Circular RNAs in Pregnancy and the Placenta
Abstract
:1. Introduction
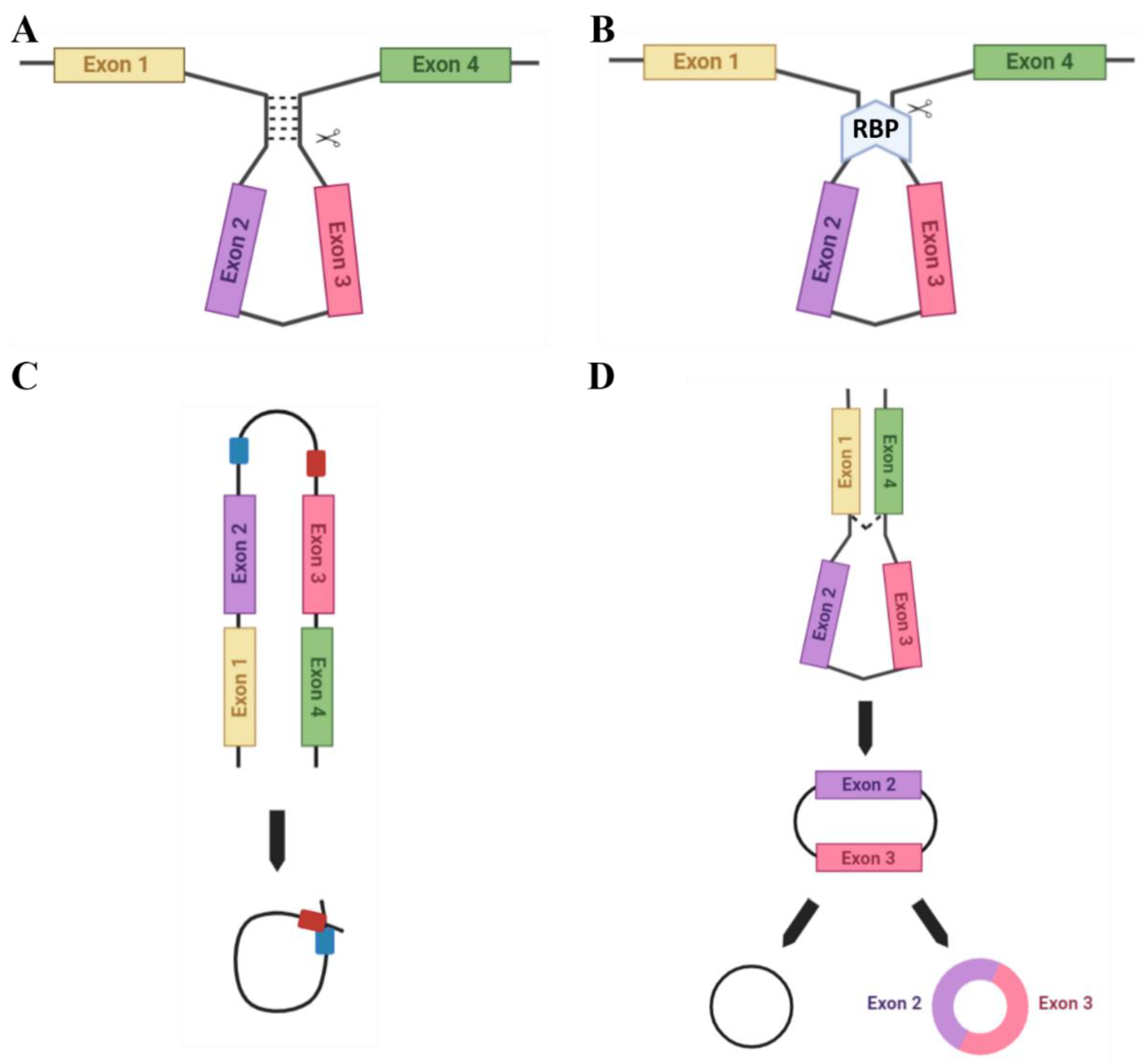
2. circRNA Biogenesis
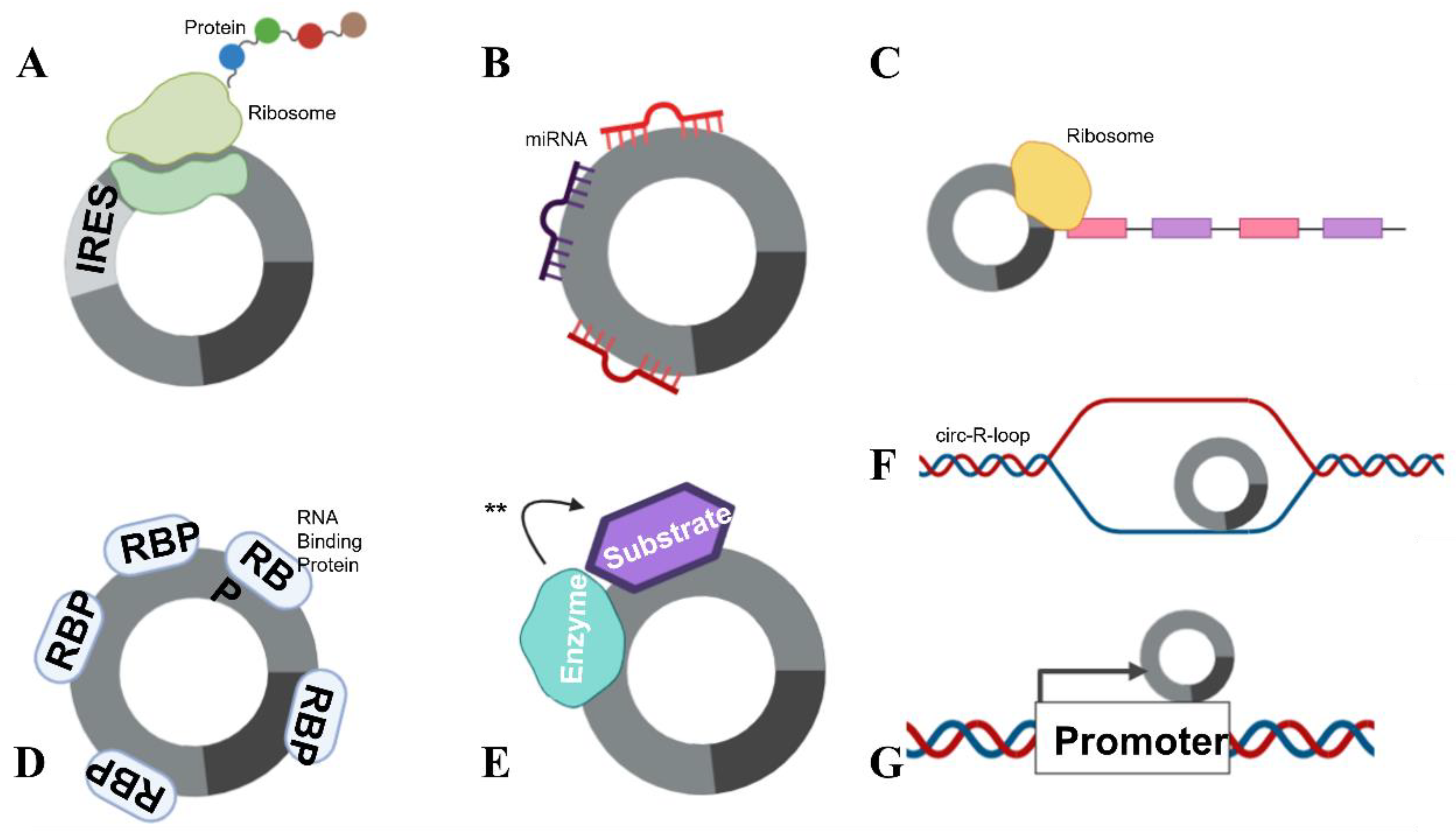
3. circRNA Function
4. The Role of circRNAs in Pregnancy
4.1. circRNAs in Preeclampsia
4.2. circRNAs in Gestational Diabetes Mellitus
4.3. circRNAs in Other Pregnancy Complications
4.4. Limitations of circRNA Research
5. The Potential Importance of circRNAs in the Placenta: What We Can Apply from Our Knowledge of Cancer
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sõber, S.; Reiman, M.; Kikas, T.; Rull, K.; Inno, R.; Vaas, P.; Teesalu, P.; Marti, J.M.L.M.; Pirkko, M.; Laan, M. Extensive shift in placental transcriptome profile in preeclampsia and placental origin of adverse pregnancy outcomes. Sci. Rep. 2015, 5, 13336. [Google Scholar] [CrossRef] [PubMed]
- Majewska, M.; Lipka, A.; Paukszto, L.; Jastrzebski, J.; Szeszko, K.; Gowkielewicz, M.; Lepiarczyk, E.; Jozwik, M.; Majewski, M. Placenta Transcriptome Profiling in Intrauterine Growth Restriction (IUGR). Int. J. Mol. Sci. 2019, 20, 1510. [Google Scholar] [CrossRef] [PubMed]
- Rizzetto, M.; Canese, M.; Arico, S.; Crivelli, O.; Trepo, C.; Bonino, F.; Verme, G. Immunofluorescence detection of new antigen-antibody system (delta/antidelta) associated to hepatitis B virus in liver and in serum of HBsAg carriers. Gut 1977, 18, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Hsu, M.; Coca-Prados, M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 1979, 280, 339–340. [Google Scholar] [CrossRef] [PubMed]
- Nigro, J.; Cho, K.; Fearon, E.; Kern, S.; Ruppert, J.; Oliner, J.; Kinzler, K.; Vogelstein, B. Scrambled exons. Cell 1991, 64, 607–613. [Google Scholar] [CrossRef]
- Ozsolak, F.; Milos, P. RNA sequencing: Advances, challenges and opportunities. Nat. Rev. Genet. 2011, 12, 87–98. [Google Scholar] [CrossRef]
- Jeck, W.; Sharpless, N. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014, 32, 423–461. [Google Scholar] [CrossRef]
- Salzman, J.; Gawad, C.; Wang, P.; Lacayo, N.; Brown, P. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef]
- Guo, J.; Agarwal, V.; Guo, H.; Bartel, D. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014, 15, 409. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.; Gregersen, L.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Chen, L.; Yang, L. Regulation of circRNA biogenesis. RNA Biol. 2015, 12, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.; Jensen, T.; Clausen, B.; Bramsen, J.; Finsen, B.; Damgaard, C. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Floris, G.; Zhang, L.; Follesa, P.; Sun, T. Regulatory Role of Circular RNAs and Neurological Disorders. Mol. Neurobiol. 2017, 54, 5156–5165. [Google Scholar] [CrossRef]
- Starke, S.; Jost, I.; Rossbach, O.; Schneider, T.; Schreiner, S.; Hung, L.; Bindereif, A. Exon circularization requires canonical splice signals. Cell Rep. 2015, 10, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 2014, 56, 55–56. [Google Scholar] [CrossRef] [PubMed]
- Quan, G.; Li, J. Circular RNAs: Biogenesis, expression and their potential roles in reproduction. J. Ovarian Res. 2018, 11, 9. [Google Scholar] [CrossRef]
- Chen, I.; Chen, C.; Chuang, T. Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip. Rev. RNA 2015, 6, 563–579. [Google Scholar] [CrossRef]
- Conn, S.J.; Pillman, K.A.; Toubia, J.; Conn, V.M.; Salmanidis, M.; Phillips, C.A.; Roslan, S.S.; Andreas, W.; Gregory, P.A.; Goodall, G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015, 160, 1125–1134. [Google Scholar] [CrossRef]
- Ebbesen, K.; Hansen, T.; Kjems, J. Insights into circular RNA biology. RNA Biol. 2017, 14, 1035–1045. [Google Scholar] [CrossRef]
- De La Mata, M.; Alonso, C.; Kadener, S.; Fededa, J.; Blaustein, M.; Pelisch, F.; Cramer, P.; Bentley, D.; Kornblihtt, A. A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell 2003, 2, 525–532. [Google Scholar] [CrossRef]
- Ip, J.; Schmidt, D.; Pan, Q.; Ramani, A.; Fraser, A.; Odom, D.; Blencowe, B. Global impact of RNA polymerase II elongation inhibition on alternative splicing regulation. Genome Res. 2011, 21, 390–401. [Google Scholar] [CrossRef] [PubMed]
- Khodor, Y.; Rodriguez, J.; Abruzzi, K.; Tang, C.; Marr, M.; Rosbash, M. Nascent-seq indicates widespread cotranscriptional pre-mRNA splicing in drosophila. Genes Dev. 2011, 23, 2502–2512. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, X.O.; Chen, T.; Xiang, J.F.; Yin, Q.F.; Xing, Y.H.; Zhu, S.; Yang, L.; Chen, L.L. Circular intronic long noncoding RNAs. Mol. Cell 2013, 51, 792–806. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, A.; Memczak, S.; Wyler, E.; Torti, F.; Porath, H.; Orejuela, M.; Piechotta, M.; Levanon, E.; Landthaler, M.; Dieterich, C.; et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015, 10, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Jeck, W.; Sorrentino, J.; Wang, K.; Slevin, M.; Burd, C.; Liu, J. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.; Linton, L.; Birren, B.; Nusbaum, C.; Zody, M.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef]
- Capel, B.; Swain, A.; Nicolis, S.; Hacker, A.; Walter, M.; Koopman, P.; Goodfellow, P.; Lovell-Badge, R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 1993, 73, 1019–1030. [Google Scholar] [CrossRef]
- Qu, S.; Yang, X.; Li, X.; Wang, J.; Gao, Y.; Shang, R.; Sun, W. Circular RNA: A new star of non-coding RNAs. Cancer Lett. 2015, 365, 141–148. [Google Scholar] [CrossRef]
- Mumtaz, P.; Taban, Q.; Dar, M.; Mir, S.; Haq, Z.; Zargar, S.; Shah, R.; Ahmad, S. Deep Insights in Circular RNAs: From biogenesis to therapeutics. Biol. Proced. Online 2020, 22, 10. [Google Scholar] [CrossRef]
- Kos, A.; Dijkema, R.; Arnberg, A.; van der Meide, P.; Schellekens, H. The hepatitis delta (delta) virus possesses a circular RNA. Nature 1986, 323, 558–560. [Google Scholar] [CrossRef]
- Hansen, T.; Wiklund, E.; Bramsen, J.; Villadsen, S.; Statham, A.; Clark, S.; Kjems, J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011, 30, 4414–4422. [Google Scholar] [CrossRef] [PubMed]
- Jens, M.; Rajewsky, N. Competition between target sites of regulators shapes post-transcriptional gene regulation. Nat. Rev. Genet. 2015, 16, 113–126. [Google Scholar] [CrossRef]
