Synaptic Plasticity Dysfunctions in the Pathophysiology of 22q11 Deletion Syndrome: Is There a Role for Astrocytes?
Abstract
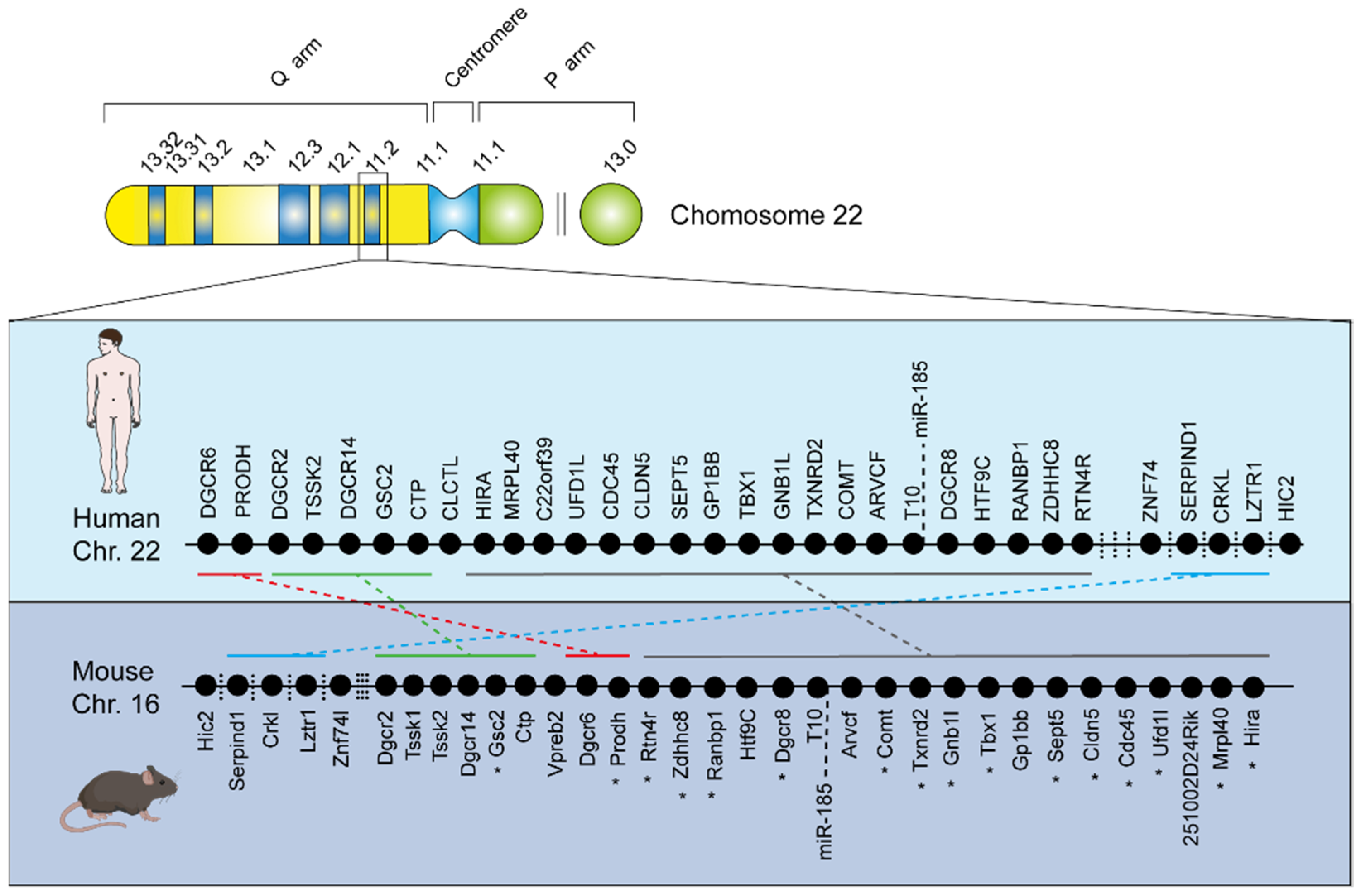
:1. Introduction
2. Synaptic Plasticity and the Pathophysiology of 22q11DS
3. Role of Mitochondrial Genes in the Dysfunctions of Synaptic Plasticity Associated with 22q11 DS
4. Astrocytes Can Modulate Synaptic Plasticity: Do They Play a Role in the Pathophysiology of 22q11DS?
5. Conclusions and Future Directions
6. Strengths and limitations and Search Strategy
7. Search Strategy and Selection Criteria
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gur, R.E.; Bassett, A.S.; McDonald-Mcginn, D.M.; Bearden, C.E.; Chow, E.; Emanuel, B.S.; Owen, M.; Swillen, A.; Van den Bree, M.; Vermeesch, J. A neurogenetic model for the study of schizophrenia spectrum disorders: The International 22q11.2 Deletion Syndrome Brain Behavior Consortium. Mol. Psychiatry 2017, 22, 1664–1672. [Google Scholar] [CrossRef] [PubMed]
- Maynard, T.M.; Haskell, G.T.; Lieberman, J.A.; Lamantia, A.-S. 22q11 DS: Genomic mechanisms and gene function in DiGeorge/velocardiofacial syndrome. Int. J. Dev. Neurosci. 2002, 20, 407–419. [Google Scholar] [CrossRef]
- McDonald-McGinn, D.M.; Sullivan, K.E.; Marino, B.; Philip, N.; Swillen, A.; Vorstman, J.A.S.; Zackai, E.H.; Emanuel, B.S.; Vermeesch, J.; Morrow, B.E. 22Q11.2 Deletion Syndrome. Nat. Rev. Dis. Primers 2015, 1, 1–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karayiorgou, M.; Simon, T.J.; Gogos, J.A. 22q11.2 microdeletions: Linking DNA structural variation to brain dysfunction and schizophrenia. Nat. Rev. Neurosci. 2010, 11, 402–416. [Google Scholar] [CrossRef]
- Carlson, C.; Sirotkin, H.; Pandita, R.; Goldberg, R.; McKie, J.; Wadey, R.; Patanjali, S.R.; Weissman, S.M.; Anyane-Yeboa, K.; Warburton, D. Molecular definition of 22q11 deletions in 151 velo-cardio-facial syndrome patients. Am. J. Hum. Genet. 1997, 61, 620–629. [Google Scholar] [CrossRef] [Green Version]
- Drew, L.J.; Crabtree, G.W.; Markx, S.; Stark, K.L.; Chaverneff, F.; Xu, B.; Mukai, J.; Fenelon, F.; Hsu, P.K.; Gogos, J.A.; et al. The 22q11.2 microdeletion: Fifteen years of insights into the genetic and neural complexity of psychiatric disorders. Int. J. Dev. Neurosci. 2011, 29, 259–281. [Google Scholar] [CrossRef] [Green Version]
- Karayiorgou, M.; Morris, M.A.; Morrow, B.; Shprintzen, R.J.; Goldberg, R.; Borrow, J.; Gos, A.; Nestadt, G.; Wolyniec, P.S.; Lasseter, V.K. Schizophrenia susceptibility associated with interstitial deletions of chromosome 22q11. Proc. Natl. Acad. Sci. USA 1995, 92, 7612–7616. [Google Scholar] [CrossRef] [Green Version]
- Murphy, K.C. Schizophrenia and velo-cardio-facial syndrome. Lancet 2002, 359, 426–430. [Google Scholar] [CrossRef]
- Bassett, A.S.; Chow, E.W.C. Chromosomal Abnormalities and Schizophrenia and a senior psychiatric genetics researcher. Am. J. Med. Genet. 2000, 97, 45–51. [Google Scholar] [CrossRef]
- Schneider, M.; Debbané, M.; Bassett, A.S.; Chow, E.W.C.; Fung, W.L.A.; Van Den Bree, M.B.M.; Owen, M.; Murphy, K.C.; Niarchou, M.; Kates, W.R.; et al. Psychiatric disorders from childhood to adulthood in 22q11.2 deletion syndrome: Results from the international consortium on brain and behavior in 22q11.2 deletion syndrome. Am. J. Psychiatry 2014, 171, 627–639. [Google Scholar] [CrossRef]
- Jolin, E.M.; Weller, R.A.; Jessani, N.R.; Zackai, E.H.; McDonald-McGinn, D.M.; Weller, E.B. Affective disorders and other psychiatric diagnoses in children and adolescents with 22q11.2 Deletion Syndrome. J. Affect. Disord. 2009, 119, 177–180. [Google Scholar] [CrossRef] [PubMed]
- Vorstman, J.A.S.; Morcus, M.E.J.; Duijff, S.N.; Klaassen, P.W.J.; Heineman-De Boer, J.A.; Beemer, F.A.; Swaab, H.; Kahn, R.S.; Van Engeland, H. The 22q11.2 deletion in children: High rate of autistic disorders and early onset of psychotic symptoms. J. Am. Acad. Child Adolesc. Psychiatry 2006, 45, 1104–1113. [Google Scholar] [CrossRef] [PubMed]
- Gothelf, D.; Schneider, M.; Green, T.; Debbané, M.; Frisch, A.; Glaser, B.; Zilkha, H.; Schaer, M.; Weizman, A.; Eliez, S. Risk factors and the evolution of psychosis in 22q11.2 deletion syndrome: A longitudinal 2-site study. J. Am. Acad. Child Adolesc. Psychiatry 2013, 52, 112–1203. [Google Scholar] [CrossRef] [PubMed]
- McDonald-McGinn, D.M.; Sullivan, K.E. Chromosome 22q11.2 deletion syndrome (DiGeorge syndrome/velocardiofacial syndrome). Medicine 2011, 90, 1–18. [Google Scholar] [CrossRef]
- Murphy, K.C.; Jones, L.A.; Owen, M.J. High rates of schizophrenia in adults with velo-cardio-facial syndrome. Arch. Gen. Psychiatry 1999, 56, 940–945. [Google Scholar] [CrossRef] [Green Version]
- Debbané, M.; Glaser, B.; David, M.K.; Feinstein, C.; Eliez, S. Psychotic symptoms in children and adolescents with 22q11.2 deletion syndrome: Neuropsychological and behavioral implications. Schizophr. Res. 2006, 84, 187–193. [Google Scholar] [CrossRef]
- Hooper, S.R.; Curtiss, K.; Schoch, K.; Keshavan, M.S.; Allen, A.; Shashi, V. A Longitudinal Examination of the Psychoeducational, Neurocognitive, and Psychiatric Functioning in Children with 22q11.2 Deletion Syndrome. Res. Dev. Disabil. 2013, 34, 1758. [Google Scholar] [CrossRef] [Green Version]
- Simon, T.; Takarae, Y.; DeBoer, T.; McDonald-McGinn, D.; Zackai, E.; Ross, J. Overlapping numerical cognition impairments in children with chromosome 22q11.2 deletion or Turner syndromes. Neuropsychologia 2008, 46, 82–94. [Google Scholar] [CrossRef] [Green Version]
