Structural Requirements for the Binding of a Peptide to Prohibitins on the Cell Surface of Monocytes/Macrophages
Abstract
:1. Introduction
2. Results
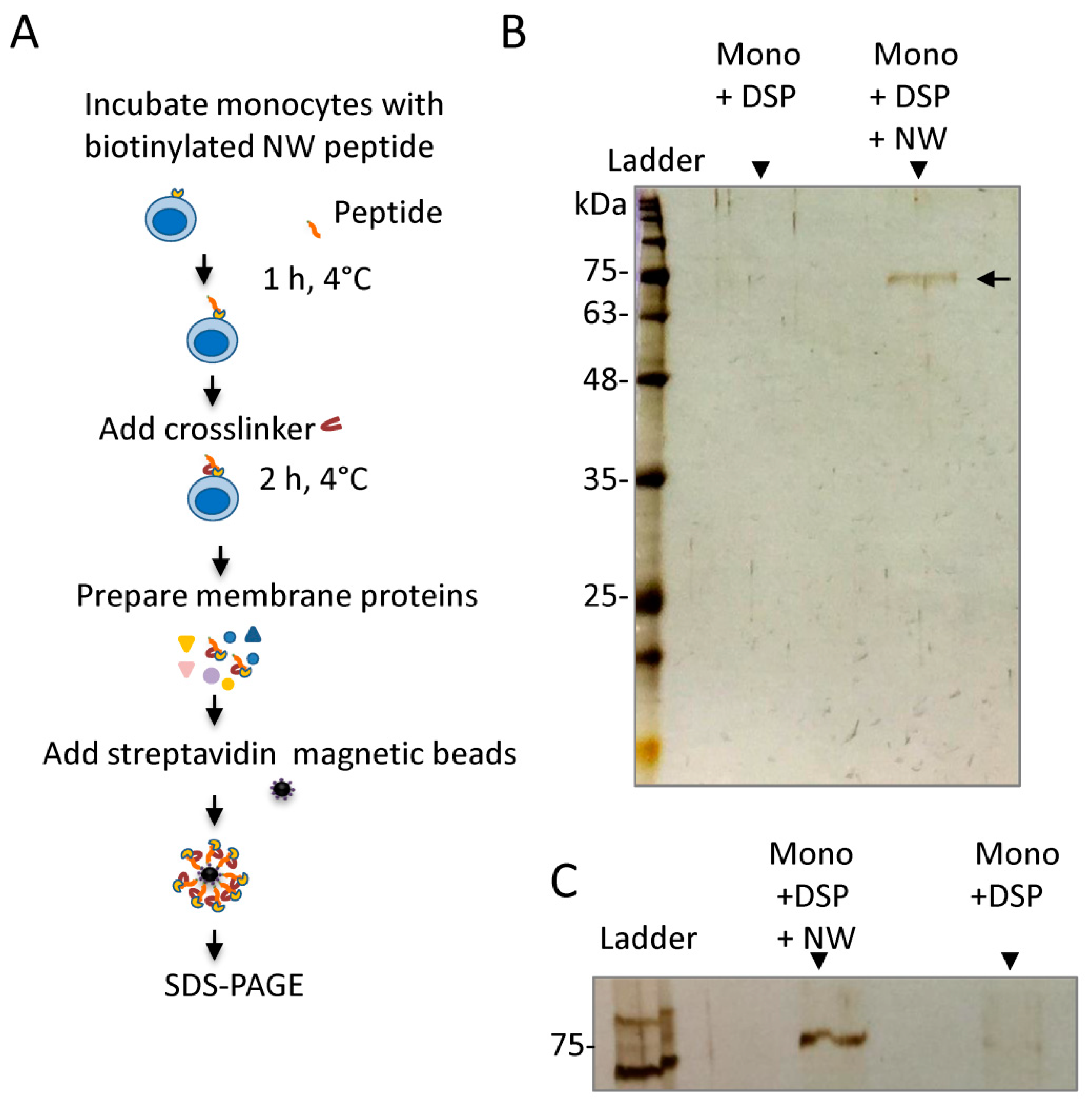
2.1. Cross-Linking Experiments Identified Modified Albumin as an Interacting Protein
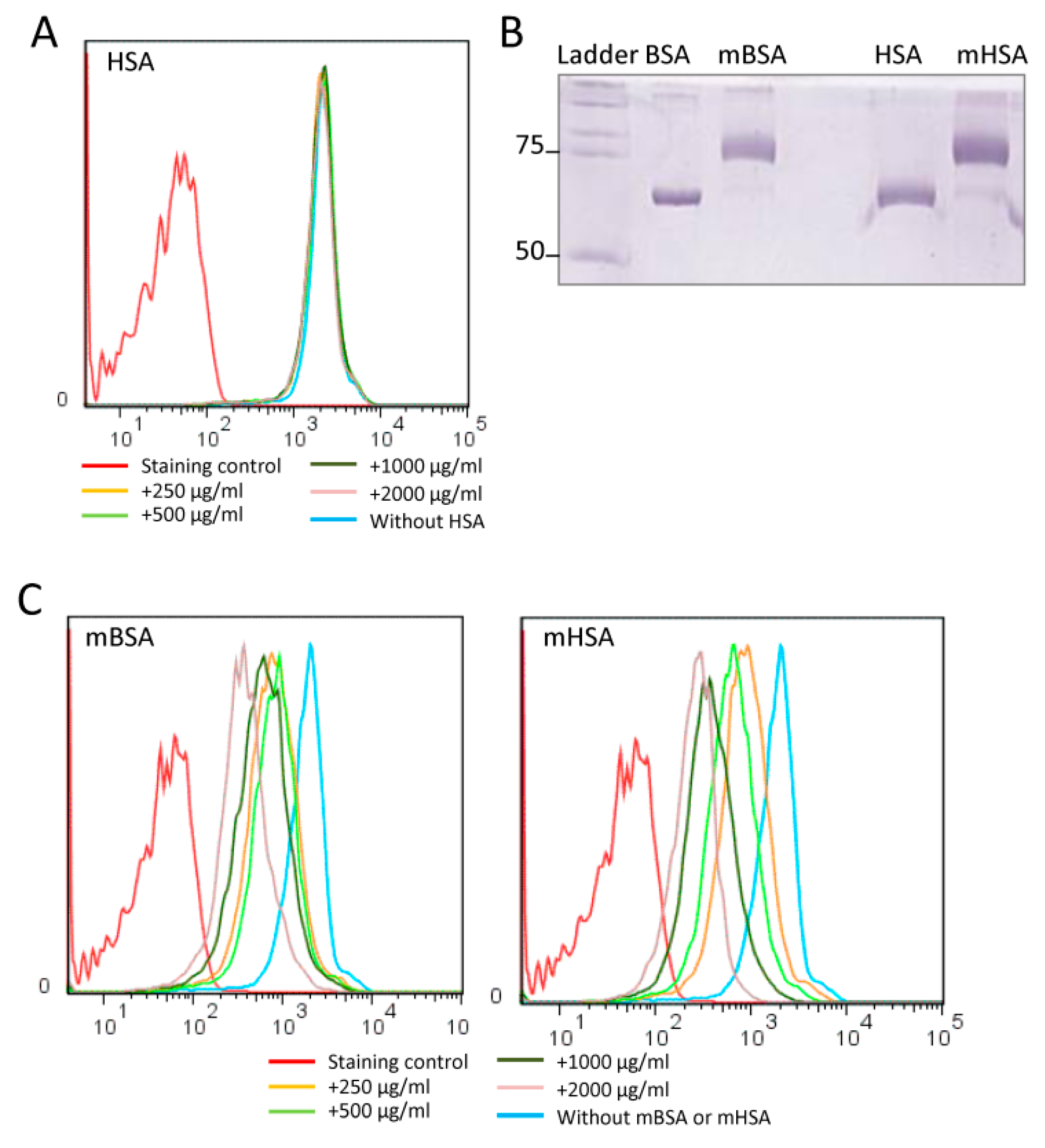
2.2. Maleic-Anhydride-Modified Albumin Competes with the NW Peptide Binding to Monocytes
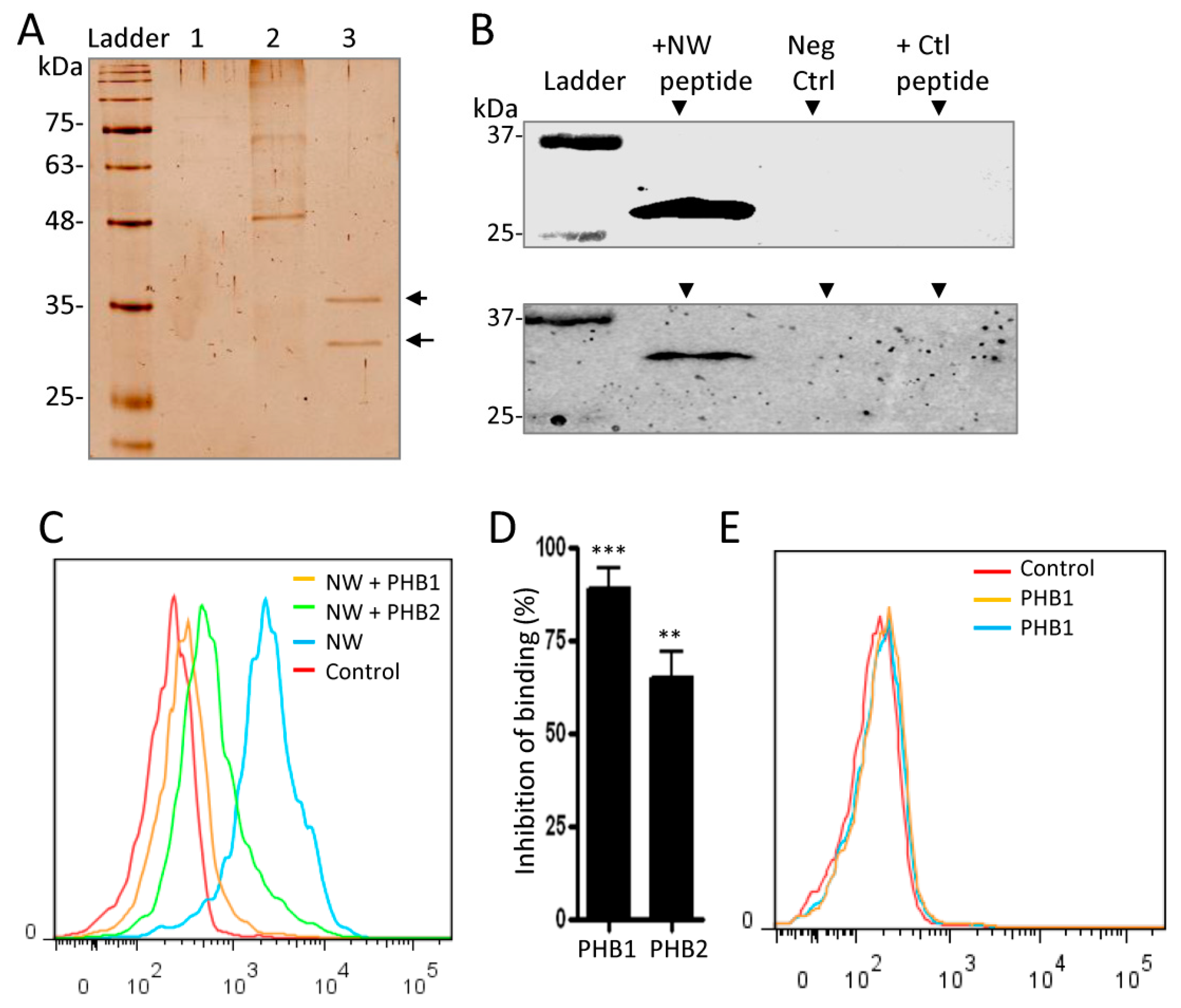
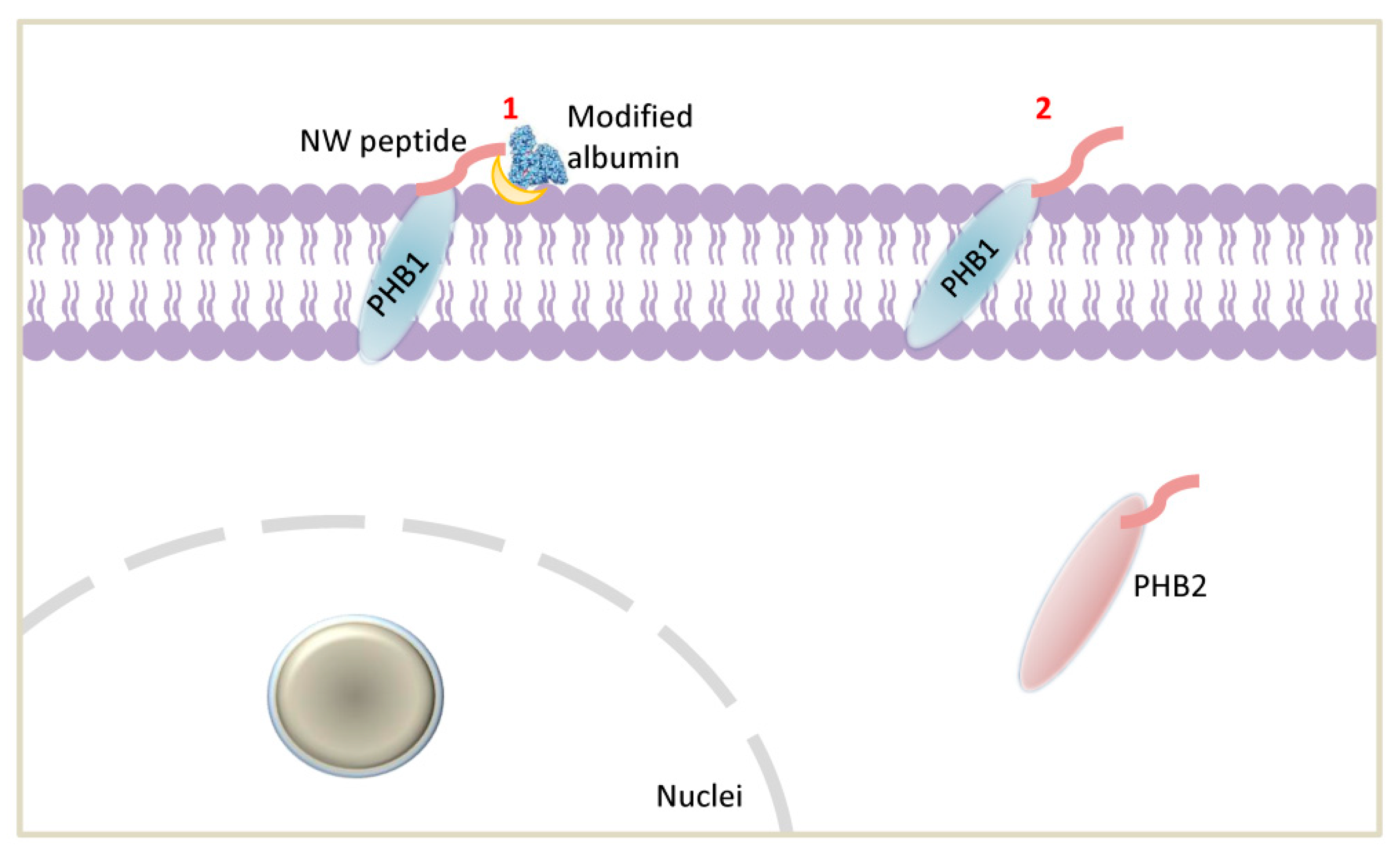
2.3. Cell Surface Prohibitins May Function as a Receptor for the NW Peptide
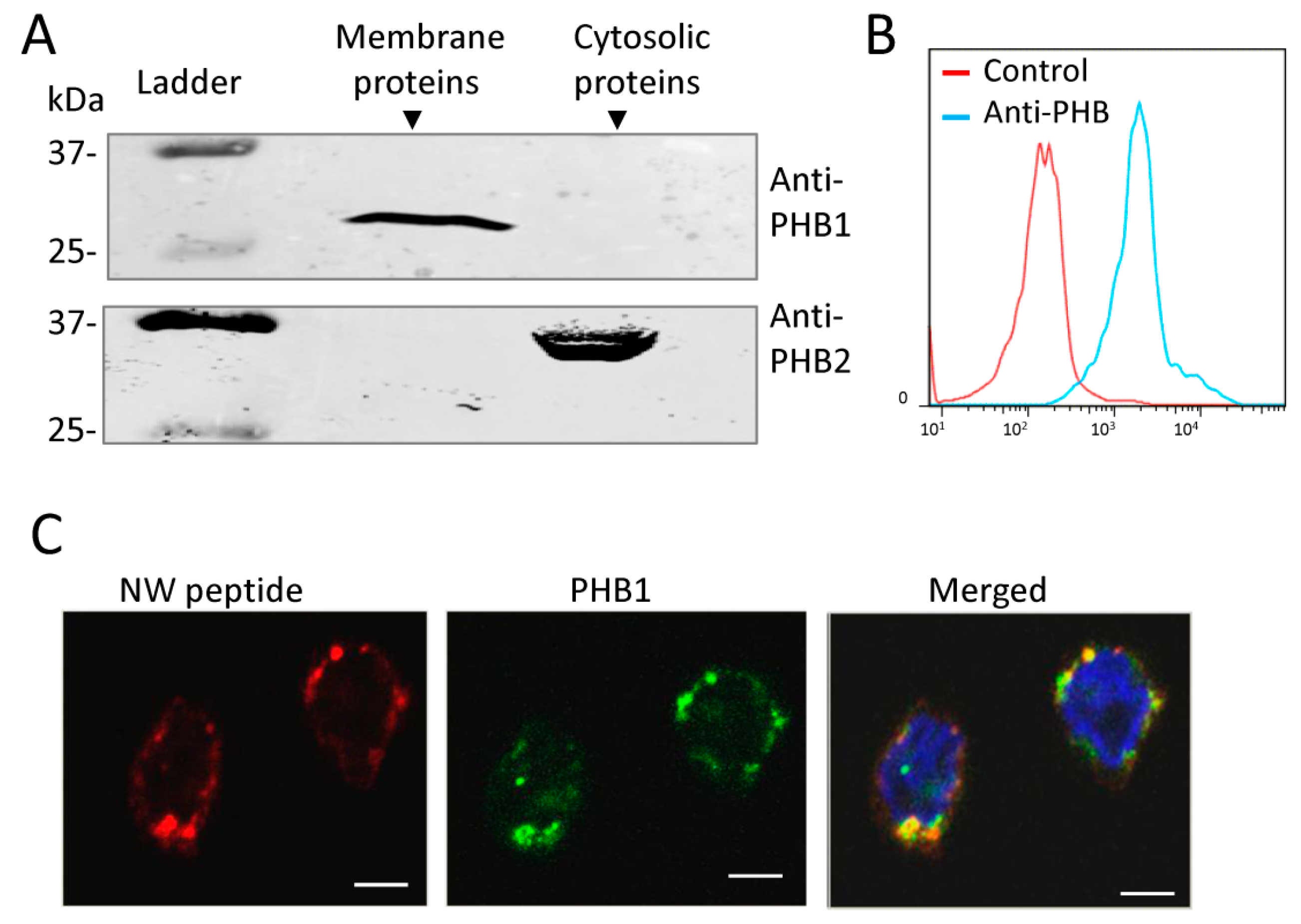
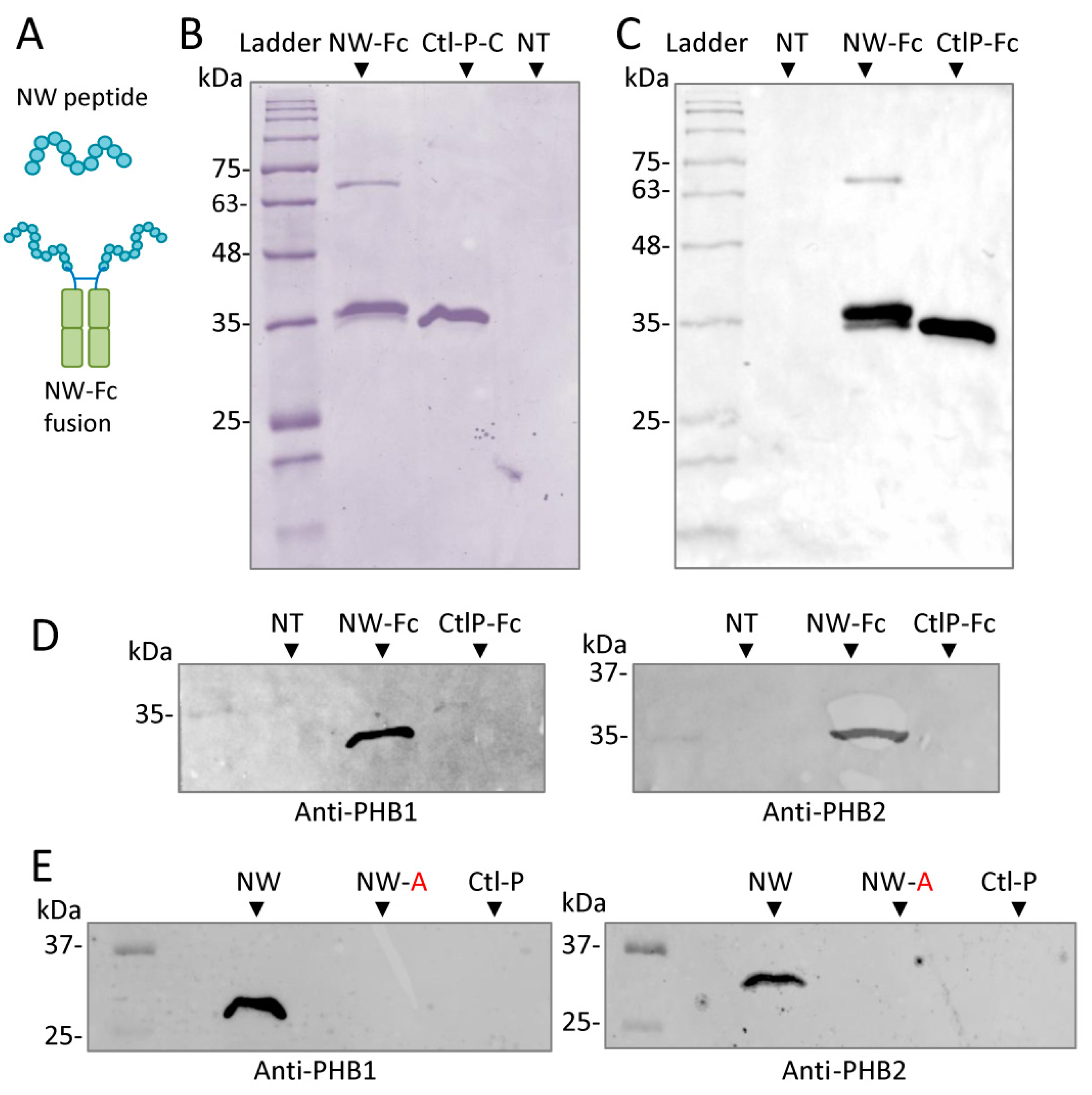
2.4. Prohibitin 1 Is a Potential Cell Surface Receptor for the NW Peptide on Monocytes
2.5. NW Peptide Interacts with Intracellular Prohibitins
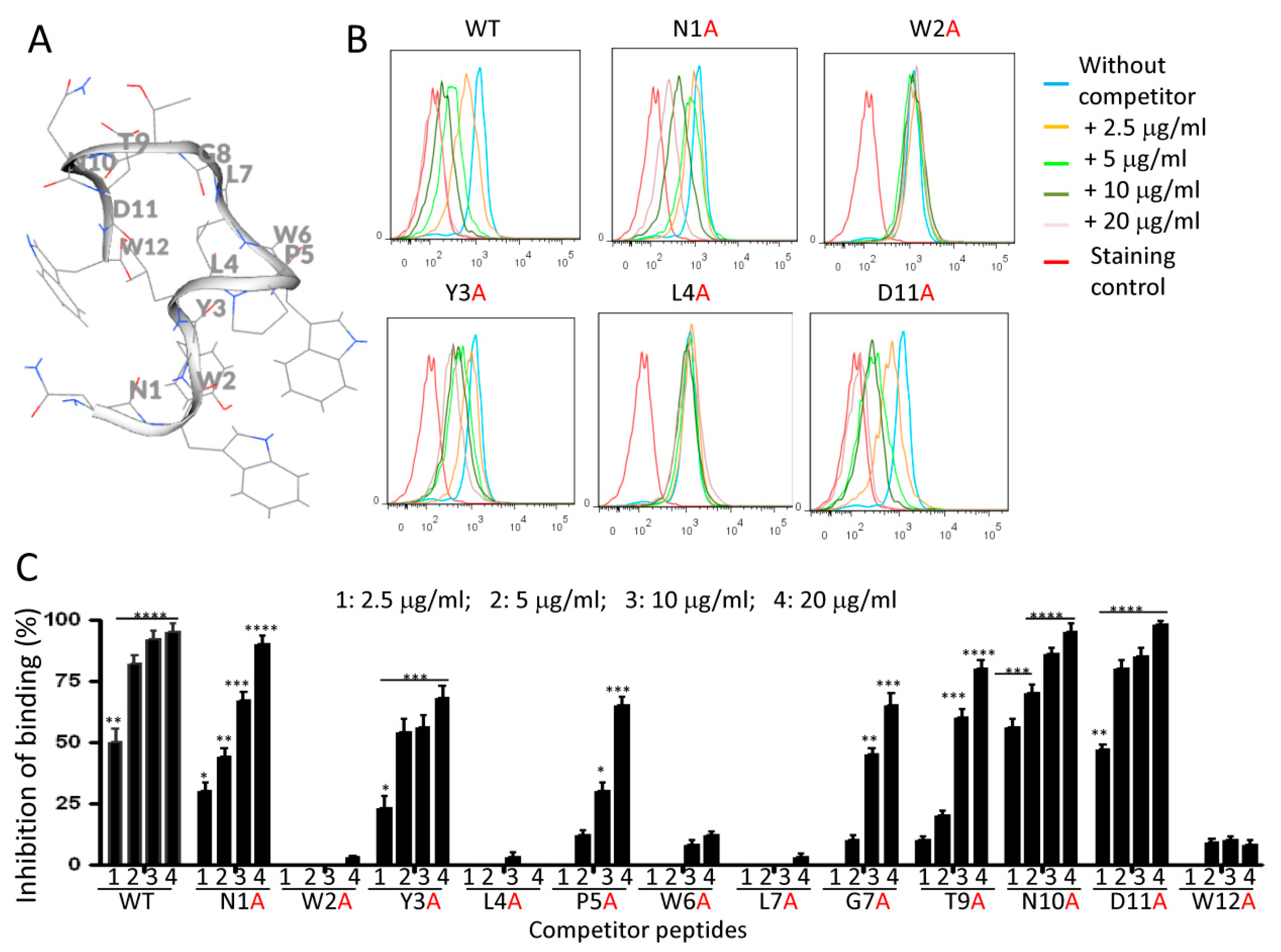
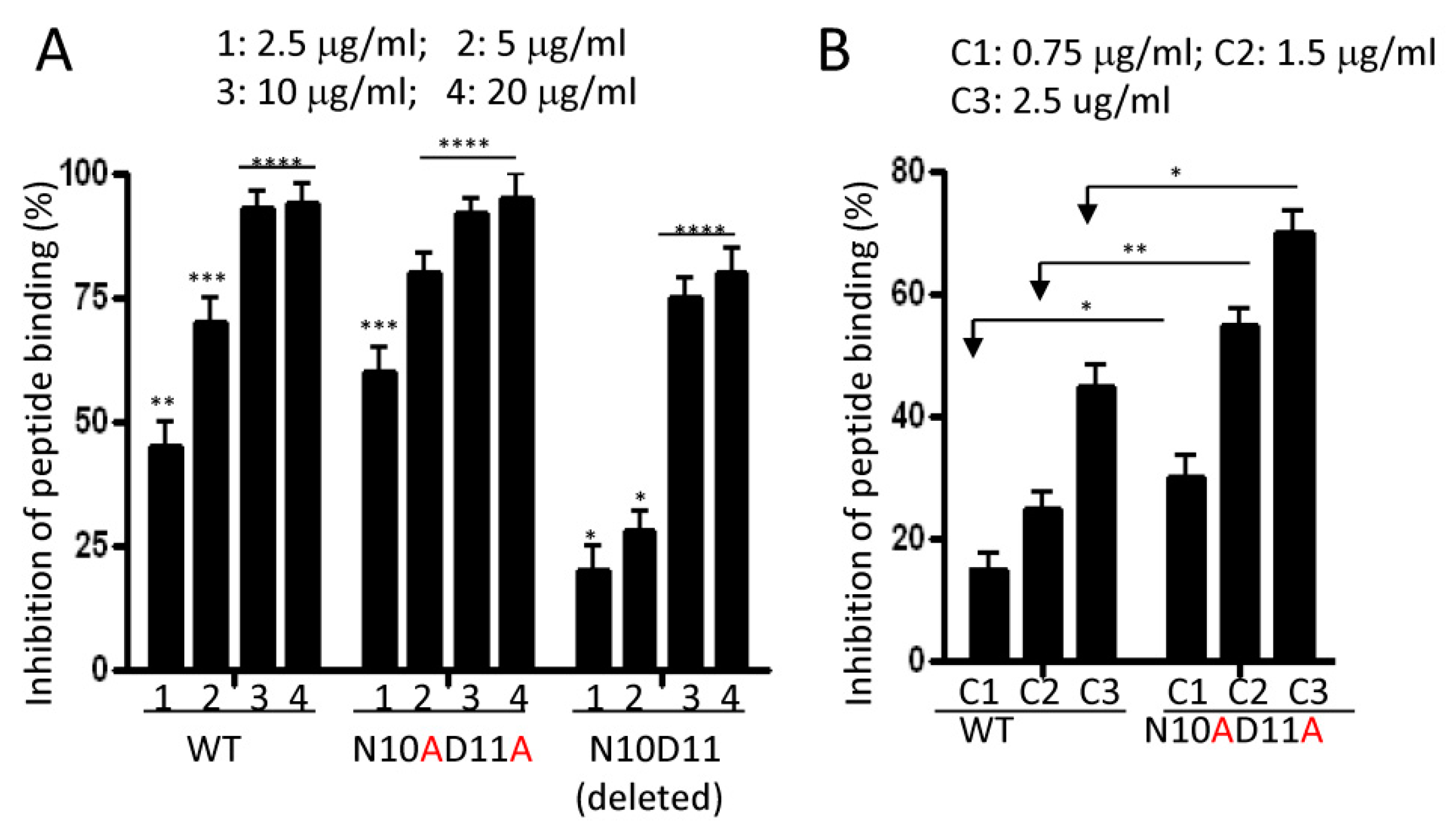
2.6. Effects of Alanine Mutations on Peptide Binding to Monocytes
3. Discussion
4. Materials and Methods
4.1. Monocyte Purification
4.2. Cross-Linking and Pull-Down Assays
4.3. Analysis of the Captured Proteins by Western Blotting
4.4. Chemical Modification of Albumin with Anhydride
4.5. Analysis of Albumin Effects on NW Peptide Binding to Monocytes
4.6. Analysis of the Prohibition Effects on NW Peptide Binding to Monocytes
4.7. Confocal Microscopy Analysis
4.8. Cloning and Expression of Peptide-Fc Fusions
4.9. Pull-Down Assays Using Peptide–Fc Fusions
4.10. Analine-Scanning Mutagenesis
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| A | Alanine |
| BSA | Bovine serum albumin |
| CtlP–Fc | Control-peptide–Fc fusion |
| DSP | Dithiobis(succinimidyl propionate) |
| D | Aspartic acid |
| F | Phenylalanine |
| HPLC | High-performance liquid chromatography |
| HSA | Human serum albumin |
