Overexpression of Nucleolin and Associated Genes in Prostate Cancer
Abstract
1. Introduction
2. Results
2.1. Baseline Patient Characteristics
2.2. Nucleolin Is Overexpressed in Prostate Cancer
2.3. NCL Expression Is Not a Prognosis Biomarker of PCa
2.4. Genes Positively Correlated with NCL Have Function in Cell Division and Ribosome Biogenesis
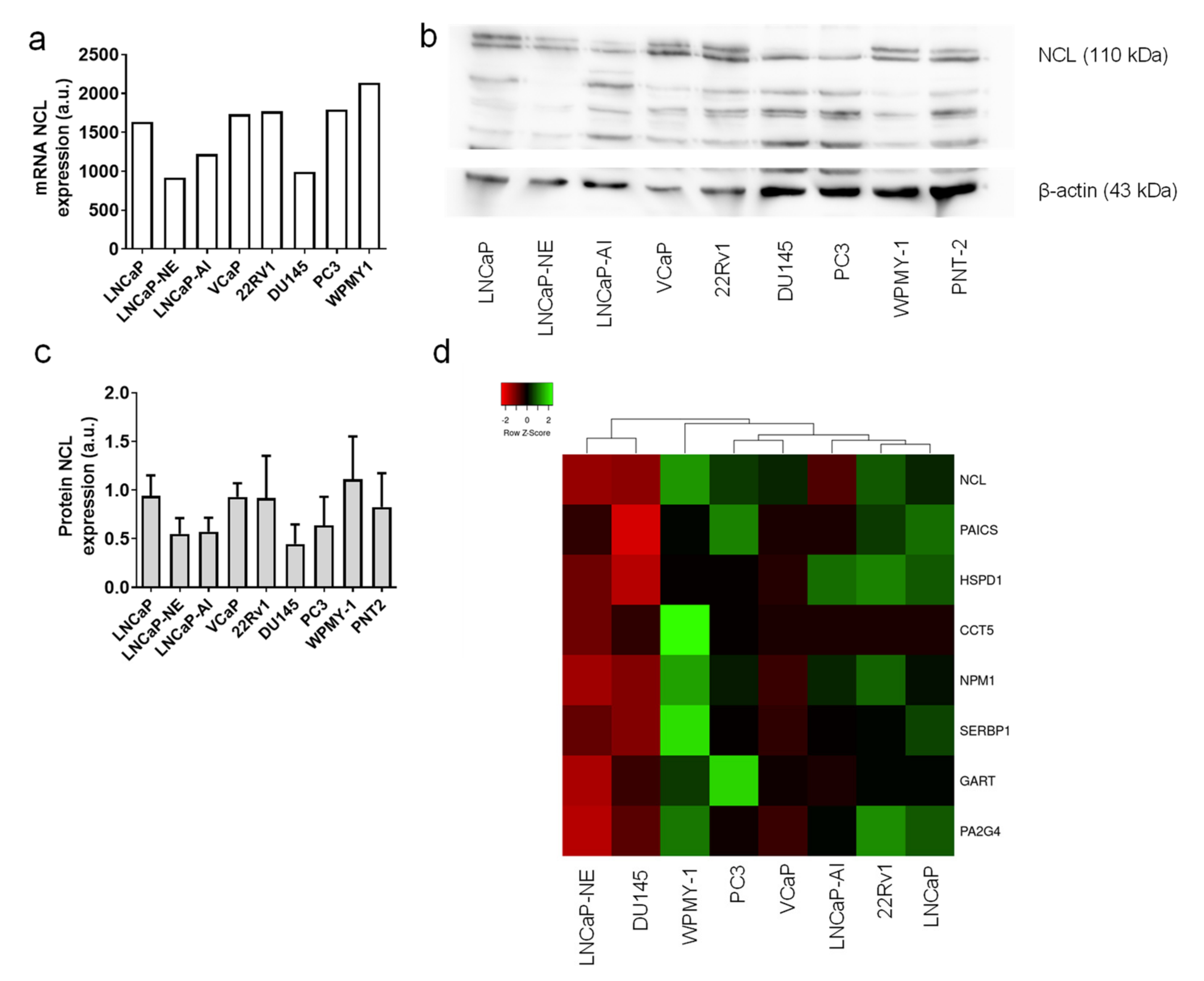
2.5. Tumor Cell Lines Highly Expressed NCL
3. Discussion
4. Materials and Methods
4.1. Human Prostate Cancer Specimens
4.2. RNA Microarray and Transcriptomic Data
4.3. Bioinformatic Analysis
4.4. Human Prostate Cancer Tissues and Immunohistochemistry
4.5. Cell Culture
4.6. Western Blot
4.7. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Tannock, I.F.; de Wit, R.; Berry, W.R.; Horti, J.; Pluzanska, A.; Chi, K.N.; Oudard, S.; Theodore, C.; James, N.D.; Turesson, I.; et al. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N. Engl. J. Med. 2004, 351, 1502–1512. [Google Scholar] [CrossRef]
- Scher, H.I.; Fizazi, K.; Saad, F.; Taplin, M.E.; Sternberg, C.N.; Miller, K.; de Wit, R.; Mulders, P.; Chi, K.N.; Shore, N.D.; et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N. Engl. J. Med. 2012, 367, 1187–1197. [Google Scholar] [CrossRef]
- Ryan, C.J.; Smith, M.R.; de Bono, J.S.; Molina, A.; Logothetis, C.J.; de Souza, P.; Fizazi, K.; Mainwaring, P.; Piulats, J.M.; Ng, S.; et al. Abiraterone in metastatic prostate cancer without previous chemotherapy. N. Engl. J. Med. 2013, 368, 138–148. [Google Scholar] [CrossRef]
- de Bono, J.S.; Oudard, S.; Ozguroglu, M.; Hansen, S.; Machiels, J.P.; Kocak, I.; Gravis, G.; Bodrogi, I.; Mackenzie, M.J.; Shen, L.; et al. Prednisone plus cabazitaxel or mitoxantrone for metastatic castration-resistant prostate cancer progressing after docetaxel treatment: A randomised open-label trial. Lancet 2010, 376, 1147–1154. [Google Scholar] [CrossRef]
- Fizazi, K.; Shore, N.; Tammela, T.L.; Ulys, A.; Vjaters, E.; Polyakov, S.; Jievaltas, M.; Luz, M.; Alekseev, B.; Kuss, I.; et al. Nonmetastatic, Castration-Resistant Prostate Cancer and Survival with Darolutamide. N. Engl. J. Med. 2020, 383, 1040–1049. [Google Scholar] [CrossRef]
- Parker, C.C.; Pascoe, S.; Chodacki, A.; O’Sullivan, J.M.; Germa, J.R.; O’Bryan-Tear, C.G.; Haider, T.; Hoskin, P. A randomized, double-blind, dose-finding, multicenter, phase 2 study of radium chloride (Ra 223) in patients with bone metastases and castration-resistant prostate cancer. Eur. Urol. 2013, 63, 189–197. [Google Scholar] [CrossRef]