- Simon, T.J.; Bish, J.P.; Bearden, C.E.; Ding, L.; Ferrante, S.; Nguyen, V.; Gee, J.G.; McDonald-McGinn, D.M.; Zackai, E.H.; Emanuel, S.E. A multilevel analysis of cognitive dysfunction and psychopathology associated with chromosome 22q11.2 deletion syndrome in children. Dev. Psychopathol. 2005, 17, 753–784. [Google Scholar] [CrossRef] [Green Version]
- Amelsvoort, T.; Van Henry, J.; Morris, R.; Owen, M.; Linszen, D.; Murphy, K.; Murphy, D. Cognitive deficits associated with schizophrenia in velo-cardio-facial syndrome. Schizophr. Res. 2004, 70, 223–232. [Google Scholar] [CrossRef]
- Weinberger, R.; Yi, J.; Calkins, M.; Guri, Y.; McDonald-McGinn, D.M.; Emanuel, B.S.; Zackai, E.H.; Ruparel, K.; Carmel, M.; Michaelovsky, E. Neurocognitive profile in psychotic versus nonpsychotic individuals with 22q11.2 deletion syndrome. Eur. Neuropsychopharmacol. J. Eur. Coll. Neuropsychopharmacol. 2016, 26, 1610–1618. [Google Scholar] [CrossRef] [PubMed]
- Fiksinski, A.M.; Breetvelt, E.J.; Lee, Y.J.; Boot, E.; Butcher, N.; Palmer, L.; Chow, E.W.C.; Kahn, R.S.; Vorstman, J.A.S.; Bassett, A.S. Neurocognition and adaptive functioning in a genetic high risk model of schizophrenia. Psychol. Med. 2019, 49, 1047–1054. [Google Scholar] [CrossRef] [PubMed]
- Vorstman, J.A.S.; Breetvelt, E.J.; Duijff, S.N.; Eliez, S.; Schneider, M.; Jalbrzikowski, M.; Armando, M.; Vicari, S.; Shashi, V.; Hooper, S.R. Cognitive decline preceding the onset of psychosis in patients with 22q11.2 deletion syndrome. JAMA Psychiatry 2015, 72, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Fénelon, K.; Mukai, J.; Xu, B.; Hsu, P.K.; Drew, L.J.; Karayiorgou, M.; Fischbach, G.D.; Macdermott, A.B.; Gogos, J.A. Deficiency of Dgcr8, a gene disrupted by the 22q11.2 microdeletion, results in altered short-term plasticity in the prefrontal cortex. Proc. Natl. Acad. Sci. USA 2011, 108, 4447–4452. [Google Scholar] [CrossRef] [Green Version]
- Tripathi, A.; Spedding, M.; Schenker, E.; Didriksen, M.; Cressant, A.; Jay, T.M. Cognition- and circuit-based dysfunction in a mouse model of 22q11.2 microdeletion syndrome: Effects of stress. Transl. Psychiatry 2020, 10, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Kimber, W.L.; Hsieh, P.; Hirotsune, S.; Yuva-Paylor, L.; Sutherland, H.F.; Chen, A.; Ruiz-Lozano, P.; Hoogstraten-Miller, S.L.; Chien, K.R.; Paylor, R.; et al. Deletion of 150 kb in the Minimal Digeorge/Velocardiofacial Syndrome Critical Region in Mouse. Hum. Mol. Genet. 1999, 8, 2229–2237. [Google Scholar] [CrossRef]
- Meechan, D.W.; Maynard, T.M.; Fernandez, A.; Karpinski, B.A.; Rothblat, L.A.; LaMantia, A.S. Modeling a model: Mouse genetics, 22q11.2 Deletion Syndrome, and disorders of cortical circuit development. Prog. Neurobiol. 2015, 130, 1–28. [Google Scholar] [CrossRef] [Green Version]
- Morris, R.G.M. Elements of a neurobiological theory of hippocampal function: The role of synaptic plasticity, synaptic tagging and schemas. Eur. J. Neurosci. 2006, 23, 2829–2846. [Google Scholar] [CrossRef]
- Martin, S.J.; Grimwood, P.D.; Morris, R.G.M. Synaptic Plasticity and Memory: An Evaluation of the Hypothesis. Annu. Rev. Neurosci. 2000, 23, 649–711. [Google Scholar] [CrossRef] [Green Version]
- Citri, A.; Malenka, R.C. Synaptic Plasticity: Multiple Forms, Functions, and Mechanisms. Neuropsychopharmacology 2008, 33, 18–41. [Google Scholar] [CrossRef] [Green Version]
- Marissal, T.; Salazar, R.F.; Bertollini, C.; Mutel, S.; De Roo, M.; Rodriguez, I.; Müller, D.; Carleton, A. Restoring wild-type-like CA1 network dynamics and behavior during adulthood in a mouse model of schizophrenia. Nat. Neurosci. 2018, 21, 1412–1420. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.; Carvalho, F.; Eliez, S.; Caroni, P. Long-Lasting Rescue of Network and Cognitive Dysfunction in a Genetic Schizophrenia Model. Cell 2019, 178, 1387–1402.e14. [Google Scholar] [PubMed]
- Kimura, H.; Fujita, Y.; Kawabata, T.; Ishizuka, K.; Wang, C.; Iwayama, Y.; Okahisa, Y.; Kushima, I.; Morikawa, M.; Uno, Y. A novel rare variant R292H in RTN4R affects growth cone formation and possibly contributes to schizophrenia susceptibility. Transl. Psychiatry 2017, 7, e1214. [Google Scholar] [PubMed] [Green Version]
- Budel, S.; Padukkavidana, T.; Liu, B.P.; Feng, Z.; Hu, F.; Johnson, S.; Lauren, J.; Park, J.H.; McGee, A.W.; Liao, J. Genetic variants of Nogo-66 receptor with possible association to schizophrenia block myelin inhibition of axon growth. J. Neurosci. 2008, 28, 13161–13172. [Google Scholar] [PubMed] [Green Version]
- Crabtree, G.; Gogos, J.A. Synaptic plasticity, neural circuits and the emerging role of altered short-term information processing in schizophrenia. Front. Synaptic Neurosci. 2014, 6, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Earls, L.R.; Bayazitov, I.T.; Fricke, R.G.; Berry, R.B.; Illingworth, E.; Mittleman, G.; Zakharenko, S.S. Dysregulation of presynaptic calcium and synaptic plasticity in a mouse model of 22q11 deletion syndrome. J. Neurosci. 2010, 30, 15843–15855. [Google Scholar] [CrossRef]
- Haydon, P.G. GLIA: Listening and talking to the synapse Septal cholinergic neuromodulation tunes the astrocyte-dependent gating of hippocampal NMDA receptors to wakefulness View project. Nat. Rev. Neurosci. 2001, 2, 185–193. [Google Scholar] [CrossRef]
- Barker, A.J.; Ullian, E.M. Astrocytes and synaptic plasticity. The Neuroscientist: A Review Journal Bringing Neurobiology. Neurol. Psychiatry 2010, 16, 40–50. [Google Scholar]