| L | Leucine |
| mBSA | Maleic-anhydride-modified bovine serum albumin |
| mHSA | Maleic-anhydride-modified human serum albumin |
| NT | Non-transfected |
| PBMCs | Peripheral mononuclear cells |
| PHB | Prohibitin |
| SDS-PAGE | Sodium dodecyl-sulfate polyacrylamide gel electrophoresis |
| TAMs | Tumor-associated macrophages |
| TME | Tumor microenvironment |
| VDAC1 | Voltage-dependent anion selective channel 1 |
| V | Valine |
| W | Tryptophan |
| WT | Wild type |
| Y | Tyrosine |
References
- Pan, C.; Liu, H.; Robins, E.; Song, W.; Liu, D.; Li, Z.; Zheng, L. Next-generation immuno-oncology agents: Current momentum shifts in cancer immunotherapy. J. Hematol. Oncol. 2020, 13, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, P.; Allison, J.P. The future of immune checkpoint therapy. Science 2015, 348, 56–61. [Google Scholar] [CrossRef]
- Beatty, G.L.; Gladney, W.L. Immune escape mechanisms as a guide for cancer immunotherapy. Clin Cancer Res. 2015, 21, 687–692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rizvi, N.A.; Hellmann, M.D.; Snyder, A.; Kvistborg, P.; Makarov, V.; Havel, J.J.; Lee, W.; Yuan, J.; Wong, P.; Ho, T.S.; et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015, 348, 124–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franklin, R.A.; Liao, W.; Sarkar, A.; Kim, M.V.; Bivona, M.R.; Liu, K.; Pamer, E.G.; Li, M.O. The cellular and molecular origin of tumor-associated macrophages. Science 2014, 344, 921–925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malfitano, A.M.; Pisanti, S.; Napolitano, F.; Di Somma, S.; Martinelli, R.; Portella, G. Tumor-Associated Macrophage Status in Cancer Treatment. Cancers 2020, 12, 1987. [Google Scholar] [CrossRef] [PubMed]