- Bissels, U.; Wild, S.; Tomiuk, S.; Holste, A.; Hafner, M.; Tuschl, T.; Bosio, A. Absolute quantification of microRNAs by using a universal reference. RNA 2009, 15, 2375–2384. [Google Scholar] [CrossRef] [PubMed]
- Chao, C.; Chan, D.; Kuo, A.; Leder, P. The mouse formin (Fmn) gene: Abundant circular RNA transcripts and gene-targeted deletion analysis. Mol. Med. 1998, 4, 614–628. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Zhang, X.; Wu, X.; Guo, H.; Hu, Y.; Tang, Y.; Huang, Y. Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome Biol. 2015, 16, 148. [Google Scholar] [CrossRef] [PubMed]
- Szabo, L.; Morey, R.; Palpant, N.; Wang, P.; Afari, N.; Jiang, C.; Parast, M.; Murry, C.; Laurent, L.; Salzman, J. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 2015, 16, 126. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, H.; Long, Y.; Li, W. Circular RNA in blood corpuscles combined with plasma protein factor for early prediction of pre-eclampsia. BJOG Int. J. Obstet. Gynaecol. 2016, 123, 2113–2118. [Google Scholar] [CrossRef]
- Qian, Y.; Lu, Y.; Rui, C.; Qian, Y.; Cai, M.; Jia, R. Potential significance of circular RNA in human placental tissue for patients with preeclampsia. Cell Physiol. Biochem. 2016, 39, 1380–1390. [Google Scholar] [CrossRef]
- Qian, Y.; Wang, X.; Ruan, H.; Rui, C.; Mao, P.; Cheng, Q.; Jia, R. Circular RNAs expressed in chorionic villi are probably involved in the occurrence of recurrent spontaneous abortion. Biomed. Pharmacother. 2017, 88, 1154–1162. [Google Scholar] [CrossRef]
- Cheng, J.; Huang, J.; Yuan, S.; Zhou, S.; Yan, W.; Shen, W. Circular RNA expression profiling of human granulosa cells during maternal aging reveals novel transcripts associated with assisted reproductive technology outcomes. PLoS ONE 2017, 12, e0177888. [Google Scholar] [CrossRef]
- Zhang, S.; Ding, Y.; He, J.; Zhang, J.; Liu, X.; Chen, X.; Su, Y.; Wang, Y.; Gao, R. Altered expression patterns of circular RNAs between implantation sites and interimplantation sites in early pregnant mice. J. Cell Physiol. 2018, 234, 9862–9872. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Rao, H.; Chen, W.; Luo, X.; Tong, C.; Qi, H. Profiles of circular RNAs in human placenta and their potential roles related to preeclampsia. Biol. Reprod. 2018, 98, 705–712. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Lash, G.; Zhao, X.; Long, Y.; Guo, C.; Yang, H. CircRNA-0004904, CircRNA-0001855, and PAPP-A: Potential Novel Biomarkers for the Prediction of Preeclampsia. Cell Physiol. Biochem. 2018, 46, 2576–2586. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Wang, H.; Wu, X.; Long, W.; Zheng, F.; Kong, J.; Yu, B. The profile analysis of circular RNAs in human placenta of preeclampsia. Exp. Biol. Med. 2018, 243, 1109–1117. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Ao, J.; Li, X.; Zhang, H.; Wu, J.; Cheng, W. Competing endogenous RNA expression profiling in pre-eclampsia identifies hsa_circ_0036877 as a potential novel blood biomarker for early pre-eclampsia. Clin. Epigenet. 2018, 10, 48. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Zheng, L.; Huang, J.; Kong, H.; Chang, Y.; Wang, F.; Xin, H. CircTRNC18 inhibits trophoblast cell migration and epithelial–mesenchymal transition by regulating miR-762/Grhl2 pathway in pre-eclampsia. RNA Biol. 2019, 16, 1563–1573. [Google Scholar] [CrossRef]
- Wang, H.; She, G.; Zhou, W.; Liu, K.; Miao, J.; Yu, B. Expression profile of circular RNAs in placentas of women with gestational diabetes mellitus. Endocr. J. 2019, 22, 431–441. [Google Scholar] [CrossRef]
- Cao, M.; Zhang, L.; Lin, Y.; Li, Z.; Xu, J.; Shi, Z.; Chen, Z.; Ma, J.; Wen, J. Circular RNA expression profiles in umbilical cord blood exosomes from normal and gestational diabetes mellitus patients. Biosci. Rep. 2020, 40, BSR20201946. [Google Scholar] [CrossRef]
- Oudejans, C.; Manders, V.; Visser, A.; Keijser, R.; Min, N.; Poutsma, A.; Mulders, J.; van den Berkmortel, T.; Wigman, D.-J.; Blanken, B. Circular RNA sequencing of maternal platelets: A novel tool for the identification of pregnancy-specific biomarkers. Clin. Chem. 2021, 67, 508–517. [Google Scholar] [CrossRef]
- Yang, H.; Ye, W.; Chen, R.; Zeng, F.; Long, Y.Z.; Zhang, X.; Ma, J.; Gan, Q.; Rehemutula, R.; Zhu, C. Circulating expression of Hsa_circRNA_102893 contributes to early gestational diabetes mellitus detection. Sci. Rep. 2020, 10, 19046. [Google Scholar] [CrossRef]
- Wang, D.; Na, Q.; Song, G.; Wang, Y.; Wang, Y. The Role of circRNA-SETD2/miR-519a/PTEN Axis in Fetal Birth Weight through Regulating Trophoblast Proliferation. BioMed Res. Int. 2020, 2020, 9809632. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhou, W.; She, G.; Yu, B.; Sun, L. Downregulation of hsa_circ_00052.43 induces trophoblast cell dysfunction and inflammation via the β-catenin and NF-κB pathways. Reprod. Biol. Endocrinol. 2020, 18, 51. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, H.; Zhang, Y.; Shi, J.; Chen, R.; Xiao, X. CircSFXN1 regulates the behaviour of trophoblasts and likely mediates preeclampsia. Placenta 2020, 101, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Zhang, X.; Li, T.; Xie, R.; Zhou, J.; Luo, Y.; Yang, C. CircZDHHC20 represses the proliferation, migration and invasion in trophoblast cells by miR-144/GRHL2 axis. Cancer Cell Int. 2020, 20, 19. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Niu, X.; Li, Q.; Zhao, Y.; Chen, X.; Sun, H. Circ_0085296 suppresses trophoblast cell proliferation, invasion, and migration via modulating miR-144/E-cadherin axis. Placenta 2020, 97, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Qi, T.; Zhang, D.; Shi, X.; Li, M.; Xu, H. Decreased circUBAP2 expression is associated with preeclampsia by limiting trophoblast cell proliferation and migration. Reprod. Sci. 2021, 28, 2237–2245. [Google Scholar] [CrossRef]
- Li, X.; Yang, R.; Xu, Y.; Zhang, Y. Circ_0001438 participates in the pathogenesis of preeclampsia via the circ_0001438/miR-942/NLRP3 regulatory network. Placenta 2021, 104, 40–50. [Google Scholar] [CrossRef]
- Ma, B.; Zhao, H.; Gong, L.; Xiao, X.; Zhou, Q.; Lu, H.; Cui, Y.; Xu, H.; Wu, S.; Tang, Y.; et al. Differentially expressed circular RNAs and the competing endogenous RNA network associated with preeclampsia. Placenta 2021, 103, 232–241. [Google Scholar] [CrossRef]