- Ben Achour, S.; Pascual, O. Glia: The many ways to modulate synaptic plasticity. Neurochem. Int. 2010, 57, 440–445. [Google Scholar] [CrossRef]
- Sancho, L.; Contreras, M.; Allen, N.J. Glia as sculptors of synaptic plasticity. Neurosci. Res. 2021, 167, 17–29. [Google Scholar] [CrossRef]
- De Pittà, M.; Brunel, N.; Volterra, A. Astrocytes: Orchestrating synaptic plasticity? Neuroscience 2016, 323, 43–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Araque, A.; Carmignoto, G.; Haydon, P.G.; Oliet, S.H.R.; Robitaille, R.; Volterra, A. Gliotransmitters Travel in Time and Space. Neuron 2014, 81, 728–739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zoghbi, H.Y.; Bear, M.F. Synaptic dysfunction in neurodevelopmental disorders associated with autism and intellectual disabilities. Cold Spring Harb. Perspect. Biol. 2012, 4, a009886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niklasson, L.; Rasmussen, P.; Óskarsdóttir, S.; Gillberg, C. Autism, ADHD, mental retardation and behavior problems in 100 individuals with 22q11 deletion syndrome. Res. Dev. Disabil. 2009, 30, 763–773. [Google Scholar] [CrossRef]
- Geschwind, D.H.; Levitt, P. Autism spectrum disorders: Developmental disconnection syndromes. Curr. Opin. Neurobiol. 2007, 17, 103–111. [Google Scholar] [CrossRef]
- Weinberger, D.R. Implications of Normal Brain Development for the Pathogenesis of Schizophrenia. Arch. Gen. Psychiatry 1987, 44, 660–669. [Google Scholar] [CrossRef]
- Gourovitch, M.L.; Goldberg, T.E.; Weinberger, D.R. Verbal fluency deficits in patients with schizophrenia: Semantic fluency is differentially impaired as compared with phonologic fluency. Neuropsychology 1996, 10, 573–577. [Google Scholar] [CrossRef]
- Meechan, D.W.; Maynard, T.M.; Tucker, E.S.; LaMantia, A.S. Three phases of DiGeorge/22q11 deletion syndrome pathogenesis during brain development: Patterning, proliferation, and mitochondrial functions of 22q11 genes. Int. J. Dev. Neurosci. 2011, 29, 283–294. [Google Scholar] [CrossRef] [Green Version]
- Eliez, S.; Schmitt, J.E.; White, C.D.; Reiss, A.L. Children and adolescents with velocardiofacial syndrome: A volumetric MRI study. Am. J. Psychiatry 2000, 157, 409–415. [Google Scholar] [CrossRef]
- Bassett, A.S.; Chow, E.W.C.; AbdelMalik, P.; Gheorghiu, M.; Husted, J.; Weksberg, R. The schizophrenia phenotype in 22q11 deletion syndrome. Am. J. Psychiatry 2003, 160, 1580–1586. [Google Scholar] [CrossRef] [Green Version]
- Kates, W.R.; Burnette, C.P.; Bessette, B.A.; Folley, B.S.; Strunge, L.; Jabs, E.W.; Pearlson, G.D. Frontal and caudate alterations in velocardiofacial syndrome (deletion at chromosome 22q11.2). J. Child Neurol. 2004, 19, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Ching, C.R.K.; Gutman, B.A.; Sun, D.; Reina, J.V.; Ragothaman, A.; Isaev, D.; Zavaliangos-Petropulu, A.; Lin, A.; Jonas, R.K.; Kushan, L. Mapping subcortical brain alterations in 22q11.2 deletion syndrome: Effects of deletion size and convergence with idiopathic neuropsychiatric illness. Am. J. Psychiatry 2020, 177, 589. [Google Scholar] [CrossRef] [PubMed]
- Rogdaki, M.; Gudbrandsen, M.; McCutcheon, R.A.; Blackmore, C.E.; Brugger, S.; Ecker, C.; Craig, M.C.; Daly, E.; Murphy, D.G.M.; Howes, O. Magnitude and heterogeneity of brain structural abnormalities in 22q11.2 deletion syndrome: A meta-analysis. Mol. Psychiatry 2020, 25, 1704–1717. [Google Scholar] [CrossRef] [PubMed]
- Kebets, V.; Favre, P.; Houenou, J.; Polosan, M.; Perroud, N.; Aubry, J.-M.; Van De Ville, D.; Piguet, C. Fronto-limbic neural variability as a transdiagnostic correlate of emotion dysregulation. Transl. Psychiatry 2021, 11, 1–8. [Google Scholar] [CrossRef]
- Ottet, M.C.; Schaer, M.; Cammoun, L.; Schneider, M.; Debbané, M.; Thiran, J.P.; Eliez, S. Reduced Fronto-Temporal and Limbic Connectivity in the 22q11.2 Deletion Syndrome: Vulnerability Markers for Developing Schizophrenia? PLoS ONE 2013, 8, e58429. [Google Scholar] [CrossRef] [Green Version]
- Pelgrim, T.A.D.; Bossong, M.G.; Cuiza, A.; Alliende, L.M.; Mena, C.; Tepper, A.; Ramirez-Mahaluf, J.P.; Iruretagoyena, B.; Ornstein, C.; Fritsch, R. Abnormal nodal and global network organization in resting state functional MRI from subjects with the 22q11 deletion syndrome. Sci. Rep. 2021, 11, 1–9. [Google Scholar] [CrossRef]
- Fernandez, A.; Meechan, D.W.; Karpinski, B.A.; Paronett, E.M.; Bryan, C.A.; Rutz, H.L.; Radin, E.A.; Lubin, N.; Bonner, E.R.; Popratiloff, A. Mitochondrial Dysfunction Leads to Cortical Under-Connectivity and Cognitive Impairment. Neuron 2019, 102, 1127–1142.e3. [Google Scholar] [CrossRef]
- Puech, A.; Saint-Jore, B.; Funke, B.; Gilbert, D.J.; Sirotkin, H.; Copeland, N.G.; Jenkins, N.A.; Kucherlapati, R.; Morrow, B.; Skoultchi, A.I. Comparative mapping of the human 22q11 chromosomal region and the orthologous region in mice reveals complex changes in gene organization. Proc. Natl. Acad. Sci. USA 1997, 94, 14608. [Google Scholar] [CrossRef] [Green Version]
- Lindsay, E.A.; Botta, A.; Jurecic, V.; Carattini-Rivera, S.; Cheah, Y.-C.; Rosenblatt, H.M.; Bradley, A.; Baldini, A. Congenital heart disease in mice deficient for the DiGeorge syndrome region. Nature 1999, 401, 379–383. [Google Scholar] [CrossRef]
- Merscher, S.; Funke, B.; Epstein, J.A.; Heyer, J.; Puech, A.; Lu, M.M.; Xavier, R.J.; Demay, M.B.; Russel, R.G.; Factor, S. TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell 2001, 104, 619–629. [Google Scholar] [CrossRef] [Green Version]