- Qian, B.Z.; Pollard, J.W. Macrophage diversity enhances tumor progression and metastasis. Cell 2010, 141, 39–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murray, P.J.; Allen, J.; Biswas, S.K.; Fisher, E.; Gilroy, D.; Goerdt, S.; Gordon, S.; A Hamilton, J.; Ivashkiv, L.B.; Lawrence, T.; et al. Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity 2014, 41, 14–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poh, A.R.; Ernst, M. Targeting Macrophages in Cancer: From Bench to Bedside. Front. Oncol. 2018, 8, 49. [Google Scholar] [CrossRef] [Green Version]
- Pollard, J.W. Tumour-educated macrophages promote tumour progression and metastasis. Nat. Rev. Cancer 2004, 4, 71–78. [Google Scholar] [CrossRef]
- Lahmar, Q.; Keirsse, J.; Laoui, D.; Movahedi, K.; Van Overmeire, E.; Van Ginderachter, J.A. Tissue-resident versus monocyte-derived macrophages in the tumor microenvironment. Biochim. Biophys. Acta 2016, 1865, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Anfray, C.; Ummarino, A.; Andón, F.T.; Allavena, P. Current Strategies to Target Tumor-Associated-Macrophages to Improve Anti-Tumor Immune Responses. Cells 2019, 9, 46. [Google Scholar] [CrossRef] [Green Version]
- Arlauckas, S.P.; Garris, C.S.; Kohler, R.H.; Kitaoka, M.; Cuccarese, M.F.; Yang, K.S.; Miller, M.A.; Carlson, J.C.; Freeman, G.J.; Anthony, R.M.; et al. In vivo imaging reveals a tumor-associated macrophage-mediated resistance pathway in anti-PD-1 therapy. Sci. Transl. Med. 2017, 9, 9. [Google Scholar] [CrossRef] [Green Version]
- Gordon, S.R.; Maute, R.L.; Dulken, B.W.; Hutter, G.; George, B.M.; McCracken, M.N.; Gupta, R.; Tsai, J.; Sinha, R.; Corey, D.; et al. PD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunity. Nature 2017, 545, 495–499. [Google Scholar] [CrossRef] [PubMed]