- Saad, F.; Cella, D.; Basch, E.; Hadaschik, B.A.; Mainwaring, P.N.; Oudard, S.; Graff, J.N.; McQuarrie, K.; Li, S.; Hudgens, S.; et al. Effect of apalutamide on health-related quality of life in patients with non-metastatic castration-resistant prostate cancer: An analysis of the SPARTAN randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 2018, 19, 1404–1416. [Google Scholar] [CrossRef]
- Sartor, O.; de Bono, J.; Chi, K.N.; Fizazi, K.; Herrmann, K.; Rahbar, K.; Tagawa, S.T.; Nordquist, L.T.; Vaishampayan, N.; El-Haddad, G.; et al. Lutetium-177-PSMA-617 for Metastatic Castration-Resistant Prostate Cancer. N. Engl. J. Med. 2021, 385, 1091–1103. [Google Scholar] [CrossRef]
- Ceder, Y.; Bjartell, A.; Culig, Z.; Rubin, M.A.; Tomlins, S.; Visakorpi, T. The Molecular Evolution of Castration-resistant Prostate Cancer. Eur. Urol. Focus 2016, 2, 506–513. [Google Scholar] [CrossRef]
- Saldana, C.; Majidipur, A.; Beaumont, E.; Huet, E.; de la Taille, A.; Vacherot, F.; Firlej, V.; Destouches, D. Extracellular Vesicles in Advanced Prostate Cancer: Tools to Predict and Thwart Therapeutic Resistance. Cancers 2021, 13, 3791. [Google Scholar] [CrossRef]
- Borer, R.A.; Lehner, C.F.; Eppenberger, H.M.; Nigg, E.A. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell 1989, 56, 379–390. [Google Scholar] [CrossRef]
- Berger, C.M.; Gaume, X.; Bouvet, P. The roles of nucleolin subcellular localization in cancer. Biochimie 2015, 113, 78–85. [Google Scholar] [CrossRef]
- Ugrinova, I.; Petrova, M.; Chalabi-Dchar, M.; Bouvet, P. Multifaceted Nucleolin Protein and Its Molecular Partners in Oncogenesis. Adv. Protein Chem. Struct. Biol. 2018, 111, 133–164. [Google Scholar] [CrossRef]
- Galzio, R.; Rosati, F.; Benedetti, E.; Cristiano, L.; Aldi, S.; Mei, S.; D’Angelo, B.; Gentile, R.; Laurenti, G.; Cifone, M.G.; et al. Glycosilated nucleolin as marker for human gliomas. J. Cell Biochem. 2012, 113, 571–579. [Google Scholar] [CrossRef]
- Christian, S.; Pilch, J.; Akerman, M.E.; Porkka, K.; Laakkonen, P.; Ruoslahti, E. Nucleolin expressed at the cell surface is a marker of endothelial cells in angiogenic blood vessels. J. Cell Biol. 2003, 163, 871–878. [Google Scholar] [CrossRef]
- Gilles, M.E.; Maione, F.; Cossutta, M.; Carpentier, G.; Caruana, L.; Di Maria, S.; Houppe, C.; Destouches, D.; Shchors, K.; Prochasson, C.; et al. Nucleolin Targeting Impairs the Progression of Pancreatic Cancer and Promotes the Normalization of Tumor Vasculature. Cancer Res. 2016, 76, 7181–7193. [Google Scholar] [CrossRef]
