 | 6.85 | Chr3 | 7056576 | 7062155 | 1 |
| ShCDPK11 | Solhab03g089800 | 1620 | 539 | 60,876.4 | 6.98 | Chr3 | 7284876 | 7289824 | 1 |
| ShCDPK12 | Solhab03g289900 | 1698 | 565 | 63,959 | 9.39 | Chr3 | 67058383 | 67066282 | 1.2 |
| ShCDPK13 | Solhab03g297700 | 1662 | 553 | 62,883.1 | 6.84 | Chr3 | 67914192 | 67922776 | 1 |
| ShCDPK14 | Solhab04g006300 | 1746 | 581 | 64,561.9 | 5.56 | Chr4 | 747800 | 753899 | 1 |
| ShCDPK15 | Solhab04g100300 | 1527 | 508 | 57,230.7 | 4.88 | Chr4 | 20036567 | 20048558 | 1 |
| ShCDPK16 | Solhab04g295200 | 1590 | 529 | 59,692.7 | 5.17 | Chr4 | 69050936 | 69056009 | 1 |
| ShCDPK17 | Solhab05g000900 | 1512 | 503 | 56,429.8 | 4.77 | Chr5 | 201594 | 206779 | 1 |
| ShCDPK18 | Solhab06g061000 | 1611 | 536 | 61,044.4 | 6.02 | Chr6 | 4514265 | 4517937 | 1 |
| ShCDPK19 | Solhab06g118900 | 1506 | 501 | 56,383.9 | 5.78 | Chr6 | 9089226 | 9093227 | 1 |
| ShCDPK20 | Solhab07g020200 | 1566 | 521 | 57,818.7 | 7.05 | Chr7 | 1368173 | 1372887 | 1 |
| ShCDPK21 | Solhab08g229700 | 1551 | 516 | 57,751 | 6.02 | Chr8 | 65522651 | 65527398 | 1 |
| ShCDPK22 | Solhab09g271700 | 1590 | 529 | 59,620.4 | 6.39 | Chr9 | 85410171 | 85418586 | 1 |
| ShCDPK23 | Solhab10g042100 | 1863 | 620 | 69,779.1 | 5.57 | Chr10 | 3099146 | 3104072 | 1 |
| ShCDPK24 | Solhab10g043000 | 1908 | 635 | 69,576.7 | 5 | Chr10 | 3180473 | 3185006 | 1 |
| ShCDPK25 | Solhab10g069000 | 1515 | 504 | 57,051.6 | 6.06 | Chr10 | 5261738 | 5265333 | 1 |
| ShCDPK26 | Solhab10g082100 | 1635 | 544 | 60,419.3 | 5.44 | Chr10 | 6302023 | 6305715 | 1 |
| ShCDPK27 | Solhab10g101400 | 1674 | 557 | 62,198.8 | 5.59 | Chr10 | 8753228 | 8760508 | 1 |
| ShCDPK28 | Solhab11g059000 | 1602 | 533 | 59,654.8 | 6.41 | Chr11 | 5422926 | 5428709 | 1 |
| ShCDPK29 | Solhab11g068700 | 1590 | 529 | 59,533 | 5.97 | Chr11 | 6787516 | 6794660 | 1 |
| ShCDPK30 | Solhab11g157600 | 1518 | 505 | 56,832.4 | 5.4 | Chr11 | 56529976 | 56534049 | 1 |
| ShCDPK31 | Solhab11g234700 | 3204 | 1067 | 120,659.3 | 7.4 | Chr11 | 65019137 | 65028821 | 1 |
| ShCDPK32 | Solhab12g006400 | 1608 | 535 | 59,607.1 | 5.31 | Chr12 | 430517 | 433905 | 1 |
| ShCDPK33 | Solhab12g260400 | 1578 | 525 | 58,321 | 6.79 | Chr12 | 69390749 | 69393879 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Zhang, H.; Liang, S.; Chen, X.; Liu, J.; Zhang, Y.; Wang, A. Identification of CDPK Gene Family in Solanum habrochaites and Its Function Analysis under Stress. Int. J. Mol. Sci. 2022, 23, 4227. https://doi.org/10.3390/ijms23084227
Li Y, Zhang H, Liang S, Chen X, Liu J, Zhang Y, Wang A. Identification of CDPK Gene Family in Solanum habrochaites and Its Function Analysis under Stress. International Journal of Molecular Sciences. 2022; 23(8):4227. https://doi.org/10.3390/ijms23084227
Chicago/Turabian StyleLi, Yingying, Haixin Zhang, Sibo Liang, Xiuling Chen, Jiayin Liu, Yao Zhang, and Aoxue Wang. 2022. "Identification of CDPK Gene Family in Solanum habrochaites and Its Function Analysis under Stress" International Journal of Molecular Sciences 23, no. 8: 4227. https://doi.org/10.3390/ijms23084227
APA StyleLi, Y., Zhang, H., Liang, S., Chen, X., Liu, J., Zhang, Y., & Wang, A. (2022). Identification of CDPK Gene Family in Solanum habrochaites and Its Function Analysis under Stress. International Journal of Molecular Sciences, 23(8), 4227. https://doi.org/10.3390/ijms23084227