- Pinto, A.T.; Pinto, M.L.; Cardoso, A.P.; Monteiro, C.; Maia, A.F.; Castro, P.; Figueira, R.; Monteiro, A.; Marques, M.; Mareel, M.; et al. Ionizing radiation modulates human macrophages towards a pro-inflammatory phenotype preserving their pro-invasive and pro-angiogenic capacities. Sci. Rep. 2016, 6, 18765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scodeller, P.; Asciutto, E.K. Targeting Tumors Using Peptides. Molecules 2020, 25, 808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dmitrieva, M.D.; Voitova, A.A.; Dymova, M.A.; Richter, V.A.; Kuligina, E.V. Tumor-Targeting Peptides Search Strategy for the Delivery of Therapeutic and Diagnostic Molecules to Tumor Cells. Int. J. Mol. Sci. 2020, 22, 314. [Google Scholar] [CrossRef] [PubMed]
- Regberg, J.; Srimanee, A.; Langel, U. Applications of cell-penetrating peptides for tumor targeting and future cancer therapies. Pharmaceuticals 2012, 5, 991–1007. [Google Scholar] [CrossRef] [Green Version]
- Lingasamy, P.; Tobi, A.; Kurm, K.; Kopanchuk, S.; Sudakov, A.; Salumäe, M.; Rätsep, T.; Asser, T.; Bjerkvig, R.; Teesalu, T. Tumor-penetrating peptide for systemic targeting of Tenascin-C. Sci. Rep. 2020, 10, 5809. [Google Scholar] [CrossRef] [Green Version]
- Sioud, M.; Skorstad, G.; Mobergslien, A.; Sæbøe-Larssen, S. A novel peptide carrier for efficient targeting of antigens and nucleic acids to dendritic cells. FASEB J. 2013, 27, 3272–3283. [Google Scholar] [CrossRef]
- Sioud, M. Phage Display Libraries: From Binders to Targeted Drug Delivery and Human Therapeutics. Mol. Biotechnol. 2019, 61, 286–303. [Google Scholar] [CrossRef] [PubMed]