- Wise, J.F.; Berkova, Z.; Mathur, R.; Zhu, H.; Braun, F.K.; Tao, R.H.; Sabichi, A.L.; Ao, X.; Maeng, H.; Samaniego, F. Nucleolin inhibits Fas ligand binding and suppresses Fas-mediated apoptosis in vivo via a surface nucleolin-Fas complex. Blood 2013, 121, 4729–4739. [Google Scholar] [CrossRef]
- Di Segni, A.; Farin, K.; Pinkas-Kramarski, R. Identification of nucleolin as new ErbB receptors- interacting protein. PLoS ONE 2008, 3, e2310. [Google Scholar] [CrossRef]
- Farin, K.; Schokoroy, S.; Haklai, R.; Cohen-Or, I.; Elad-Sfadia, G.; Reyes-Reyes, M.E.; Bates, P.J.; Cox, A.D.; Kloog, Y.; Pinkas-Kramarski, R. Oncogenic synergism between ErbB1, nucleolin, and mutant Ras. Cancer Res. 2011, 71, 2140–2151. [Google Scholar] [CrossRef]
- Schokoroy, S.; Juster, D.; Kloog, Y.; Pinkas-Kramarski, R. Disrupting the oncogenic synergism between nucleolin and Ras results in cell growth inhibition and cell death. PLoS ONE 2013, 8, e75269. [Google Scholar] [CrossRef]
- Shibata, Y.; Muramatsu, T.; Hirai, M.; Inui, T.; Kimura, T.; Saito, H.; McCormick, L.M.; Bu, G.; Kadomatsu, K. Nuclear targeting by the growth factor midkine. Mol. Cell Biol. 2002, 22, 6788–6796. [Google Scholar] [CrossRef]
- Said, E.A.; Courty, J.; Svab, J.; Delbe, J.; Krust, B.; Hovanessian, A.G. Pleiotrophin inhibits HIV infection by binding the cell surface-expressed nucleolin. FEBS J. 2005, 272, 4646–4659. [Google Scholar] [CrossRef]
- Huang, Y.; Shi, H.; Zhou, H.; Song, X.; Yuan, S.; Luo, Y. The angiogenic function of nucleolin is mediated by vascular endothelial growth factor and nonmuscle myosin. Blood 2006, 107, 3564–3571. [Google Scholar] [CrossRef]
- Tawfic, S.; Goueli, S.A.; Olson, M.O.; Ahmed, K. Androgenic regulation of phosphorylation and stability of nucleolar protein nucleolin in rat ventral prostate. Prostate 1994, 24, 101–106. [Google Scholar] [CrossRef]
- Drabik, A.; Ciolczyk-Wierzbicka, D.; Dulinska-Litewka, J.; Bodzon-Kulakowska, A.; Suder, P.; Silberring, J.; Laidler, P. A comparative study of glycoproteomes in androgen-sensitive and -independent prostate cancer cell lines. Mol. Cell Biochem. 2014, 386, 189–198. [Google Scholar] [CrossRef][Green Version]
- Tate, A.; Isotani, S.; Bradley, M.J.; Sikes, R.A.; Davis, R.; Chung, L.W.; Edlund, M. Met-Independent Hepatocyte Growth Factor-mediated regulation of cell adhesion in human prostate cancer cells. BMC Cancer 2006, 6, 197. [Google Scholar] [CrossRef]