- Paylor, R.; McIlwain, K.L.; McAninch, R.; Nellis, A.; Yuva-Paylor, L.A.; Baldini, A.; Lindsay, E.A. Mice deleted for the DiGeorge/velocardiofacial syndrome region show abnormal sensorimotor gating and learning and memory impairments. Hum. Mol. Genet. 2001, 10, 2645–2650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stark, K.L.; Xu, B.; Bagchi, A.; Lai, W.S.; Liu, H.; Hsu, R.; Wan, X.; Pavlidis, P.; Mills, A.A.; Karayiorgou, M. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat. Genet. 2008, 40, 751–760. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.J.; Fanselow, M.S. Modality-specific retrograde amnesia of fear. Science 1992, 256, 675–677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phillips, R.G.; LeDoux, J.E. Differential contribution of amygdala and hippocampus to cued and contextual fear conditioning. Behav. Neurosci. 1992, 106, 274–285. [Google Scholar] [CrossRef]
- Mukai, J.; Tamura, M.; Fénelon, K.; Rosen, A.M.; Spellman, T.J.; Kang, R.; MacDermott, A.B.; Karayiorgou, M.; Gordon, J.A.; Gogos, J.A. Molecular Substrates of Altered Axonal Growth and Brain Connectivity in a Mouse Model of Schizophrenia. Neuron 2015, 86, 680–695. [Google Scholar] [CrossRef] [Green Version]
- Paterlini, M.; Zakharenko, S.S.; Lai, W.S.; Qin, J.; Zhang, H.; Mukai, J.; Westphal, K.G.C.; Olivier, B.; Sulzer, D.; Pavlidis, P. Transcriptional and behavioral interaction between 22q11.2 orthologs modulates schizophrenia-related phenotypes in mice. Nat. Neurosci. 2005, 8, 1586–1594. [Google Scholar] [CrossRef]
- Pulver, A.E.; Nestadt, G.; Goldberg, R.; Shprintzen, R.J.; Lamacz, M.; Wolyniec, P.S.; Morrow, B.; Karayiorgou, M.; Antonarakis, S.E.; Housman, D. Psychotic illness in patients diagnosed with velo-cardio-facial syndrome and their relatives. J. Nerv. Ment. Dis. 1994, 182, 476–478. [Google Scholar] [CrossRef]
- Chow, E.W.C.; Watson, M.; Young, D.A.; Bassett, A.S. Neurocognitive profile in 22q11 deletion syndrome and schizophrenia. Schizophr. Res. 2006, 87, 270–278. [Google Scholar] [CrossRef] [Green Version]
- Lewis, D.A. Cortical circuit dysfunction and cognitive deficits in schizophrenia—Implications for preemptive interventions. Eur. J. Neurosci. 2012, 35, 1871–1878. [Google Scholar] [CrossRef] [Green Version]
- Heckers, S.; Rauch, S.L.; Goff, D.; Savage, C.R.; Schacter, D.L.; Fischman, A.J.; Alpert, N.M. Impaired recruitment of the hippocampus during conscious recollection in schizophrenia. Nat. Neurosci. 1998, 1, 318–323. [Google Scholar] [CrossRef]
- Tamminga, C.A.; Stan, A.D.; Wagner, A.D. The hippocampal formation in schizophrenia. Am. J. Psychiatry 2010, 167, 1178–1193. [Google Scholar] [CrossRef] [PubMed]
- Fénelon, K.; Xu, B.; Lai, C.S.; Mukai, J.; Markx, S.; Stark, K.L.; Hsu, P.K.; Gan, W.B.; Fischbach, G.D.; MacDermott, A.B.; et al. The Pattern of Cortical Dysfunction in a Mouse Model of a Schizophrenia-Related Microdeletion. J. Neurosci. 2013, 33, 14825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Devaraju, P.; Yu, J.; Eddins, D.; Mellado-Lagarde, M.M.; Earls, L.R.; Westmoreland, J.J.; Quarato, G.; Green, D.R.; Zakharenko, S.S. Haploinsufficiency of the 22q11.2 microdeletion gene Mrpl40 disrupts short-term synaptic plasticity and working memory through dysregulation of mitochondrial calcium. Mol. Psychiatry 2017, 22, 1313–1326. [Google Scholar] [CrossRef] [PubMed]
- Howland, J.G.; Wang, Y.T. Synaptic plasticity in learning and memory: Stress effects in the hippocampus. Prog. Brain Res. 2008, 169, 145–158. [Google Scholar] [PubMed]
- Neves, G.; Cooke, S.F.; Bliss, T.V.P. Synaptic plasticity, memory and the hippocampus: A neural network approach to causality. Nat. Rev. Neurosci. 2008, 9, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Mukai, J.; Liu, H.; Burt, R.A.; Swor, D.E.; Lai, W.S.; Karayiorgou, M.; Gogos, J.A. Evidence that the gene encoding ZDHHC8 contributes to the risk of schizophrenia. Nat. Genet. 2004, 36, 725–731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, W.Y.; Shi, Y.Y.; Zheng, Y.L.; Zhao, X.Z.; Zhang, G.J.; Chen, S.Q.; Yang, P.D.; He, L. Case-control study and transmission disequilibrium test provide consistent evidence for association between schizophrenia and genetic variation in the 22q11 gene ZDHHC8. Hum. Mol. Genet. 2004, 13, 2991–2995. [Google Scholar] [CrossRef]
- Fukata, Y.; Fukata, M. Protein palmitoylation in neuronal development and synaptic plasticity. Nat. Rev. Neurosci. 2010, 11, 161–175. [Google Scholar] [CrossRef]
- El-Husseini, A.E.D.; Bredt, D.S. Protein palmitoylation: A regulator of neuronal development and function. Nat. Rev. Neurosci. 2002, 3, 791–802. [Google Scholar] [CrossRef]
- Eryilmaz, H.; Tanner, A.S.; Ho, N.F.; Nitenson, A.Z.; Silverstein, N.J.; Petruzzi, L.J.; Goff, D.C.; Manoach, D.S.; Roffman, J.L. Disrupted Working Memory Circuitry in Schizophrenia: Disentangling fMRI Markers of Core Pathology vs. Other Aspects of Impaired Performance. Neuropsychopharmacol. Off. Publ. Am. Coll. Neuropsychopharmacol. 2016, 41, 2411–2420. [Google Scholar] [CrossRef] [Green Version]
- Gilks, W.P.; Hill, M.; Gill, M.; Donohoe, G.; Corvin, A.P.; Morris, D.W. Functional investigation of a schizophrenia GWAS signal at the CDC42 gene. World J. Biol. Psychiatry Off. J. World Fed. Soc. Biol. Psychiatry 2012, 13, 550–554. [Google Scholar] [CrossRef] [PubMed]