- Tang, R.; Zhang, Z.; Han, W. CircLRRK1 targets miR-223-3p to inhibit the proliferation, migration and invasion of trophoblast cells by regulating the PI3K/AKT signaling pathway. Placenta 2021, 104, 110–118. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, S.; Wu, Y.; Li, Z.; Wang, D.; Cai, S.; Wang, Z. The role of circular RNA circ_0008285 in gestational diabetes mellitus by regulating the biological functions of trophoblasts. Biol. Res. 2021, 54, 14. [Google Scholar] [CrossRef]
- Dai, W.; Liu, X. Circular RNA 0004904 promotes autophagy and regulates the fused in sarcoma/vascular endothelial growth factor axis in preeclampsia. Int. J. Mol. Med. 2021, 47, 1–10. [Google Scholar] [CrossRef]
- Ping, Z.; Ai, L.; Shen, H.; Zhang, X.; Jiang, H.; Song, Y. Identification and comparison of circular RNAs in preeclampsia. PeerJ 2021, 9, e11299. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zeng, M.; Tang, F.; Chen, J.; Cao, D.; Tang, Z.-n. Circ-PNPT1 contributes to gestational diabetes mellitus (GDM) by regulating the function of trophoblast cells through miR-889-3p/PAK1 axis. Diabetol. Metab. Syndr. 2021, 13, 1–14. [Google Scholar] [CrossRef]
- Tang, M.; Bai, L.; Wan, Z.; Wan, S.; Xiang, Y.; Qian, Y.; Cui, L.; You, J.; Hu, X.; Qu, F. circRNA-DURSA regulates trophoblast apoptosis via miR-760-HIST1H2BE axis in unexplained recurrent spontaneous abortion. Mol. Ther. Nucleic Acids 2021, 26, 1433–1445. [Google Scholar] [CrossRef]
- Yao, P.; Hu, G.; Niu, H. Hsa_circ_0074371 Regulates Proliferation, Apoptosis, Migration, and Invasion via the miR-582-3p/LRP6 Axis in Trophoblast Cells. Biochem. Genet. 2021, 60, 267–285. [Google Scholar] [CrossRef] [PubMed]
- Fan, Z.; Wang, Q.; Deng, H. Circ_0011460 upregulates HTRA1 expression by sponging miR-762 to suppress HTR8/SVneo cell growth, migration, and invasion. Am. J. Reprod. Immunol. 2021, 86, e13485. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Li, Q. Circular RNA circ_0111277 serves as ceRNA, targeting the miR-424-5p/NFAT5 axis to regulate the proliferation, migration, and invasion of trophoblast cells in preeclampsia. Reprod. Sci. 2021, 29, 923–935. [Google Scholar] [CrossRef]
- Zhou, F.; Liu, H.; Zhang, R.; Sun, Y. Circ_0007121 Facilitates Trophoblastic Cell Proliferation, Migration, and Invasion via the Regulation of the miR-421/ZEB1 Axis in Preeclampsia. Reprod. Sci. 2022, 29, 100–109. [Google Scholar] [CrossRef]
- Huang, Y.; Liang, B.; Chen, X. Exosomal circular RNA circ_0074673 regulates the proliferation, migration, and angiogenesis of human umbilical vein endothelial cells via the microRNA-1200/MEOX2 axis. Bioengineered 2021, 12, 6782–6792. [Google Scholar] [CrossRef]
- Shan, L.; Hou, X. Circular RNA hsa_circ_0026552 inhibits the proliferation, migration and invasion of trophoblast cells via the miR-331-3p/TGF-βR1 axis in pre-eclampsia. Mol. Med. Rep. 2021, 24, 798. [Google Scholar] [CrossRef]
- Jiang, B.; Zhang, J.; Sun, X.; Yang, C.; Cheng, G.; Xu, M.; Li, S.; Wang, L. Circulating exosomal hsa_circRNA_0039480 is highly expressed in gestational diabetes mellitus and may be served as a biomarker for early diagnosis of GDM. J. Transl. Med. 2022, 20, 5. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Tang, Y.; Sun, Q.; Guan, G.; Wu, X.; Shi, F.; Zhou, Z.; Yang, W. Circular RNA FOXP1 relieves trophoblastic cell dysfunction in recurrent pregnancy loss via the miR-143-3p/S100A11 cascade. Bioengineered 2021, 12, 9081–9093. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Zheng, X.; Liu, Y.; Shen, J.; Ye, M.; Zhang, Y. Hsa_circRNA_102682 is closely related to lipid metabolism in gestational diabetes mellitus. J. Gynaecol. Endocrinol. 2022, 38, 50–54. [Google Scholar] [CrossRef] [PubMed]
- She, W.; Li, T.; Liu, Y.; Liu, X. CircRNA circVEGFC is Highly Expressed in Gestational Diabetes Mellitus (GDM) and It is Correlated with Multiple Adverse Events. Diabetes Metab. Syndr. Obes. 2021, 14, 4409. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.; Mao, Q. Circ_0037078 promotes trophoblast cell proliferation, migration, invasion and angiogenesis by miR-576-5p/IL1RAP axis. Am. J. Reprod. Immunol. 2022, 87, e13507. [Google Scholar] [CrossRef]
- Mao, Q.; Zou, H. Circular RNA circ_0032962 promotes trophoblast cell progression as ceRNA to target PBX3 via sponging miR-326 in preeclampsia. Reprod. Biol. 2021, 21, 100571. [Google Scholar] [CrossRef]
- Wang, L.; Wang, X.; Chen, X.; Wu, D.; Cen, H.; Mao, D.; Mo, Y.; Zheng, L. The relationship between hsa_circ_0051326 and HLA-G expression in the blood of patients with pre-eclampsia. Ginekol. Pol. 2021, in press. [Google Scholar] [CrossRef]
- Shu, C.; Xu, P.; Han, J.; Han, S.; He, J. Upregulation of circRNA hsa_circ_0008726 in Pre-eclampsia Inhibits Trophoblast Migration, Invasion, and EMT by Regulating miR-345-3p/RYBP Axis. Reprod. Sci. 2021, in press. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, H.; Long, Y.; Zhang, Y.; Chen, R.; Shi, J.; Chen, J. circRNA N6-methyladenosine methylation in preeclampsia and the potential role of N6-methyladenosine-modified circPAPPA2 in trophoblast invasion. Sci. Rep. 2021, 11, 24357. [Google Scholar] [CrossRef]
- Wang, W.; Liu, J.; Pan, E. CircHIPK3 contributes to human villous trophoblast growth, migration and invasion via modulating the pathway of miR-346/KCMF1. Placenta 2021, 118, 46–54. [Google Scholar] [CrossRef]
- Li, J.; Han, J.; Zhao, A.; Zhang, G. CircPAPPA Regulates the Proliferation, Migration, Invasion, Apoptosis, and Cell Cycle of Trophoblast Cells Through the miR-3127-5p/HOXA7 Axis. Reprod. Sci. 2022, 29, 1215–1225. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Shi, J.; Zheng, L. Identification of Circular RNA circ_0017068 as a Regulator of Proliferation and Apoptosis in Trophoblast Cells by miR-330-5p/XIAP Axis. Reprod. Sci. 2022, in press. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, H.; Zhang, Y.; Shi, J.; Long, Y. A Novel Circular RNA CircBRAP May Be Used as an Early Predictor of Preeclampsia and Its Potential Mechanism. Reprod. Sci. 2022, in press. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Gong, Y.; Zhong, L.; Ding, X.; Yang, Z.; Su, X.; Chen, M.; Zhang, F.; Yang, L. Circular RNA expression profile and competing endogenous RNA regulatory network in preeclampsia. Placenta 2022, 19, 32–38. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, Y.; Zhang, J.; Du, X.; Li, Q.; Pan, Z. circSLC41A1 Resists Porcine Granulosa Cell Apoptosis and Follicular Atresia by Promoting SRSF1 through miR-9820-5p Sponging. Int. J. Mol. Sci. 2022, 23, 1509. [Google Scholar] [CrossRef]