- Sioud, M.; Pettersen, S.; Ailte, I.; Fløisand, Y. Targeted Killing of Monocytes/Macrophages and Myeloid Leukemia Cells with Pro-Apoptotic Peptides. Cancers 2019, 11, 1088. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.H.; Kuo, Y.H.; Hong, R.L.; Wu, H.C. α-Enolase-binding peptide enhances drug delivery efficiency and therapeutic efficacy against colorectal cancer. Sci. Transl. Med. 2015, 7, 290ra291. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.X.; Kim, H.Y. Effective identification of Akt interacting proteins by two-step chemical crosslinking, co-immunoprecipitation and mass spectrometry. PLoS ONE 2013, 8, e61430. [Google Scholar] [CrossRef] [Green Version]
- Mishra, S.; Murphy, L.C.; Nyomba, B.L.; Murphy, L.J. Prohibitin: A potential target for new therapeutics. Trends Mol. Med. 2005, 11, 192–197. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, B.; He, Q.Y. Significance of prohibitin domain family in tumorigenesis and its implication in cancer diagnosis and treatment. Cell Death Dis. 2018, 9, 580. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Moulik, S.; Murphy, L.J. Prohibitin binds to C3 and enhances complement activation. Mol. Immunol. 2007, 44, 1897–1902. [Google Scholar] [CrossRef]
- Cunningham, B.C.; Wells, J.A. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science 1989, 244, 1081–1085. [Google Scholar] [CrossRef]
- Marcelino, A.M.; Gierasch, L.M. Roles of beta-turns in protein folding: From peptide models to protein engineering. Biopolymers 2008, 89, 380–391. [Google Scholar] [CrossRef] [Green Version]
- Ayo, A.; Laakkonen, P. Peptide-Based Strategies for Targeted Tumor Treatment and Imaging. Pharmaceutics 2021, 13, 481. [Google Scholar] [CrossRef]