- Lai, W.S.; Xu, B.; Westphal, K.G.C.; Paterlini, M.; Olivier, B.; Pavlidis, P.; Karayiorgou, M.; Gogos, J.A. Akt1 deficiency affects neuronal morphology and predisposes to abnormalities in prefrontal cortex functioning. Proc. Natl. Acad. Sci. USA 2006, 103, 16906–16911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewis, B.P.; Shih, I.H.; Jones-Rhoades, M.W.; Bartel, D.P.; Burge, C.B. Prediction of mammalian microRNA targets. Cell 2003, 115, 787–798. [Google Scholar] [CrossRef] [Green Version]
- Kolfschoten, I.G.M.; Regazzi, R. Technology Insight: Small, noncoding RNA molecules as tools to study and treat endocrine diseases. Nat. Clin. Pract. Endocrinol. Metab. 2007, 3, 827–834. [Google Scholar] [CrossRef]
- Han, J.; Lee, Y.; Yeom, K.H.; Kim, Y.K.; Jin, H.; Kim, V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef] [Green Version]
- Schratt, G. microRNAs at the synapse. Nat. Rev. Neurosci. 2009, 10, 842–849. [Google Scholar] [CrossRef]
- Diamantopoulou, A.; Sun, Z.; Mukai, J.; Xu, B.; Fenelon, K.; Karayiorgou, M.; Gogos, J.A. Loss-of-function mutation in Mirta22/Emc10 rescues specific schizophrenia-related phenotypes in a mouse model of the 22q11.2 deletion. Proc. Natl. Acad. Sci. USA 2017, 114, E6127–E6136. [Google Scholar] [CrossRef] [Green Version]
- Xu, B.; Hsu, P.K.; Stark, K.L.; Karayiorgou, M.; Gogos, J.A. Derepression of a neuronal inhibitor due to miRNA dysregulation in a schizophrenia-related microdeletion. Cell 2013, 152, 262–275. [Google Scholar] [CrossRef] [Green Version]
- Sivagnanasundaram, S.; Fletcher, D.; Hubank, M.; Illingworth, E.; Skuse, D.; Scambler, P. Differential gene expression in the hippocampus of the Df1/+ mice: A model for 22q11.2 deletion syndrome and schizophrenia. Brain Res. 2007, 1139, 48–59. [Google Scholar] [CrossRef]
- Elgersma, Y.; Sweatt, J.D.; Giese, K.P. Mouse Genetic Approaches to Investigating Calcium/Calmodulin-Dependent Protein Kinase II Function in Plasticity and Cognition. J. Neurosci. 2004, 24, 8410. [Google Scholar] [CrossRef] [Green Version]
- Cammarota, M.; Bevilaqua, L.R.M.; Viola, H.; Kerr, D.S.; Reichmann, B.; Teixeira, V.; Bulla, M.; Izquierdo, I.; Medina, J.H. Participation of CaMKII in neuronal plasticity and memory formation. Cell. Mol. Neurobiol. 2002, 22, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Colbran, R.J.; Brown, A.M. Calcium/calmodulin-dependent protein kinase II and synaptic plasticity. Curr. Opin. Neurobiol. 2004, 14, 318–327. [Google Scholar] [PubMed]
- Yamagata, K.; Andreasson, K.I.; Sugiura, H.; Maru, E.; Dominique, M.; Irie, Y.; Miki, N.; Hayashi, Y.; Yoshioka, M.; Kaneko, K. Arcadlin Is a Neural Activity-regulated Cadherin Involved in Long Term Potentiation. J. Biol. Chem. 1999, 274, 19473–19479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Philip, N.; Acevedo, S.F.; Skoulakis, E.M.C. Conditional rescue of olfactory learning and memory defects in mutants of the 14-3-3zeta gene leonardo. J. Neurosci. Off. J. Soc. Neurosci. 2001, 21, 8417–8425. [Google Scholar] [CrossRef] [Green Version]
- Butler, M.G.; Rafi, S.K.; Hossain, W.; Stephan, D.A.; Manzardo, A.M. Whole exome sequencing in females with autism implicates novel and candidate genes. Int. J. Mol. Sci. 2015, 16, 1312–1335. [Google Scholar] [CrossRef] [Green Version]
- Clements, C.C.; Wenger, T.L.; Zoltowski, A.R.; Bertollo, J.R.; Miller, J.S.; De Marchena, A.B.; Mitteer, L.A.; Carey, J.C.; Yerys, B.E.; Zackai, E.H. Critical region within 22q11.2 linked to higher rate of autism spectrum disorder. Mol. Autism 2017, 8, 58. [Google Scholar] [CrossRef] [Green Version]
- Middleton, F.A.; Peng, L.; Lewis, D.A.; Levitt, P.; Mirnics, K. Altered Expression of 14-3-3 Genes in the Prefrontal Cortex of Subjects with Schizophrenia. Neuropsychopharmacology 2005, 30, 974–983. [Google Scholar] [CrossRef] [Green Version]
- Meechan, D.W.; Rutz, H.L.H.; Fralish, M.S.; Maynard, T.M.; Rothblat, L.A.; Lamantia, A.S. Cognitive ability is associated with altered medial frontal cortical circuits in the LgDel mouse model of 22q11.2DS. Cereb. Cortex 2015, 25, 1143–1151. [Google Scholar] [CrossRef] [Green Version]
- Piskorowski, R.A.; Nasrallah, K.; Diamantopoulou, A.; Mukai, J.; Hassan, S.I.; Siegelbaum, S.A.; Siegelbaum, S.A.; Gogos, J.A.; Chevaleyre, V. Age-Dependent Specific Changes in Area CA2 of the Hippocampus and Social Memory Deficit in a Mouse Model of the 22q11.2 Deletion Syndrome. Neuron 2016, 89, 163–176. [Google Scholar] [CrossRef] [Green Version]
- Sigurdsson, T.; Stark, K.L.; Karayiorgou, M.; Gogos, J.A.; Gordon, J.A. Impaired hippocampal–prefrontal synchrony in a genetic mouse model of schizophrenia. Nature 2010, 464, 763–767. [Google Scholar]
- Maynard, T.M.; Meechan, D.W.; Dudevoir, M.L.; Gopalakrishna, D.; Peters, A.Z.; Heindel, C.C.; Sugimoto, T.J.; Wu, Y.; Lieberman, J.A.; LaMantia, A.S. Mitochondrial localization and function of a subset of 22q11 deletion syndrome candidate genes. Mol. Cell. Neurosci. 2008, 39, 439–451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hara, Y.; Yuk, F.; Puri, R.; Janssen, W.G.M.; Rapp, P.R.; Morrison, J.H. Presynaptic mitochondrial morphology in monkey prefrontal cortex correlates with working memory and is improved with estrogen treatment. Proc. Natl. Acad. Sci. USA 2014, 111, 486–491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmad, T.; Aggarwal, K.; Pattnaik, B.; Mukherjee, S.; Sethi, T.; Tiwari, B.K.; Kumar, M.; Micheal, A.; Mabalirajan, U.; Ghosh, B. Computational classification of mitochondrial shapes reflects stress and redox state. Cell Death Dis. 2013, 4, e461. [Google Scholar] [PubMed] [Green Version]
- Arnér, E.S.J.; Holmgren, A. Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 2000, 267, 6102–6109. [Google Scholar] [CrossRef] [PubMed]