- Pandey, P.; Rout, P.; Das, A.; Gorospe, M.; Panda, A. RPAD (RNase R treatment, polyadenylation, and poly(A)+ RNA depletion) method to isolate highly pure circular RNA. Methods 2019, 155, 41–48. [Google Scholar] [CrossRef]
- Xiao, M.; Wilusz, J. An improved method for circular RNA purification using RNase R that efficiently removes linear RNAs containing G-quadruplexes or structured 3’ ends. Nucleic Acids Res. 2019, 41, 8755–8769. [Google Scholar] [CrossRef]
- Holtan, S.; Creedon, D.; Haluska, P.; Markovic, S. Cancer and pregnancy: Parallels in growth, invasion, and immune modulation and implications for cancer therapeutic agents. Mayo Clin. Proc. 2009, 84, 985–1000. [Google Scholar] [CrossRef]
- Costanzo, V.; Bardelli, A.; Siena, S.; Abrignani, S. Exploring the links between cancer and placenta development. Open Biol. 2018, 8, 180081. [Google Scholar] [CrossRef]
- Bischof, P.; Campana, A. A putative role for oncogenes in trophoblast invasion? Mol. Hum. Reprod. 2000, 15, 51–58. [Google Scholar]
- Kim, D.; Xing, T.; Yang, Z.; Dudek, R.; Lu, Q.; Chen, Y. Epithelial mesenchymal transition in embryonic development, tissue repair and cancer: A comprehensive overview. J. Clin. Med. 2017, 7, 1. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Hammer, A. Immunological regulation of trophoblast invasion. J. Reprod. Immunol. 2011, 90, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Demir, R.; Kaufmann, P.; Castellucci, M.; Erbengi, T.; Kotowski, A. Fetal vasculogenesis and angiogenesis in human placental villi. Acta Anat. (Basel) 1989, 136, 190–203. [Google Scholar] [CrossRef] [PubMed]
- Morrish, D.; Bhardwaj, D.; Dabbagh, L.; Marusyk, H.; Siy, O. Epidermal growth factor induces differentiation and secretion of human chorionic gonadotropin and placental lactogen in normal human placenta. J. Clin. Endocrinol. Metab. 1987, 65, 1282–1290. [Google Scholar] [CrossRef]
- Normanno, N.; Bianco, C.; De Luca, A.; Salomon, D. The role of EGF-related peptides in tumor growth. Front. Biosci. 2001, 6, 685–707. [Google Scholar] [CrossRef]
- Xi, J.; Wang, Y.; Long, X.; Ma, Y. Mangiferin potentiates neuroprotection by isoflurane in neonatal hypoxic brain injury by reducing oxidative stress and activation of phosphatidylinositol-3-kinase/Akt/mammalian target of rapamycin (PI3K/Akt/mTOR) signaling. Med. Sci. Monit. 2018, 24, 7459–7468. [Google Scholar] [CrossRef]
- Nicholson, K.; Anderson, N. The protein kinase B/Akt signalling pathway in human malignancy. Cell Signal. 2002, 14, 381–395. [Google Scholar] [CrossRef]
- Marzusch, K.; Ruckh, P.; Horny, H.; Dietle, J.; Kaiserling, E. Expression of the p53 Tumour Suppressor Gene in Human Placenta: An Immunohistochemical Study. Placenta 1995, 16, 101–104. [Google Scholar] [CrossRef]
- Lane, D. Mutation of the p53 protein: Common steps found in the majority of human cancers. In Accomplishments in Cancer Research; Fortner, J.G., Ed.; Lippincott Co: Philadelphia, PA, USA, 1990; pp. 252–266. [Google Scholar]
- Forbes, K.; Westwood, M.; Baker, P.N.; Aplin, J.D. Insulin-like growth factor I and II regulate the life cycle of trophoblast in the developing human placenta. Am. J. Physiol. Cell Physiol. 2008, 294, 1313–1322. [Google Scholar] [CrossRef]
- Jones, J.; Clemmons, D. Insulin-like growth factors and their binding proteins: Biological actions. Endocr. Rev. 1995, 16, 3–35. [Google Scholar] [PubMed]
- Dufourny, B.; Alblas, J.; van Teeffelen, H.; van Schaik, F.; van der Burg, B.; Steenbergh, P. Mitogenic signaling of insulin-like growth factor I in MCF-7 human breast cancer cells requires phosphatidylinositol 3-kinase and is independent of mitogen-activated protein kinase. J. Biol. Chem. 1997, 272, 31163–31171. [Google Scholar] [CrossRef] [PubMed]
- Buckbinder, L.; Talbott, R.; Velasco Miguel, S.; Takenaka, I.; Faha, B.; Seizinger, B. Induction of the growth inhibitor IGF-binding protein 3 by p53. Nature 1995, 377, 646–649. [Google Scholar] [CrossRef] [PubMed]
- Gorivodsky, M.; Torchinsky, A.; Zemliak, I.; Savion, S.; Fein, A.; Toder, V. TGF beta 2 mRNA expression and pregnancy failure in mice. Am. J. Reprod. Immunol. 1999, 42, 124–133. [Google Scholar] [PubMed]
- Akhurst, R.; Derynck, R. TGF-β signaling in cancer—A double-edged sword. Trends Cell Biol. 2001, 11, 44–51. [Google Scholar]
- Ahmed, A.; Dunk, C.; Kniss, D.; Wilkes, M. Role of VEGF receptor (Flt-1) in mediating calcium dependant nitric oxide release and limiting DNA synthesis in human trophoblast cells. Lab. Invest. 1997, 76, 779–791. [Google Scholar]
- Senger, D.; Galli, S.; Dvorak, A.; Perruzzi, C.; Harvey, V.; Dvorak, H. Tumor cells secrete a vascular permeability factor that promotes accumulation of ascites fluid. Science 1983, 219, 983–985. [Google Scholar] [CrossRef]
- Gangloff, Y.; Mueller, M.; Dann, S.; Svoboda, P.; Sticker, M.; Spetz, J.; Um, S.; Brown, E.; Cereghini, S.; Thomas, G.; et al. Disruption of the mouse mTOR gene leads to early postimplantation lethality and prohibits embryonic stem cell development. Mol. Cell 2004, 24, 9508–9516. [Google Scholar] [CrossRef]
- Martin, P.; Sutherland, A. Exogenous amino acids regulate trophectoderm differentiation in the mouse blastocyst through an mTOR-dependent pathway. Dev. Biol. 2001, 240, 182–193. [Google Scholar] [CrossRef]
- Faivre, S.; Kroemer, G.; Raymond, E. Current development of mTOR inhibitors as anticancer agents. Nat. Rev. Drug Discov. 2006, 5, 671–688. [Google Scholar] [CrossRef]
- Nadeau, V.; Guillemette, S.; Belanger, L.; Jacob, O.; Roy, S.; Charron, J. Map2k1 and Map2k2 genes contribute to the normal development of syncytiotrophoblasts during placentation. Development 2009, 136, 1363–1374. [Google Scholar] [CrossRef]