- Thurber, G.M.; Schmidt, M.M.; Wittrup, K.D. Antibody tumor penetration: Transport opposed by systemic and antigen-mediated clearance. Adv. Drug Deliv. Rev. 2008, 60, 1421–1434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saw, P.E.; Song, E.W. Phage display screening of therapeutic peptide for cancer targeting and therapy. Protein Cell 2019, 10, 787–807. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Santino, F.; Giacomini, D.; Gentilucci, L. Integrin-Targeting Peptides for the Design of Functional Cell-Responsive Biomaterials. Biomedicines 2020, 8, 307. [Google Scholar] [CrossRef] [PubMed]
- Andrieu, J.; Re, F.; Russo, L.; Nicotra, F. Phage-displayed peptides targeting specific tissues and organs. J. Drug Target. 2019, 27, 555–565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohamed, R.; Degac, J.; Helms, V. Composition of Overlapping Protein-Protein and Protein-Ligand Interfaces. PLoS ONE 2015, 10, e0140965. [Google Scholar] [CrossRef] [PubMed]
- Yurugi, H.; Tanida, S.; Ishida, A.; Akita, K.; Toda, M.; Inoue, M.; Nakada, H. Expression of prohibitins on the surface of activated T cells. Biochem. Biophys. Res. Commun. 2012, 420, 275–280. [Google Scholar] [CrossRef]
- Terashima, M.; Kim, K.M.; Adachi, T.; Nielsen, P.J.; Reth, M.; Köhler, G.; Lamers, M.C. The IgM antigen receptor of B lymphocytes is associated with prohibitin and a prohibitin-related protein. EMBO J. 1994, 13, 3782–3792. [Google Scholar] [CrossRef]
- Kolonin, M.G.; Saha, P.K.; Chan, L.; Pasqualini, R.; Arap, W. Reversal of obesity by targeted ablation of adipose tissue. Nat. Med. 2004, 10, 625–632. [Google Scholar] [CrossRef]
- Sharma, A.; Qadri, A. Vi polysaccharide of Salmonella typhi targets the prohibitin family of molecules in intestinal epithelial cells and suppresses early inflammatory responses. Proc. Natl. Acad. Sci. USA 2004, 101, 17492–17497. [Google Scholar] [CrossRef] [Green Version]