- Shin, S.H.; Lee, G.Y.; Lee, M.; Kang, J.; Shin, H.W.; Chun, Y.S.; Park, J.W. Aberrant expression of CITED2 promotes prostate cancer metastasis by activating the nucleolin-AKT pathway. Nat. Commun. 2018, 9, 4113. [Google Scholar] [CrossRef]
- Sheetz, T.; Mills, J.; Tessari, A.; Pawlikowski, M.; Braddom, A.E.; Posid, T.; Zynger, D.L.; James, C.; Embrione, V.; Parbhoo, K.; et al. NCL Inhibition Exerts Antineoplastic Effects against Prostate Cancer Cells by Modulating Oncogenic MicroRNAs. Cancers 2020, 12, 1861. [Google Scholar] [CrossRef]
- Girvan, A.C.; Teng, Y.; Casson, L.K.; Thomas, S.D.; Juliger, S.; Ball, M.W.; Klein, J.B.; Pierce, W.M., Jr.; Barve, S.S.; Bates, P.J. AGRO100 inhibits activation of nuclear factor-kappaB (NF-kappaB) by forming a complex with NF-kappaB essential modulator (NEMO) and nucleolin. Mol. Cancer Ther. 2006, 5, 1790–1799. [Google Scholar] [CrossRef]
- Reyes-Reyes, E.M.; Teng, Y.; Bates, P.J. A new paradigm for aptamer therapeutic AS1411 action: Uptake by macropinocytosis and its stimulation by a nucleolin-dependent mechanism. Cancer Res. 2010, 70, 8617–8629. [Google Scholar] [CrossRef]
- Taghdisi, S.M.; Danesh, N.M.; Ramezani, M.; Yazdian-Robati, R.; Abnous, K. A Novel AS1411 Aptamer-Based Three-Way Junction Pocket DNA Nanostructure Loaded with Doxorubicin for Targeting Cancer Cells in Vitro and in Vivo. Mol. Pharm. 2018, 15, 1972–1978. [Google Scholar] [CrossRef]
- Yang, S.; Ren, Z.; Chen, M.; Wang, Y.; You, B.; Chen, W.; Qu, C.; Liu, Y.; Zhang, X. Nucleolin-Targeting AS1411-Aptamer-Modified Graft Polymeric Micelle with Dual pH/Redox Sensitivity Designed To Enhance Tumor Therapy through the Codelivery of Doxorubicin/TLR4 siRNA and Suppression of Invasion. Mol. Pharm. 2018, 15, 314–325. [Google Scholar] [CrossRef]
- Xu, G.; Qin, M.; Mukundan, A.; Siddiqui, J.; Takada, M.; Vilar-Saavedra, P.; Tomlins, S.A.; Kopelman, R.; Wang, X. Prostate cancer characterization by optical contrast enhanced photoacoustics. Proc. SPIE Int. Soc. Opt. Eng. 2016, 9708, 127–132. [Google Scholar] [CrossRef]
- Hamma-Kourbali, Y.; Bermek, O.; Bernard-Pierrot, I.; Karaky, R.; Martel-Renoir, D.; Frechault, S.; Courty, J.; Delbe, J. The synthetic peptide P111-136 derived from the C-terminal domain of heparin affin regulatory peptide inhibits tumour growth of prostate cancer PC-3 cells. BMC Cancer 2011, 11, 212. [Google Scholar] [CrossRef]