- Plotnikov, A.; Zehorai, E.; Procaccia, S.; Seger, R. The MAPK cascades: Signaling components, nuclear roles and mechanisms of nuclear translocation. Biochim. Biophys. Acta 2011, 1813, 1619–1633. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, J.; Germeyer, A.; Huppertz, B.; Jeschke, U.; Knofler, M.; Moser, G. Governing the invasive trophoblast: Current aspects on intra- and extra-cellular regulation. Am. J. Reprod. Immunol. 2010, 63, 492–505. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.; Snowden, J.; Zeidler, M.; Danson, S. The role of JAK/STAT signalling in the pathogenesis, prognosis and treatment of solid tumours. Br. J. Cancer 2015, 113, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Vainio, S.; Heikkila, M.; Kispert, A.; Chin, N.; McMahon, A. Female development in mammals is regulated by Wnt-4 signalling. Nature 1999, 397, 405–409. [Google Scholar] [CrossRef]
- Nusse, R.; van Ooyen, A.; Cox, D.; Fung, Y.; Varmus, H. Mode of proviral activation of a putative mammary oncogene (int-1) on mouse chromosome 15. Nature 1984, 307, 131–136. [Google Scholar] [CrossRef]
- Lindstrom, T.; Bennett, P. The role of nuclear factor kappa B in human labour. Reproduction 2005, 130, 569–581. [Google Scholar] [CrossRef]
- Pikarsky, E.; Porat, R.; Stein, I.; Abramovitch, R.; Amit, S.; Kasem, S.; Gutkovich-Pyest, E.; Urieli-Shoval, S.; Galun, E.; Ben-Neriah, Y. NF-kappaB functions as a tumour promoter in inflammation-associated cancer. Nature 2004, 431, 461–466. [Google Scholar] [CrossRef]
- Pique-Regi, R.; Romero, R.; Tarca, A.L.; Sendler, E.D.; Xu, Y.; Garcia-Flores, V.; Leng, Y.; Luca, F.; Hassan, S.S.; Gomez-Lopez, N. Single cell transcriptional signatures of the human placenta in term and preterm parturition. Elife 2019, 8, e52004. [Google Scholar] [CrossRef]
- Suryawanshi, H.; Morozov, P.; Straus, A.; Sahasrabudhe, N.; Max, K.E.; Garzia, A.; Kustagi, M.; Tuschl, T.; Williams, Z. A single-cell survey of the human first-trimester placenta and decidua. Sci. Adv. 2018, 4, eaau4788. [Google Scholar] [CrossRef]
- Gong, S.; Gaccioli, F.; Dopierala, J.; Sovio, U.; Cook, E.; Volders, P.-J.; Martens, L.; Kirk, P.D.; Richardson, S.; Smith, G. The RNA landscape of the human placenta in health and disease. Nat. Commun. 2021, 12, 2639. [Google Scholar] [CrossRef] [PubMed]
- Buckberry, S.; Bianco-Miotto, T.; Bent, S.J.; Clifton, V.; Shoubridge, C.; Shankar, K.; Roberts, C.T. Placental transcriptome co-expression analysis reveals conserved regulatory programs across gestation. BMC Genet. 2017, 18, 10. [Google Scholar] [CrossRef] [PubMed]
- Mikheev, A.; Nabekura, T.; Kaddoumi, A.; Bammler, T. Profiling gene expression in human placentae of different gestational ages: An OPRU Network and UW SCOR Study. Reprod. Sci. 2008, 15, 866–877. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Huang, M.; Lv, M.; He, Y.; Duan, C.; Zhang, L. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Placenta 2017, 403, 305–317. [Google Scholar] [CrossRef]
- Zeman, M.; Cimprich, K. Causes and consequences of replication stress. Nat. Cell Biol. 2014, 16, 2–9. [Google Scholar] [CrossRef]
- Conn, V.M.; Hugouvieux, V.; Nayak, A.; Conos, S.A.; Capovilla, G.; Cildir, G.; Jourdain, A.; Tergaonkar, V.; Schmid, M.; Zubieta, C.; et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat. Plants 2017, 3, 17053. [Google Scholar] [CrossRef]
- Weigelt, C.; Sehgal, R.; Tain, L.; Cheng, J.; Eßer, J.; Pahl, A.; Dieterich, C.; Gronke, S.; Partridge, L. An Insulin-Sensitive Circular RNA that Regulates Lifespan in Drosophila. Mol. Cell. 2020, 79, 268–279. [Google Scholar] [CrossRef]
- Knupp, D.; Miura, P. CircRNA accumulation: A new hallmark of aging? Mech. Ageing Dev. 2018, 173, 71–79. [Google Scholar] [CrossRef]
- Maiti, K.; Sultana, Z.; Aitken, R.J.; Morris, J.; Park, F.; Andrew, B.; Riley, S.; Smith, R. Evidence that fetal death is associated with placental aging. Am. J. Obstet. Gynecol. 2017, 217, 441.e1–441.e14. [Google Scholar] [CrossRef]

| Author | Year | Tissue | Pregnancy Status | Key Findings | Limitations |
|---|---|---|---|---|---|
| Fan X, et al. [35] | 2015 | Mouse—oocytes and preimplantation embryos | Uncomplicated (assumed) | Detected 2891 circRNAs from 1316 host genes. A majority of these circRNAs are unique to the preimplantation stage and a large proportion of them exhibit dynamic expression patterns during this developmental process. | Sequencing using the SUPeR-seq method could possibly limit depth of circRNA sequencing due to no enrichment using RNase R. |
| Szabo L, et al. [36] | 2015 | Fetal tissue—unspecified, various species | Uncomplicated (assumed) | Developed an algorithm to compare established data sets in human, rat and mouse tissue and cell lines, with their generated circRNA data from RNA-seq on fetal tissue. | This study assumes a degree of conservation between species. |
| Zhang YG, et al. [37] | 2016 | Human maternal red blood cells | PE vs. uncomplicated | The levels of circ_101222 in red blood cells of patients with PE were significantly higher than in healthy women. Using ENG in combination with circ_101222 improved confidence for the prediction of PE. | |
| Qian Y, et al. [38] | 2016 | Human placenta | PE vs. preterm birth (PTB) | 143 circRNAs were up-regulated and 158 were down-regulated in PE samples compared with preterm. | Use of microarray is not as comprehensive as sequencing techniques. PTB placenta is used as a gestational age control but PTB can occur for multiple reasons and is a pathology of pregnancy. Caution on interpretation is required. |
| Qian Y, et al. [39] | 2017 | Human placenta—chorionic villi | Recurrent spontaneous abortion (RSA) vs uncomplicated | 594 aberrantly expressed circRNAs between gestational age-matched RSA and healthy placentae. Of these, 335 circRNAs were up-regulated and 259 were down-regulated. | No validation or investigation into results or mechanisms. |
| Cheng J, et al. [40] | 2017 | Human granulosa cells | Non-pregnant Advanced age (AA ≥ 38 years) vs. young age (YA ≤ 30 years) | 46 upregulated and 11 downregulated circRNAs in AA samples compared with YA. | Use of microarray is not as thorough as sequencing techniques. |
| Zhang S, et al. [41] | 2018 | Mouse—endometrium | Uncomplicated (assumed) | Used microarray to find that 101 upregulated and 75 downregulated circRNAs at implantation sites compared with interimplantation sites. Four randomly selected circRNAs were validated for their expression using qRT-PCR | Use of microarray is not as comprehensive as sequencing techniques. |