- Garg, R.; Qadri, A. Hemoglobin transforms anti-inflammatory Salmonella typhi virulence polysaccharide into a TLR-2 agonist. J. Immunol. 2010, 184, 5980–5987. [Google Scholar] [CrossRef] [Green Version]
- Wintachai, P.; Wikan, N.; Kuadkitkan, A.; Jaimipuk, T.; Ubol, S.; Pulmanausahakul, R.; Auewarakul, P.; Kasinrerk, W.; Weng, W.-Y.; Panyasrivanit, M.; et al. Identification of prohibitin as a Chikungunya virus receptor protein. J. Med. Virol. 2012, 84, 1757–1770. [Google Scholar] [CrossRef] [PubMed]
- Freed, E.O. HIV-1 and the host cell: An intimate association. Trends Microbiol. 2004, 12, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, W.; Brown, L.E.; Qiu, C.; Lajkiewicz, N.; Zhao, T.; Zhou, J.; Porco, J.A., Jr.; Wang, T.T. A Novel Class of Small Molecule Compounds that Inhibit Hepatitis C Virus Infection by Targeting the Prohibitin-CRaf Pathway. EBioMedicine 2015, 2, 1600–1606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farooq, A.V.; Valyi-Nagy, T.; Shukla, D. Mediators and mechanisms of herpes simplex virus entry into ocular cells. Curr. Eye Res. 2010, 35, 445–450. [Google Scholar] [CrossRef]
- Nemerow, G.; Flint, J. Lessons learned from adenovirus (1970-2019). FEBS Lett. 2019, 593, 3395–3418. [Google Scholar] [CrossRef] [Green Version]
- Fukuhara, H.; Mwaba, M.H.; Maenaka, K. Structural characteristics of measles virus entry. Curr. Opin. Virol. 2020, 41, 52–58. [Google Scholar] [CrossRef]
- Sharma, A.; Vasanthapuram, R.; Venkataswamy, M.; Desai, A. Prohibitin 1/2 mediates Dengue-3 entry into human neuroblastoma (SH-SY5Y) and microglia (CHME-3) cells. J. Biomed. Sci. 2020, 27, 55. [Google Scholar] [CrossRef] [Green Version]
- Sakamoto, S.; Hamachi, I. Recent Progress in Chemical Modification of Proteins. Anal. Sci. 2019, 35, 5–27. [Google Scholar] [CrossRef] [Green Version]