- Gokhale, A.; Hartwig, C.; Freeman, A.A.H.; Bassell, J.L.; Zlatic, S.A.; Savas, C.S.; Vadlamudi, T.; Abudulai, F.; Pham, T.T.; Crocker, A. Systems analysis of the 22q11.2 microdeletion syndrome converges on a mitochondrial interactome necessary for synapse function and behavior. J. Neurosci. 2019, 39, 3561–3581. [Google Scholar] [CrossRef]
- Catalina-Rodriguez, O.; Kolukula, V.K.; Tomita, Y.; Preet, A.; Palmieri, F.; Wellstein, A.; Byers, S.; Giaccia, A.J.; Glasgow, E.; Albanese, C. The mitochondrial citrate transporter, CIC, is essential for mitochondrial homeostasis. Oncotarget 2012, 3, 1220–1235. [Google Scholar] [CrossRef] [Green Version]
- Gokhale, A.; Lee, C.E.; Zlatic, S.A.; Freeman, A.A.H.; Shearing, N.; Hartwig, C.; Ogunbona, O.; Bassell, J.L.; Wynne, M.E.; Werner, E. Mitochondrial Proteostasis Requires Genes Encoded in a Neurodevelopmental Syndrome Locus. J. Neurosci. Off. J. Soc. Neurosci. 2021, 41, 6596–6616. [Google Scholar]
- Amunts, A.; Brown, A.; Bai, X.C.; Llácer, J.L.; Hussain, T.; Emsley, P.; Long, F.; Murshudov, G.; Scheres, S.H.W.; Ramakrishnan, V. Structure of the yeast mitochondrial large ribosomal subunit. Science 2014, 343, 1485–1489. [Google Scholar] [CrossRef] [Green Version]
- Tang, M.; Lu, L.; Huang, Z.; Chen, L. Palmitoylation signaling: A novel mechanism of mitochondria dynamics and diverse pathologies. Acta Biochim. Biophys. Sin. 2018, 50, 831–833. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Heath, S.C.; Sobin, C.; Roos, J.L.; Galke, B.L.; Blundell, M.L.; Lenane, M.; Robertson, B.; Wijsman, E.M.; Rapoport, J.L. Genetic variation at the 22q11 PRODH2/DGCR6 locus presents an unusual pattern and increases susceptibility to schizophrenia. Proc. Natl. Acad. Sci. USA 2002, 99, 3717–3722. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Abecasis, G.R.; Heath, S.C.; Knowles, A.; Demars, S.; Chen, Y.J.; Roos, J.L.; Rapoport, J.L.; Gogos, J.A.; Karayiorgou, M. Genetic variation in the 22q11 locus and susceptibility to schizophrenia. Proc. Natl. Acad. Sci. USA 2002, 99, 16859–16864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bard, F.; Casano, L.; Mallabiabarrena, A.; Wallace, E.; Saito, K.; Kitayama, H.; Guizzunti, G.; Hu, Y.; Wendler, F.; Dasgupta, R. Functional genomics reveals genes involved in protein secretion and Golgi organization. Nature 2006, 439, 604–607. [Google Scholar] [CrossRef] [PubMed]
- Mattson, M.P.; Gleichmann, M.; Cheng, A. Mitochondria in neuroplasticity and neurological disorders. Neuron 2008, 60, 748–766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Earls, L.R.; Gaines Fricke, R.; Yu, J.; Berry, R.B.; Baldwin, L.T.; Zakharenko, S.S. Age-Dependent MicroRNA Control of Synaptic Plasticity in 22q11 Deletion Syndrome and Schizophrenia. J. Neurosci. 2012, 32, 14132. [Google Scholar] [CrossRef] [Green Version]
- Cooke, S.F.; Bliss, T.V.P. Plasticity in the human central nervous system. Brain J. Neurol. 2006, 129, 1659–1673. [Google Scholar] [CrossRef] [Green Version]
- Auld, D.S.; Robitaille, R. Glial Cells and Neurotransmission: An Inclusive View of Synaptic Function. Neuron 2003, 40, 389–400. [Google Scholar] [CrossRef] [Green Version]
- Pannasch, U.; Rouach, N. Emerging role for astroglial networks in information processing: From synapse to behavior. Trends Neurosci. 2013, 36, 405–417. [Google Scholar] [CrossRef]
- Volterra, A.; Meldolesi, J. Astrocytes, from brain glue to communication elements: The revolution continues. Nat. Rev. Neurosci. 2005, 6, 626–640. [Google Scholar] [CrossRef]
- Allen, N.J.; Barres, B.A. Signaling between glia and neurons: Focus on synaptic plasticity. Curr. Opin. Neurobiol. 2005, 15, 542–548. [Google Scholar] [CrossRef]
- Perea, G.; Navarrete, M.; Araque, A. Tripartite synapses: Astrocytes process and control synaptic information. Trends Neurosci. 2009, 32, 421–431. [Google Scholar] [CrossRef]
- López-Hidalgo, M.; Schummers, J. Cortical maps: A role for astrocytes? Curr. Opin. Neurobiol. 2014, 24, 176–189. [Google Scholar] [CrossRef] [PubMed]
- Bushong, E.A.; Martone, M.E.; Jones, Y.Z.; Ellisman, M.H. Protoplasmic astrocytes in CA1 stratum radiatum occupy separate anatomical domains. J. Neurosci. 2002, 22, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Halassa, M.M.; Fellin, T.; Takano, H.; Dong, J.-H.; Haydon, P.G. Synaptic Islands Defined by the Territory of a Single Astrocyte. J. Neurosci. 2007, 27, 6473–6477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simard, M.; Nedergaard, M. The neurobiology of glia in the context of water and ion homeostasis. Neuroscience 2004, 129, 877–896. [Google Scholar] [CrossRef] [PubMed]
- Deitmer, J.W.; Theparambil, S.M.; Ruminot, I.; Noor, S.I.; Becker, H.M. Energy Dynamics in the Brain: Contributions of Astrocytes to Metabolism and pH Homeostasis. Front. Neurosci. 2019, 13, 1301. [Google Scholar] [CrossRef] [PubMed]
- Magistretti, P.J. Role of glutamate in neuron-glia metabolic coupling. Am. J. Clin. Nutr. 2009, 90, 875S–880S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magistretti Pj Sorg, O.; Martin, J.-L. Astrocytes 1993, 243–265. Available online: https://books.google.ca/books?hl=en&lr=&id=4tpGN-_X6m4C&oi=fnd&pg=PA243&dq=Regulation+of+glycogen+metabolism+in+astrocytes:+physiological,+pharmacological+and+pathological+aspects+&ots=fc7SacoArw&sig=yxUNMa-kDtT1taLkbH2iJf6EOyk#v=onepage&q=Regulation%20of%20glycogen%20metabolism%20in%20astrocytes%3A%20physiological%2C%20pharmacological%20and%20pathological%20aspects&f=false (accessed on 1 March 2022).