| Bai Y, et al. [42] | 2018 | Human placenta | PE vs. uncomplicated | 151 circRNAs were upregulated and 149 were downregulated in PE samples compared with normal. Possible biomarker hsa_circ_0007121 had a significant predictive index | Only 3 of the 10 circRNAs for validation matched sequencing results. |
| Jiang M, et al. [43] | 2018 | Human maternal peripheral blood mononuclear cells | PE vs. uncomplicated | 884 circRNAs were downregulated and 1294 circRNAs were upregulated in PE samples compared with control. circ_0004904 and circ_0001855 combined with PAPP-A might be biomarkers for PE detection. | Use of microarray is not as thorough as sequencing techniques. As blood was collected at the time of disease, there is no way to be certain that circ_0004904 and circ_0001855 are causes of PE and not resultant. |
| Zhou W, et al. [44] | 2018 | Human placenta | PE vs. uncomplicated | Two circRNAs were up-regulated and 47 were down-regulated in PE compared with control placentae. Hsa_circRNA_3286 reduced invasion in HTR-8/SVneo cells. | RNase R enrichment for circRNAs not completed on RNA-seq samples. Only 3 of the 10 circRNAs for validation matched sequencing results. |
| Hu X, et al. [45] | 2018 | Human placenta Maternal whole blood | Severe PE vs. uncomplicated | 4569 upregulated and 3984 downregulated circRNAs between severe PE and healthy pregnancy. Identified hsa_circ_0036877 as a potential novel blood biomarker for early PE. | Use of microarray is not as thorough as sequencing techniques. Significant differences in BMI, gestational age at delivery and % caesarean sections between severe PE and control groups. |
| Shen, XY, et al. [46] | 2019 | Human placenta | PE vs. uncomplicated | circTNRC18 upregulated in PE placentae. circTNRC18 reduced trophoblast cell migration and EMT. circTNRC18 repressed miR-762 activity and elevated Grhl2 protein. | |
| Wang H, et al. [47] | 2019 | Human placenta | GDM vs. uncomplicated | Three circRNAs were upregulated and 43 were downregulated between GDM and normal placentae. Ten randomly selected circRNAs were validated for their expression using qRT-PCR. | RNase R enrichment for circRNAs not completed on RNA-seq samples. Only 3 of the 10 circRNAs for validation matched sequencing results. |
| Cao M, et al. [48] | 2020 | Human umbilical cord blood | GDM vs. uncomplicated | 229 upregulated and 278 downregulated circRNAs between GDM and healthy pregnancy. Exosome particle size was larger and exosome concentration was higher in GDM. | Use of microarray is not as thorough as sequencing techniques. RNase R enrichment for circRNAs not completed on microarray samples. |
| Oudejans C, et al. [49] | 2020 | Human maternal platelets | Uncomplicated (assumed) | Proof of concept study showing that pregnancy-specific circRNAs can be detected in first-trimester platelet RNA. | Could possibly limit depth of circRNA sequencing due to no enrichment using RNase R. |
| Yang H, et al. [50] | 2020 | Human maternal blood | GDM vs uncomplicated | Blood samples (n = 12) were collected from GDM and healthy pregnant women between 15–24 weeks’ gestation prior to RNase R treatment and circRNA microarray analysis. | Use of microarray for detection of circRNAs is not as thorough as sequencing techniques. No mention of gestational age matching between GDM and controls. |
| Wang D, et al. [51] | 2020 | Human placenta | Macrosomia vs. uncomplicated | Circ-SETD2 was upregulated in placentae of patients with fetal macrosomia. Circ-SETD2 upregulation increased proliferation and invasion in HTR-8/SVneo cells. Suggested circ-SETD2/miR-519a/PTEN axis involved in regulating trophoblasts in macrosomia. | Use of lnc-microarray for detection of circRNAs is not as thorough as sequencing techniques. |
| Wang H, et al. [52] | 2020 | Human placenta and maternal plasma | GDM vs. uncomplicated | Circ_0005243 was identified using RNase R (determining circular form). Circ_0005243 expression was downregulated in placenta and maternal plasma in GDM. Knockdown of circ_0005243 in HTR-8/SVneo cells suppressed cell proliferation and migration and increased secretion of inflammatory factors (TNF-α and IL-6). It also reduced β-catenin expression and increased nuclear NF-κB p65 nuclear translocation. | |
| Zhang Y, et al. [53] | 2020 | Human placenta and in vivo rat model | PE vs. uncomplicated | CircSFXN1 was identified using RNase R (determining circular form). CircSFXN1 was elevated in PE placenta. Knockdown of circSFXN1 promoted TEV-1 cell invasion and HUVEC angiogenesis—this effect was opposed with circSFXN1 overexpression. Pregnant rats injected with sFLT1-expressing adenovirus had in increased blood pressure and proteinuria; si-circSFXN1 reversed this. CircSFXN1 recruits sFLT1, validated by RNA-protein pulldown, RNA immunoprecipitation and dual-luciferase reporter assays. | Use of microarray is not as thorough as sequencing techniques, although the study validates these results with qRT-PCR. |
| Zhou B, et al. [54] | 2020 | Human placenta | PE vs. uncomplicated | CircZDHHC20 was identified using RNase R (determining circular form). CircZDHHC20 was up-regulated and miR-144 was down-regulated in PE placenta. CircZDHHC20 overexpression in HTR-8/SVneo cells repressed trophoblast proliferation, migration, and invasion. miR-144 regulated circZDHHC20 was inhibited by GRHL2. | |
| Zhu H, et al. [55] | 2020 | Human placenta | PE vs. uncomplicated | Circ_0085296 was identified using RNase R (determining circular form). Circ_0085296 elevated in PE placenta. Knockdown of circ_0085296 in HTR-8/SVneo cells promoted trophoblast cell proliferation, invasion, and migration. miR-144 down-regulated in PE placenta, directly bound to circ_0085296 and E-cadherin. Circ_0085296 bound to miR-144 to regulate E-cadherin. | |
| Qi T, et al. [56] | 2020 | Human placenta | PE vs. uncomplicated | CircUBAP2 (hsa_circ_0003496) was downregulated in PE placentae. CircUBAP2 knockdown suppressed HTR-8/SVneo cell proliferation and migration. CircUBAP2 sponges miR-1244 to regulate FOXM1. Cotransfection of si-circUBAP2 and a miR-1244 inhibitor partially restored cell proliferation and migration induced by circUBAP2 depletion. | |