- Sun, Z.; Wang, Q.; Jia, R.; Xia, S.; Li, Y.; Liu, Q.; Xu, W.; Xu, J.; Du, L.; Lu, L.; et al. Intranasal administration of maleic anhydride-modified human serum albumin for pre-exposure prophylaxis of respiratory syncytial virus infection. Viruses 2015, 7, 798–819. [Google Scholar] [CrossRef]
- Sioud, M.; Westby, P.; Olsen, J.K.; Mobergslien, A. Generation of new peptide-Fc fusion proteins that mediate antibody-dependent cellular cytotoxicity against different types of cancer cells. Mol. Ther.-Methods Clin. Dev. 2015, 2, 15043. [Google Scholar] [CrossRef]
- GeneCust Europe. Available online: http://www.genecust.com (accessed on 2 April 2022).


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Q.; Olberg, A.; Sioud, M. Structural Requirements for the Binding of a Peptide to Prohibitins on the Cell Surface of Monocytes/Macrophages. Int. J. Mol. Sci. 2022, 23, 4282. https://doi.org/10.3390/ijms23084282
Zhang Q, Olberg A, Sioud M. Structural Requirements for the Binding of a Peptide to Prohibitins on the Cell Surface of Monocytes/Macrophages. International Journal of Molecular Sciences. 2022; 23(8):4282. https://doi.org/10.3390/ijms23084282
Chicago/Turabian StyleZhang, Qindong, Anniken Olberg, and Mouldy Sioud. 2022. "Structural Requirements for the Binding of a Peptide to Prohibitins on the Cell Surface of Monocytes/Macrophages" International Journal of Molecular Sciences 23, no. 8: 4282. https://doi.org/10.3390/ijms23084282
APA StyleZhang, Q., Olberg, A., & Sioud, M. (2022). Structural Requirements for the Binding of a Peptide to Prohibitins on the Cell Surface of Monocytes/Macrophages. International Journal of Molecular Sciences, 23(8), 4282. https://doi.org/10.3390/ijms23084282