- Magistretti, P.J.; Allaman, I. Brain energy metabolism. In Neuroscience in the 21st Century: From Basic to Clinical; Springer: New York, NY, USA, 2013; pp. 1591–1620. [Google Scholar]
- Bélanger, M.; Allaman, I.; Magistretti, P.J. Brain Energy Metabolism: Focus on Astrocyte-Neuron Metabolic Cooperation. Cell Metab. 2011, 14, 724–738. [Google Scholar] [CrossRef] [Green Version]
- Attwell, D.; Buchan, A.M.; Charpak, S.; Lauritzen, M.; MacVicar, B.A.; Newman, E.A. Glial and neuronal control of brain blood flow. Nature 2010, 468, 232. [Google Scholar] [CrossRef] [Green Version]
- Bezzi, P.; Volterra, A. A neuron–glia signalling network in the active brain. Curr. Opin. Neurobiol. 2001, 11, 387–394. [Google Scholar] [CrossRef]
- Petrelli, F.; Bezzi, P. Novel insights into gliotransmitters. Curr. Opin. Pharmacol. 2016, 26, 138–145. [Google Scholar] [CrossRef] [PubMed]
- Santello, M.; Calì, C.; Bezzi, P. Gliotransmission and the Tripartite Synapse2012. Synaptic Plast. 2012, 970, 307–331. [Google Scholar]
- Bezzi, P.; Gundersen, V.; Galbete, J.L.; Seifert, G.; Steinhäuser, C.; Pilati, E.; Volterra, A. Astrocytes contain a vesicular compartment that is competent for regulated exocytosis of glutamate. Nat. Neurosci. 2004, 7, 613–620. [Google Scholar] [CrossRef]
- Jourdain, P.; Bergersen, L.H.; Bhaukaurally, K.; Bezzi, P.; Santello, M.; Domercq, M.; Matute, C.; Tonello, F.; Gundersen, V.; Volterra, A. Glutamate exocytosis from astrocytes controls synaptic strength. Nat. Neurosci. 2007, 10, 331–339. [Google Scholar] [CrossRef] [PubMed]
- Vesce, S.; Bezzi, P.; Volterra, A. The active role of astrocytes in synaptic transmission. Cell. Mol. Life Sci. 1999, 56, 991–1000. [Google Scholar] [CrossRef] [PubMed]
- Calì, C.; Marchaland, J.; Spagnuolo, P.; Gremion, J.; Bezzi, P. Chapter 20 Regulated Exocytosis from Astrocytes2009. Int. Rev. Neurobiol. 2009, 85, 261–293. [Google Scholar] [PubMed]
- Gómez-Gonzalo, M.; Zehnder, T.; Requie, L.M.; Bezzi, P.; Carmignoto, G. Insights into the release mechanism of astrocytic glutamate evoking in neurons NMDA receptor-mediated slow depolarizing inward currents. GLIA 2018, 66, 2188–2199. [Google Scholar] [CrossRef]
- Schousboe, A.; Sarup, A.; Bak, L.K.; Waagepetersen, H.S.; Larsson, O.M. Role of astrocytic transport processes in glutamatergic and GABAergic neurotransmission. Neurochem. Int. 2004, 45, 521–527. [Google Scholar] [CrossRef]
- Huang, Y.H.; Sinha, S.R.; Tanaka, K.; Rothstein, J.D.; Bergles, D.E. Astrocyte Glutamate Transporters Regulate Metabotropic Glutamate Receptor-Mediated Excitation of Hippocampal Interneurons. J. Neurosci. 2004, 24, 4551. [Google Scholar] [CrossRef]
- Tzingounis, A.V.; Wadiche, J.I. Glutamate transporters: Confining runaway excitation by shaping synaptic transmission. Nat. Rev. Neurosci. 2007, 8, 935–947. [Google Scholar] [CrossRef]
- Hirst, W.D.; Price, G.W.; Rattray, M.; Wilkin, G.P. Serotonin transporters in adult rat brain astrocytes revealed by [3H]5-HT uptake into glial plasmalemmal vesicles. Neurochem. Int. 1998, 33, 11–22. [Google Scholar] [CrossRef]
- Inazu, M.; Takeda, H.; Matsumiya, T. The role of glial monoamine transporters in the central nervous system. Nihon Shinkei Seishin Yakurigaku Zasshi 2003, 23, 171–178. [Google Scholar] [PubMed]
- Perdan-Pirkmajer, K.; Mavri, J.; Krẑan, M. Histamine (re)uptake by astrocytes: An experimental and computational study. J. Mol. Modeling 2010, 16, 1151–1158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naganuma, F.; Yoshikawa, T.; Nakamura, T.; Iida, T.; Harada, R.; Mohsen, A.S.; Miura, Y.; Yanai, K. Predominant role of plasma membrane monoamine transporters in monoamine transport in 1321N1, a human astrocytoma-derived cell line. J. Neurochem. 2014, 129, 591–601. [Google Scholar] [CrossRef] [Green Version]
- Cahoy, J.D.; Emery, B.; Kaushal, A.; Foo, L.C.; Zamanian, J.L.; Christopherson, K.S.; Xing, Y.; Lubischer, J.L.; Krieg, P.A.; Krupenko, S.A. A transcriptome database for astrocytes, neurons, and oligodendrocytes: A new resource for understanding brain development and function. J. Neurosci. 2008, 28, 264–278. [Google Scholar] [CrossRef] [Green Version]
- Cui, M.; Aras, R.; Christian, W.V.; Rappold, P.M.; Hatwar, M.; Panza, J.; Jackson-Lewis, V.; Javitch, J.A.; Ballatori, N.; Przedborski, S. The organic cation transporter-3 is a pivotal modulator of neurodegeneration in the nigrostriatal dopaminergic pathway. Proc. Natl. Acad. Sci. USA 2009, 106, 8043–8048. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Sloan, S.A.; Clarke, L.E.; Caneda, C.; Plaza, C.A.; Blumenthal, P.D.; Vogel, H.; Steinberg, G.K.; Edwards, M.S.B.; Li, G. Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron 2016, 89, 37–53. [Google Scholar] [CrossRef] [Green Version]
- Yoshikawa, T.; Naganuma, F.; Iida, T.; Nakamura, T.; Harada, R.; Mohsen, A.S.; Kasajima, A.; Sasano, H.; Yanai, K. Molecular mechanism of histamine clearance by primary human astrocytes. Glia 2013, 61, 905–916. [Google Scholar] [CrossRef]
- Petrelli, F.; Dallérac, G.; Pucci, L.; Calì, C.; Zehnder, T.; Sultan, S.; Lecca, S.; Chicca, A.; Ivanov, A.; Asensio, C.S. Dysfunction of homeostatic control of dopamine by astrocytes in the developing prefrontal cortex leads to cognitive impairments. Mol. Psychiatry 2020, 25, 732–749. [Google Scholar] [CrossRef] [Green Version]
- Baganz, N.L.; Horton, R.E.; Calderon, A.S.; Owens, W.A.; Munn, J.L.; Watts, L.T.; Koldzic-Zivanovic, N.; Jeske, N.A.; Koek, W.; Toney, G.M. Organic cation transporter 3, Keeping the brake on extracellular serotonin in serotonin-transporter-deficient mice. Proc. Natl. Acad. Sci. USA 2008, 105, 18976. [Google Scholar] [CrossRef] [Green Version]
- Bacq, A.; Balasse, L.; Biala, G.; Guiard, B.; Gardier, A.M.; Schinkel, A.; Louis, F.; Vialou, V.; Martres, M.P.; Chevarin, C. Organic cation transporter 2 controls brain norepinephrine and serotonin clearance and antidepressant response. Mol. Psychiatry 2012, 17, 926–939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giros, B.; Jaber, M.; Jones, S.R.; Wightman, R.M.; Caron, M.G. Hyperlocomotion and indifference to cocaine and amphetamine in mice lacking the dopamine transporter. Nature 1996, 379, 606–612. [Google Scholar] [CrossRef] [PubMed]
- Käenmäki, M.; Tammimäki, A.; Myöhänen, T.; Pakarinen, K.; Amberg, C.; Karayiorgou, M.; Gogos, J.A.; Männistö, P.T. Quantitative role of COMT in dopamine clearance in the prefrontal cortex of freely moving mice. J. Neurochem. 2010, 114, 1745–1755. [Google Scholar] [CrossRef] [PubMed]
- Carboni, E.; Tanda, G.L.; Frau, R.; Chiara, G.D. Blockade of the noradrenaline carrier increases extracellular dopamine concentrations in the prefrontal cortex: Evidence that dopamine is taken up in vivo by noradrenergic terminals. J. Neurochem. 1990, 55, 1067–1070. [Google Scholar] [CrossRef] [PubMed]
- Mundorf, M.L.; Joseph, J.D.; Austin, C.M.; Caron, M.G.; Wightman, R.M. Catecholamine release and uptake in the mouse prefrontal cortex. J. Neurochem. 2001, 79, 130–142. [Google Scholar] [CrossRef] [PubMed]
- Mazei, M.S.; Pluto, C.P.; Kirkbride, B.; Pehek, E.A. Effects of catecholamine uptake blockers in the caudate-putamen and subregions of the medial prefrontal cortex of the rat. Brain Res. 2002, 936, 58–67. [Google Scholar] [CrossRef]
- Eisenhofer, G.; Kopin, I.J.; Goldstein, D.S. Catecholamine Metabolism: A Contemporary View with Implications for Physiology and Medicine. Pharmacol. Rev. 2004, 56, 331–349. [Google Scholar] [CrossRef]
- Howes, O.D.; Kapur, S. The dopamine hypothesis of schizophrenia: Version III—The final common pathway. Schizophr. Bull. 2009, 35, 549–562. [Google Scholar] [CrossRef] [Green Version]
- Moncrieff, J. A critique of the dopamine hypothesis of schizophrenia and psychosis. Harv. Rev. Psychiatry 2009, 17, 214–225. [Google Scholar] [CrossRef]
- Glatt, S.J.; Faraone Ming TTsuang, S.V. Association between a Functional Catechol O-Methyltransferase Gene Polymorphism and Schizophrenia: Meta-Analysis of Case-Control and Family-Based Studies. Am. J. Psychiatry 2003, 160, 3. [Google Scholar] [CrossRef] [Green Version]
- Weinberger, D.R.; Egan, M.F.; Bertolino, A.; Callicott, J.H.; Mattay, V.S.; Lipska, B.K.; Berman, K.F.; Goldberg, T.E. Prefrontal neurons and the genetics of schizophrenia. Biol. Psychiatry 2001, 50, 825–844. [Google Scholar] [CrossRef]
- Huotari, M.; Gogos, J.A.; Karayiorgou, M.; Koponen, O.; Forsberg, M.; Raasmaja, A.; Hyttinen, J.; Männistö, P.T. Brain catecholamine metabolism in catechol-O-methyltransferase (COMT)-deficient mice. Eur. J. Neurosci. 2002, 15, 246–256. [Google Scholar] [CrossRef] [PubMed]
- Yavich, L.; Forsberg, M.M.; Karayiorgou, M.; Gogos, J.A.; Männistö, P.T. Site-Specific Role of Catechol-O-Methyltransferase in Dopamine Overflow within Prefrontal Cortex and Dorsal Striatum. J. Neurosci. 2007, 27, 10196. [Google Scholar] [CrossRef] [PubMed]
- Vijayraghavan, S.; Wang, M.; Birnbaum, S.G.; Williams, G.V.; Arnsten, A.F.T. Inverted-U dopamine D1 receptor actions on prefrontal neurons engaged in working memory. Nat. Neurosci. 2007, 10, 376–384. [Google Scholar] [CrossRef]
- Dickinson, D.; Elvevåg, B. Genes, cognition and brain through a COMT lens. Neuroscience 2009, 164, 72–87. [Google Scholar] [CrossRef] [Green Version]
- Phang, J.M.; Hu, C.A.; Valle, D. Disorders of proline and hydroxyproline metabolism. Metab. Mol. Basis Inherit. Dis. 2001, 1821–1838. Available online: https://www.researchgate.net/publication/284684119_Disorders_of_proline_and_hydroxyproline_metabolism (accessed on 1 March 2022).