- Destouches, D.; El Khoury, D.; Hamma-Kourbali, Y.; Krust, B.; Albanese, P.; Katsoris, P.; Guichard, G.; Briand, J.P.; Courty, J.; Hovanessian, A.G. Suppression of tumor growth and angiogenesis by a specific antagonist of the cell-surface expressed nucleolin. PLoS ONE 2008, 3, e2518. [Google Scholar] [CrossRef]
- Destouches, D.; Page, N.; Hamma-Kourbali, Y.; Machi, V.; Chaloin, O.; Frechault, S.; Birmpas, C.; Katsoris, P.; Beyrath, J.; Albanese, P.; et al. A simple approach to cancer therapy afforded by multivalent pseudopeptides that target cell-surface nucleoproteins. Cancer Res. 2011, 71, 3296–3305. [Google Scholar] [CrossRef]
- Grinstein, E.; Wernet, P.; Snijders, P.J.; Rosl, F.; Weinert, I.; Jia, W.; Kraft, R.; Schewe, C.; Schwabe, M.; Hauptmann, S.; et al. Nucleolin as activator of human papillomavirus type 18 oncogene transcription in cervical cancer. J. Exp. Med. 2002, 196, 1067–1078. [Google Scholar] [CrossRef]
- Ridley, L.; Rahman, R.; Brundler, M.A.; Ellison, D.; Lowe, J.; Robson, K.; Prebble, E.; Luckett, I.; Gilbertson, R.J.; Parkes, S.; et al. Multifactorial analysis of predictors of outcome in pediatric intracranial ependymoma. Neuro Oncol. 2008, 10, 675–689. [Google Scholar] [CrossRef]
- Peng, L.; Liang, J.; Wang, H.; Song, X.; Rashid, A.; Gomez, H.F.; Corley, L.J.; Abbruzzese, J.L.; Fleming, J.B.; Evans, D.B.; et al. High levels of nucleolar expression of nucleolin are associated with better prognosis in patients with stage II pancreatic ductal adenocarcinoma. Clin. Cancer Res. 2010, 16, 3734–3742. [Google Scholar] [CrossRef]
- Qiu, W.; Zhou, F.; Zhang, Q.; Sun, X.; Shi, X.; Liang, Y.; Wang, X.; Yue, L. Overexpression of nucleolin and different expression sites both related to the prognosis of gastric cancer. APMIS 2013, 121, 919–925. [Google Scholar] [CrossRef]
- Wu, D.M.; Zhang, P.; Liu, R.Y.; Sang, Y.X.; Zhou, C.; Xu, G.C.; Yang, J.L.; Tong, A.P.; Wang, C.T. Phosphorylation and changes in the distribution of nucleolin promote tumor metastasis via the PI3K/Akt pathway in colorectal carcinoma. FEBS Lett. 2014, 588, 1921–1929. [Google Scholar] [CrossRef]
- Guo, X.; Xiong, L.; Yu, L.; Li, R.; Wang, Z.; Ren, B.; Dong, J.; Li, B.; Wang, D. Increased level of nucleolin confers to aggressive tumor progression and poor prognosis in patients with hepatocellular carcinoma after hepatectomy. Diagn. Pathol. 2014, 9, 175. [Google Scholar] [CrossRef]
- Lin, Q.; Ma, X.; Hu, S.; Li, R.; Wei, X.; Han, B.; Ma, Y.; Liu, P.; Pang, Y. Overexpression of Nucleolin is a Potential Prognostic Marker in Endometrial Carcinoma. Cancer Manag. Res. 2021, 13, 1955–1965. [Google Scholar] [CrossRef]