| Li X, et al. [57] | 2021 | Human placenta | PE vs. uncomplicated | Circ_0001438 and NLRP3 were elevated in PE placenta. Knockdown of circ_0001438 promoted cell proliferation, migration and invasion but inhibited apoptosis and inflammatory responses in HTR-8/SVneo cells. Circ_0001438 bound to miR-942 to regulate NLRP3. | RNase R treatment not used—Circ_0001438 was not enriched for circular form only. |
| Ma B, et al. [58] | 2021 | Human placenta | PE vs. uncomplicated | 252 upregulated and 109 downregulated circRNAs between preeclamptic and healthy placentae; 6 circRNAs were further validated using qPCR. | No mention of gestational age matching between PE and controls. |
| Tang R, et al. [59] | 2021 | Human placenta | PE vs. uncomplicated | CircLRRK1 was identified using Actinomycin D and RNase R (determining circular form). circLRRK1 was elevated in PE placenta. Knockdown of circLRRK1 promoted cell proliferation, migration and invasion in HTR-8/SVneo cells. circLRRK1 bound to miR-223-3p to regulate PI3K/Akt signalling. | |
| Chen H, et al. [60] | 2021 | Maternal plasma | GDM vs. uncomplicated | Circ_0008285 was increased, while circ_0001173 was decreased, in GDM samples. Circ_0008285 correlated with total cholesterol and LDL-C levels. Circ_0001173 correlated with glycated haemoglobin. High glucose media promoted HTR-8/SVneo cell proliferation, invasion, and migration, while circ_0008285 knockdown exerted the opposite effect. | High glucose media contained 30 mmol/L glucose which is potentially too high for physiological relevance. |
| Dai W, et al. [61] | 2021 | Human placenta and maternal plasma | PE vs. uncomplicated | Circ_0004904 levels were elevated in PE placentae and maternal plasma. Aberrant circ_0004904 expression inhibited autophagy and induced JEG3 cell proliferation and invasion. Circ_0004904 also regulated ATG12 levels via miR-570, as well as controlling the FUS/VEGF axis in HTR-8/SVneo and JEG3 cells. | |
| Ping Z, et al. [62] | 2021 | Maternal blood | PE vs. uncomplicated | 121 differentially expressed circRNAs were upregulated and 30 downregulated in PE samples. Functional and pathway enrichment analysis was conducted using Gene Ontology and KEGG databases. | Could possibly limit depth of circRNA sequencing due to no enrichment using RNase R. No mechanistic studies conducted or PCR validation of results. |
| Zhang L, et al. [63] | 2021 | Human placenta | GDM vs. uncomplicated | Circ-PNPT1 levels were elevated in GDM placentae and high glucose (HG)-induced HTR-8/SVneo cells. HG-induced arrest of cell viability, migration, invasion and apoptosis was reversed with circ-PNPT1 knockdown. Circ-PNPT1 also sponged miR-889-3p to regulate PAK1. HTR-8/SVneo cell line experiments showed circ-PNPT1 was packaged into exosomes and internalised by surrounding cells. | Exosome studies were conducted in HTR-8/SVneo cell line, not replicated in placental explants or maternal blood. High glucose media contained 25 mmol/L glucose which is potentially too high for physiological relevance. |
| Tang M, et al. [64] | 2021 | Human placental villous tissue and embryos In vivo mouse model | Unexplained recurrent spontaneous abortion (URSA) vs. uncomplicated | Circ-0050703 (circRNA-DURSA) is downregulated in URSA placental villous tissue. In vitro, circRNA-DURSA silencing results in cell apoptosis and circRNA-DURSA competitively binds miR-760, regulating HIST1H2BE. In vivo, circRNA-DURSA silencing decreased number of embryos successfully implanted. | |
| Yao P, et al. [65] | 2021 | Human placenta | Fetal growth restriction vs. uncomplicated | Circ_0074371 and LRP6 were downregulated, and miR-582-3p was upregulated in fetal growth restriction (FGR) placentae and HTR-8/SVneo cells. Circ_0074371 sponges miR-582-3p to regulate LRP6. Circ_0074371 knockdown induced HTR-8/SVneo cell cycle arrest, apoptosis, and inhibited cell proliferation, migration, and invasion, which was reversed with a miR-582-3p inhibitor. | |
| Fan Z, et al. [66] | 2021 | Human placenta | PE vs. uncomplicated | Circ_0011460 is upregulated in PE placentae, and overexpression in HTR-8/SVneo cells suppressed proliferation, migration and invasion, and increased cell apoptosis. Circ_0011460 also sponges miR-762 and regulates HTRA1. | |
| Li C, et al. [67] | 2021 | Human placenta | PE vs. uncomplicated | Circ_0111277 and NFAT5 expression were increased in PE placentae and miR-424-5p was decreased. Circ_0111277 knockdown increased cell viability, migration, invasion, and angiogenesis in HTR-8/SVneo cells. Circ_0111277 acted as a sponge of miR-424-5p to regulate NFAT5 expression. | |
| Zhou F, et al. [68] | 2022 | Human placenta | PE vs. uncomplicated | Circ_0007121 is downregulated in PE placentae. Upregulation of circ_0007121 promotes cell proliferation, migration, invasion and EMT. Circ_0007121 also sponges miR-421 and thus regulates ZEB1 expression. | |
| Huang Y, et al. [69] | 2021 | Human umbilical cord blood—exosomes isolated | GDM vs. uncomplicated | Larger exosomes and greater number of exosomes in umbilical cord blood of GDM patients. Circ_0074673 was upregulated in exosomes from GDM and in HUVECs co-cultured with exosomes. Loss of exosomal circ_0074673 facilitated the proliferation, migration, and angiogenesis of high glucose-HUVECs via the miR- 1200/MEOX2 axis. | High glucose media contained 25 mM glucose which is potentially too high for physiological relevance. |
| Shan L, et al. [70] | 2021 | Human placenta | PE vs. uncomplicated | miR-331-3p negatively correlates with circ_0026552 relative expression, while TGF-βR1 positively correlates with circ_0026552 expression. Silencing circ_0026552 increased proliferation, migration and invasion of HTR-8/SVneo cells, which was reversed with circ_0026552 overexpression. Circ_0026552 sponges miR-331-3p to upregulate TGF-βR1 expression. | RNase R enrichment for circRNAs not completed on microarray samples. Use of microarray is not as thorough as sequencing techniques, although the study validates these results with qRT-PCR. Small number (n = 3 PE, n = 4 control) tissues used for microarray. |