- Raux, G.; Bumsel, E.; Hecketsweiler, B.; van Amelsvoort, T.; Zinkstok, J.; Manouvrier-Hanu, S.; Fantini, C.; Brévière, G.M.; Di Rosa, G.; Pustorino, G.; et al. Involvement of hyperprolinemia in cognitive and psychiatric features of the 22q11 deletion syndrome. Hum. Mol. Genet. 2007, 16, 83–91. [Google Scholar] [CrossRef] [Green Version]
- Gogos, J.A.; Santha, M.; Takacs, Z.; Beck, K.D.; Luine, V.; Lucas, L.R.; Nadler, J.V.; Karayiorgou, M. The gene encoding proline dehydrogenase modulates sensorimotor gating in mice. Nat. Genet. 1999, 21, 434–439. [Google Scholar] [CrossRef]
- Boisvert, M.M.; Erikson, G.A.; Shokhirev, M.N.; Allen, N.J. The Aging Astrocyte Transcriptome from Multiple Regions of the Mouse Brain. Cell Rep. 2018, 22, 269–285. [Google Scholar] [CrossRef] [Green Version]
- Servet, C.; Ghelis, T.; Richard, L.; Zilberstein, A.; Savoure, A. Proline dehydrogenase: A key enzyme in controlling cellular homeostasis. Front. Biosci. 2012, 17, 607. [Google Scholar] [CrossRef] [Green Version]
- Maynard, T.M.; Haskell, G.T.; Peters, A.Z.; Sikich, L.; Lieberman, J.A.; Lamantia, A.-S. A comprehensive analysis of 22q11 gene expression in the developing and adult brain. Proc. Natl. Acad. Sci. USA 2003, 100, 14433–14438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clarke, L.E.; Barres, B.A. Emerging roles of astrocytes in neural circuit development. Nat. Rev. Neurosci. 2013, 14, 311–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ullian, E.M.; Sapperstein, S.K.; Christopherson, K.S.; Barres, B.A. Control of synapse number by glia. Science 2001, 291, 657–661. [Google Scholar] [CrossRef] [PubMed]
- Dietz, A.G.; Goldman, S.A.; Nedergaard, M. Glial cells in schizophrenia: A unified hypothesis. Lancet Psychiatry 2020, 7, 272–281. [Google Scholar] [CrossRef]
- Allen, N.J.; Eroglu, C. Cell Biology of Astrocyte-Synapse Interactions. Neuron 2017, 96, 697–708. [Google Scholar] [CrossRef]
- Zehnder, T.; Petrelli, F.; Romanos, J.; de Oliveira Figueiredo, E.C.; Lewis, T.L.; Polleux, F.; Santello, M.; Bezzi, P. Mitochondrial biogenesis in developing astrocytes regulates astrocyte maturation and synapse formation. Cell Rep. 2021, 35, 108952. [Google Scholar] [CrossRef]
- Buscemi, L.; Ginet, V.; Lopatar, J.; Montana, V.; Pucci, L.; Spagnuolo, P.; Zehnder, T.; Grubišic, V.; Truttman, A.; Sala, C. Homer1 Scaffold Proteins Govern Ca2+ Dynamics in Normal and Reactive Astrocytes. Cereb. Cortex 2017, 27, 2365–2384. [Google Scholar] [CrossRef] [Green Version]
- Sun, W.; McConnell, E.; Pare, J.F.; Xu, Q.; Chen, M.; Peng, W.; Lovatt, D.; Han, X.; Smith, Y.; Nedergaard, M. Glutamate-Dependent Neuroglial Calcium Signaling Differs between Young and Adult Brain. Science 2013, 339, 197–200. [Google Scholar] [CrossRef] [Green Version]
- Petrelli, F.; Bezzi, P. mGlu5-mediated signalling in developing astrocyte and the pathogenesis of autism spectrum disorders. Curr. Opin. Neurobiol. 2018, 48, 139–145. [Google Scholar] [CrossRef]
- Yakoub, A.M.; Sadek, M. Analysis of Synapses in Cerebral Organoids. Cell Transplant. 2019, 28, 1173–1182. [Google Scholar] [CrossRef] [Green Version]
- Crockett, A.M.; Ryan, S.K.; Vasquez, A.H.; Canning, C.; Kanyuch, N.; Kebir, H.; Ceja, G.; Gesualdi, J.; Zackai, E.; McDon-ald-McGinn, D.; et al. Disruption of the blood-brain barrier in 22q11.2 deletion syndrome. Brain 2021, 144, 1351–1360. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Oliveira Figueiredo, E.C.; Bondiolotti, B.M.; Laugeray, A.; Bezzi, P. Synaptic Plasticity Dysfunctions in the Pathophysiology of 22q11 Deletion Syndrome: Is There a Role for Astrocytes? Int. J. Mol. Sci. 2022, 23, 4412. https://doi.org/10.3390/ijms23084412
de Oliveira Figueiredo EC, Bondiolotti BM, Laugeray A, Bezzi P. Synaptic Plasticity Dysfunctions in the Pathophysiology of 22q11 Deletion Syndrome: Is There a Role for Astrocytes? International Journal of Molecular Sciences. 2022; 23(8):4412. https://doi.org/10.3390/ijms23084412
Chicago/Turabian Stylede Oliveira Figueiredo, Eva Cristina, Bianca Maria Bondiolotti, Anthony Laugeray, and Paola Bezzi. 2022. "Synaptic Plasticity Dysfunctions in the Pathophysiology of 22q11 Deletion Syndrome: Is There a Role for Astrocytes?" International Journal of Molecular Sciences 23, no. 8: 4412. https://doi.org/10.3390/ijms23084412