- Masiuk, M.; Lewandowska, M.; Dobak, E.; Urasinska, E. Nucleolin and Nucleophosmin Expression in Gleason 3 and Gleason 4 Prostate Cancer With Seminal Vesicles Invasion (pT3b). Anticancer Res. 2020, 40, 1973–1979. [Google Scholar] [CrossRef]
- Kane, C.J.; Eggener, S.E.; Shindel, A.W.; Andriole, G.L. Variability in Outcomes for Patients with Intermediate-risk Prostate Cancer (Gleason Score 7, International Society of Urological Pathology Gleason Group 2–3) and Implications for Risk Stratification: A Systematic Review. Eur. Urol. Focus 2017, 3, 487–497. [Google Scholar] [CrossRef]
- Fujita, K.; Pavlovich, C.P.; Netto, G.J.; Konishi, Y.; Isaacs, W.B.; Ali, S.; De Marzo, A.; Meeker, A.K. Specific detection of prostate cancer cells in urine by multiplex immunofluorescence cytology. Hum. Pathol. 2009, 40, 924–933. [Google Scholar] [CrossRef]
- Castilla, C.; Congregado, B.; Conde, J.M.; Medina, R.; Torrubia, F.J.; Japon, M.A.; Saez, C. Immunohistochemical expression of Hsp60 correlates with tumor progression and hormone resistance in prostate cancer. Urology 2010, 76, 1017.e1–1017.e6. [Google Scholar] [CrossRef]
- Chakravarthi, B.V.; Goswami, M.T.; Pathi, S.S.; Dodson, M.; Chandrashekar, D.S.; Agarwal, S.; Nepal, S.; Hodigere Balasubramanya, S.A.; Siddiqui, J.; Lonigro, R.J.; et al. Expression and Role of PAICS, a De Novo Purine Biosynthetic Gene in Prostate Cancer. Prostate 2017, 77, 10–21. [Google Scholar] [CrossRef]
- Guo, K.; Zheng, S.; Xu, Y.; Xu, A.; Chen, B.; Wen, Y. Loss of miR-26a-5p promotes proliferation, migration, and invasion in prostate cancer through negatively regulating SERBP1. Tumour Biol. 2016, 37, 12843–12854. [Google Scholar] [CrossRef]
- Zhou, H.; Zhang, Y.; Hamburger, A.W. EBP1 inhibits translation of androgen receptor mRNA in castration resistant prostate cancer cells. Anticancer Res. 2011, 31, 3129–3135. [Google Scholar]
- Destouches, D.; Sader, M.; Terry, S.; Marchand, C.; Maille, P.; Soyeux, P.; Carpentier, G.; Semprez, F.; Ceraline, J.; Allory, Y.; et al. Implication of NPM1 phosphorylation and preclinical evaluation of the nucleoprotein antagonist N6L in prostate cancer. Oncotarget 2016, 7, 69397–69411. [Google Scholar] [CrossRef]
- Fogal, V.; Sugahara, K.N.; Ruoslahti, E.; Christian, S. Cell surface nucleolin antagonist causes endothelial cell apoptosis and normalization of tumor vasculature. Angiogenesis 2009, 12, 91–100. [Google Scholar] [CrossRef]
- Bates, P.J.; Laber, D.A.; Miller, D.M.; Thomas, S.D.; Trent, J.O. Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp. Mol. Pathol. 2009, 86, 151–164. [Google Scholar] [CrossRef]
- Pichiorri, F.; Palmieri, D.; De Luca, L.; Consiglio, J.; You, J.; Rocci, A.; Talabere, T.; Piovan, C.; Lagana, A.; Cascione, L.; et al. In vivo NCL targeting affects breast cancer aggressiveness through miRNA regulation. J. Exp. Med. 2013, 210, 951–968. [Google Scholar] [CrossRef]
- Yazdian-Robati, R.; Bayat, P.; Oroojalian, F.; Zargari, M.; Ramezani, M.; Taghdisi, S.M.; Abnous, K. Therapeutic applications of AS1411 aptamer, an update review. Int. J. Biol. Macromol. 2020, 155, 1420–1431. [Google Scholar] [CrossRef]
- Reyes-Reyes, E.M.; Salipur, F.R.; Shams, M.; Forsthoefel, M.K.; Bates, P.J. Mechanistic studies of anticancer aptamer AS1411 reveal a novel role for nucleolin in regulating Rac1 activation. Mol. Oncol. 2015, 9, 1392–1405. [Google Scholar] [CrossRef]
- Chalfin, H.J.; Verdone, J.E.; van der Toom, E.E.; Glavaris, S.; Gorin, M.A.; Pienta, K.J. Nucleolin Staining May Aid in the Identification of Circulating Prostate Cancer Cells. Clin. Genitourin. Cancer 2017, 15, e477–e481. [Google Scholar] [CrossRef]
- Pinskaya, M.; Saci, Z.; Gallopin, M.; Gabriel, M.; Nguyen, H.T.; Firlej, V.; Descrimes, M.; Rapinat, A.; Gentien, D.; Taille, A.; et al. Reference-free transcriptome exploration reveals novel RNAs for prostate cancer diagnosis. Life Sci. Alliance 2019, 2. [Google Scholar] [CrossRef]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repecka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef]
- Chapat, C.; Chettab, K.; Simonet, P.; Wang, P.; De La Grange, P.; Le Romancer, M.; Corbo, L. Alternative splicing of CNOT7 diversifies CCR4-NOT functions. Nucleic Acids Res. 2017, 45, 8508–8523. [Google Scholar] [CrossRef]
- Vallerand, D.; Massonnet, G.; Kebir, F.; Gentien, D.; Maciorowski, Z.; De la Grange, P.; Sigal-Zafrani, B.; Richardson, M.; Humbert, S.; Thuleau, A.; et al. Characterization of Breast Cancer Preclinical Models Reveals a Specific Pattern of Macrophage Polarization. PLoS ONE 2016, 11, e0157670. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009, 37, 1–13. [Google Scholar] [CrossRef]
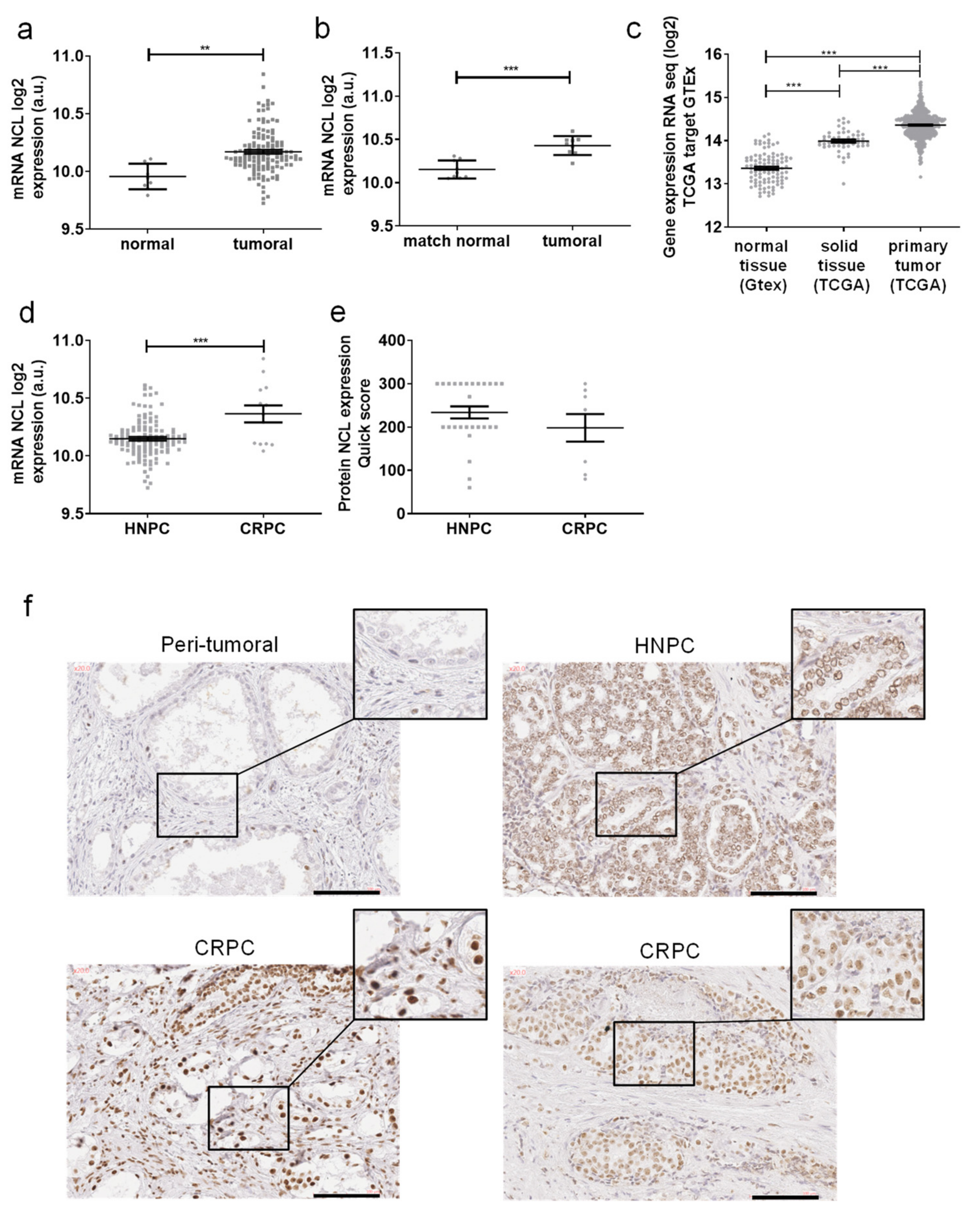
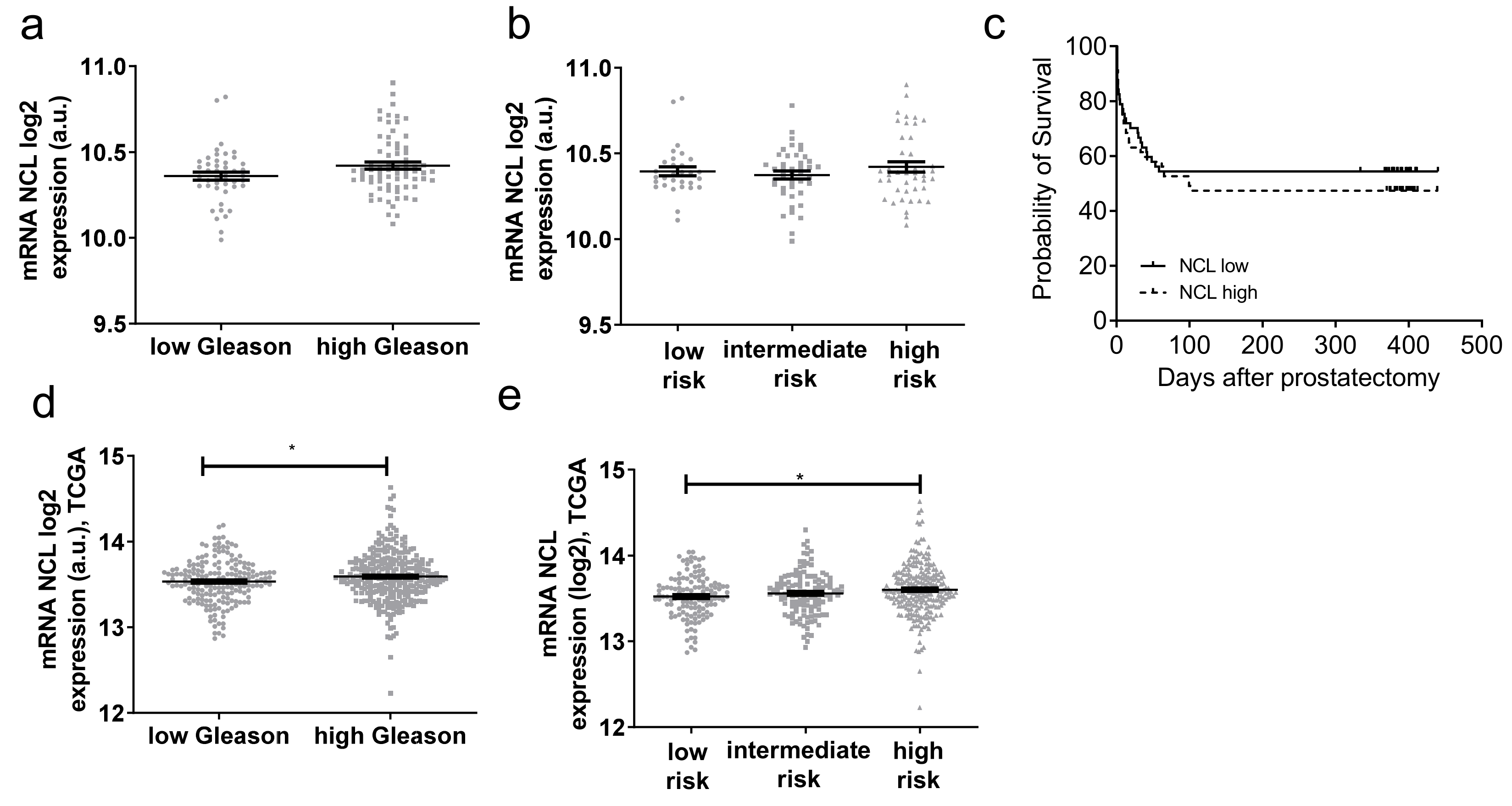
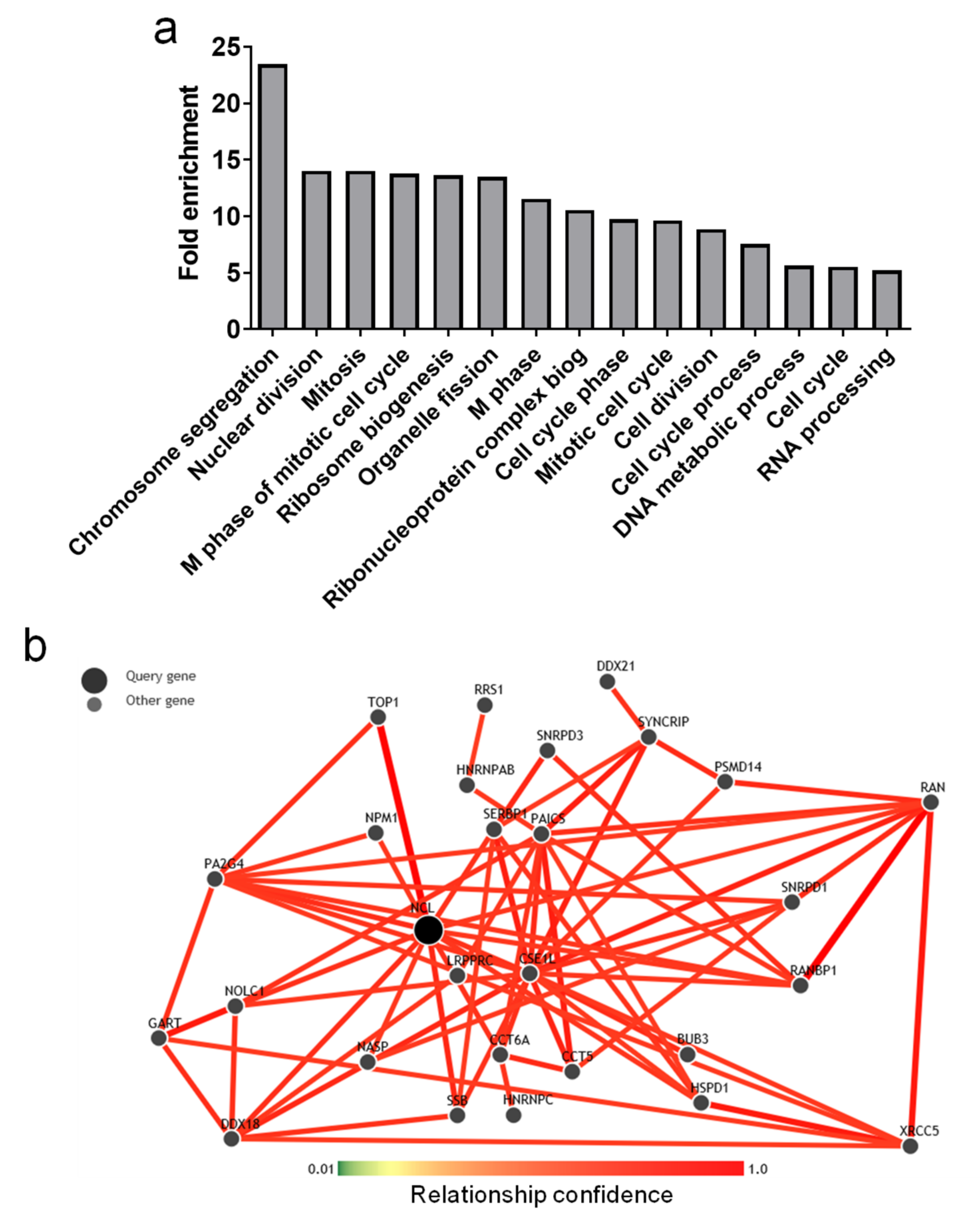
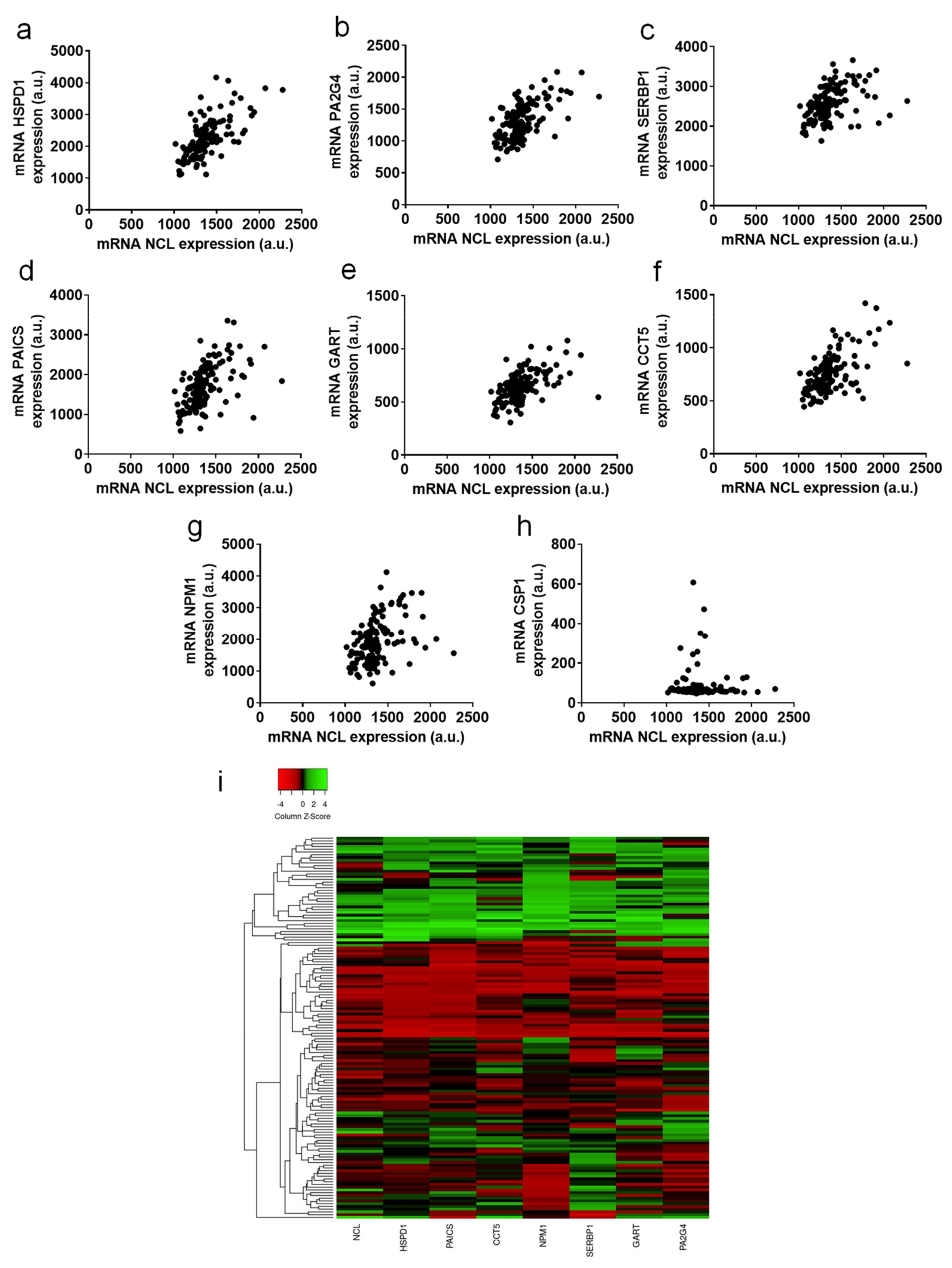
| Characteristic | Mean/Median (Range) | No. of Patients (%) |
|---|---|---|
| Age, y | 62.87/63 (44–76) | |
| PSA, ng/mL | 11.16/7 (1.7–76) | |
| Follow-up n = 113, mo | 99.3/105.7 (1.4–173.9) | |
| Pathological tumor classification | ||
| pT2a/b | 9 (7.7%) | |
| pT2c | 28 (24.1%) | |
| pT3a | 54 (46.5%) | |
| pT3b | 19 (16.3%) | |
| pT4 | 6 (5.1%) | |
| Prostatectomy gleason score | ||
| 6 | 20 (17.2%) | |
| 7 (3 + 4) | 25 (21.5%) | |
| 7 (4 + 3) | 40 (34.5%) | |
| 8 | 26 (22.4%) | |
| 9 | 5 (4.3%) | |
| Relapse n = 113 | ||
| yes | 56 | |
| no | 57 |
| TCGA | PAIR Prostate | GIANT | ||||
|---|---|---|---|---|---|---|
| Correlated Gene | Cytoband | Spearman’s Correlation | p-Value | Spearman’s Correlation | p-Value | Score |
| HSPD1 | 2q33.1 | 0.6890 | 5.62 x10-70 | 0.6844 | <0.0001 | 0.668 |
| PAICS | 4q12 | 0.6780 | 5.62 x10-67 | 0.5860 | <0.0001 | 0.595 |
| CCT5 | 5p15.2 | 0.6327 | 5.88 x10-56 | 0.5402 | <0.0001 | 0.558 |
| NPM1 | 5q35.1 | 0.6087 | 8.06 x10-51 | 0.4385 | <0.0001 | 0.657 |
| SERBP1 | 1p31.3 | 0.5227 | 1.40 x10-35 | 0.5038 | <0.0001 | 0.691 |
| GART | 21q22.11 | 0.5145 | 2.45 x10--34 | 0.5501 | <0.0001 | 0.534 |
| PA2G4 | 12q13.2 | 0.5054 | 5.20 x10--33 | 0.5700 | <0.0001 | 0.708 |
| PAICS | 4q12 | 0.6780 | 5.62 x10--67 | 0.5860 | <0.0001 | 0.595 |
| CCT5 | 5p15.2 | 0.6327 | 5.88 x10--56 | 0.5402 | <0.0001 | 0.558 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Firlej, V.; Soyeux, P.; Nourieh, M.; Huet, E.; Semprez, F.; Allory, Y.; Londono-Vallejo, A.; de la Taille, A.; Vacherot, F.; Destouches, D. Overexpression of Nucleolin and Associated Genes in Prostate Cancer. Int. J. Mol. Sci. 2022, 23, 4491. https://doi.org/10.3390/ijms23094491
Firlej V, Soyeux P, Nourieh M, Huet E, Semprez F, Allory Y, Londono-Vallejo A, de la Taille A, Vacherot F, Destouches D. Overexpression of Nucleolin and Associated Genes in Prostate Cancer. International Journal of Molecular Sciences. 2022; 23(9):4491. https://doi.org/10.3390/ijms23094491
Chicago/Turabian StyleFirlej, Virginie, Pascale Soyeux, Maya Nourieh, Eric Huet, Fannie Semprez, Yves Allory, Arturo Londono-Vallejo, Alexandre de la Taille, Francis Vacherot, and Damien Destouches. 2022. "Overexpression of Nucleolin and Associated Genes in Prostate Cancer" International Journal of Molecular Sciences 23, no. 9: 4491. https://doi.org/10.3390/ijms23094491
APA StyleFirlej, V., Soyeux, P., Nourieh, M., Huet, E., Semprez, F., Allory, Y., Londono-Vallejo, A., de la Taille, A., Vacherot, F., & Destouches, D. (2022). Overexpression of Nucleolin and Associated Genes in Prostate Cancer. International Journal of Molecular Sciences, 23(9), 4491. https://doi.org/10.3390/ijms23094491