| Jiang B, et al. [71] | 2021 | Maternal blood | GDM vs. uncomplicated | Plasma exosomal circRNA_0039480 and circRNA_0026497 were increased in GDM. circRNA_0039480 was elevated in GDM vs. normal glucose tolerance control throughout trimesters and positively correlated with OGTT during the second trimester. The combination of circRNA_0039480 and circRNA_0026497 suggested as a useful biomarker for GDM in the first trimester (AUC = 0.754, p < 0.001). | RNase R enrichment for circRNAs not completed on microarray samples. Use of microarray is not as thorough as sequencing techniques, although the study validates these results with qRT-PCR. Small number (n = 3) samples used for microarray. |
| Gao Y, et al. [72] | 2021 | Human placenta | Recurrent pregnancy loss (RPL) vs. uncomplicated | MiR–143–3p targeted S100A11 and was negatively regulated by circFOXP1. miR–143–3p competitively bound circFOXP1. circFOXP1 regulated HTR-8/SVneo cell functions through the miR–143–3p/S100A11 axis. | |
| Wu H, et al. [73] | 2022 | Maternal blood | GDM vs. uncomplicated | Circ_102682 was decreased in GDM blood samples. circRNA_102682 was significantly correlated with triglycerides, APOA1, APOB, 1-h blood glucose in the serum of GDM patients. | |
| She W, et al. [74] | 2021 | Maternal plasma | GDM vs. uncomplicated | CircVEGFC regulates glucose metabolism—higher incidence of GDM in patients with high circVEGFC levels. Elevated circVEGFC levels in GDM plasma. High circVEGFC level group showed higher incidence rates of fetal malformation and hypertension. | Correlation established but further mechanistic studies required to establish whether this is causative. |
| Zou H, et al. [75] | 2022 | Human placenta | PE vs. uncomplicated | Circ_0037078 is upregulated in PE placentae. Knockdown of circ_0037078 increases trophoblast cell proliferation, migration, invasion and angiogenesis. Circ_0037078 also sponges miR-576-5p and increases IL1RAP expression. | |
| Mao Q, et al. [76] | 2021 | Human placenta | PE vs. uncomplicated | Circ_0032962 and PBX3 levels were decreased in PE placentae and miR-326 was elevated. Circ_0032962 knockdown suppressed cell proliferation ability, migration, invasion, and EMT in HTR-8/SVneo cells. | |
| Wang L, et al. [77] | 2021 | Maternal blood | PE vs. uncomplicated | Circ_0051326 and HLA-G protein and mRNA were decreased in PE samples. There was a positive correlation between the expression of serum circ_0051326 with HLA-G mRNA. | |
| Shu C, et al. [78] | 2021 | Human placenta | PE vs. uncomplicated | Silencing circ_0008726 promoted cell migration and EMT, while circ_0008726 overexpression suppressed these processes. Circ_0008726 sponged miR-345-3p to regulate RYBP expression. Circ_0008726 was negatively correlated with miR-345-3p and positively correlated with RYBP expression levels in PE placentae. Transfection of miR-345-3p mimic or RYBP knockdown counteracted the effects of circ_0008726 overexpression on cell migration and EMT. | |
| Zhang Y, et al. [79] | 2021 | Human placenta | PE vs. uncomplicated | Increases in m6A-modified circRNAs are prevalent in PE placentae, with the main methylation changes occurring in the 3′ UTR and near the start codon. In PE, circPAPPA2 levels are decreased while m6A modification is increased. METTL14 increases circPAPPA2 m6A methylation and IGF2BP3 maintains circPAPPA2 stability. | |
| Wang W, et al. [80] | 2021 | Human placenta | PE vs. uncomplicated | In PE placentae, circHIPK3 and KCMF1 were downregulated and miR-346 was upregulated. CircHIPK3 overexpression promotes trophoblast cell proliferation, migration and invasion, as well as decreasing cell cycle arrest and apoptosis. CircHIPK3 also targets miR-346 and regulates KCMF1 expression. | |
| Li J, et al. [81] | 2022 | Human placenta | PE vs. uncomplicated | CircPAPPA positively regulates trophoblast cell proliferation, migration and invasion, and causes apoptosis and cell cycle arrest, through the miR-3127-5p/HOXA7 axis. | |
| Wang W, et al. [82] | 2022 | Human placenta | PE vs. uncomplicated | Circ_0017068 was downregulated in PE placental samples. Circ_0017068 overexpression promoted HTR-8/SVneo cell proliferation, cycle progression, and suppressed apoptosis while silencing of circ_0017068 exhibited opposite effects. Circ_0017068 targeted miR-330-5p to regulate XIAP expression, and through this regulated proliferation, cycle progression, and apoptosis. | |
| Zhang Y, et al. [83] | 2021 | Human placenta and maternal plasma | PE vs. uncomplicated | PE predictive power was greatest when plasma sFLT1 and circBRAP levels were combined with uterine pulsatility index. CircBRAP was increased in PE placentae and may regulate miR-106b to decrease TEV-1 cell proliferation, invasion and apoptosis. | |
| Yuan Y, et al. [84] | 2022 | Human placenta | PE vs. uncomplicated | 2432 circRNAs were differentially expressed between PE and control tissues. hsa_circRNA_0001687/hsa-miR-532-3p/MMP14/AXL, hsa_circ_0001513/hsa-miR-188-5p/HMGCS1 and hsa_circ_0001513/hsa_circ_0001329/hsa-miR-760/MAP1LC3B axes may contribute to PE pathogenesis. | The paper uses publicly available datasets instead of completing their own sequencing. As such, data quality cannot be ascertained. No luciferase assays to validate the potential axes listed. |
| Wang H, et al. [85] | 2022 | Pig ovaries | Atretic vs. healthy follicles | CircSCL41A1 was elevated in healthy follicles compared with atretic follicles. miR-9820-5p competitively binds circSLC41A1 to regulate SRSF1. A circSLC41A1-miR-9820-5p-SRSF1 axis regulates follicular granulosa cell apoptosis. |
| Signalling Pathway | Placenta | Cancer |
|---|---|---|
| EGFR | [95] | [96] |
| PI3K/Akt | [97] | [98] |
| P53 | [99] | [100] |
| IGF | [101,102] | [103,104] |
| TGF-β | [105] | [106] |
| VEGF | [107] | [108] |
| mTOR | [109,110] | [111] |
| MAPK | [112] | [113] |
| Jak/STAT | [114] | [115] |
| Wnt/β-catenin | [116] | [117] |
| NF-κB | [118] | [119] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arthurs, A.L.; Jankovic-Karasoulos, T.; Smith, M.D.; Roberts, C.T. Circular RNAs in Pregnancy and the Placenta. Int. J. Mol. Sci. 2022, 23, 4551. https://doi.org/10.3390/ijms23094551
Arthurs AL, Jankovic-Karasoulos T, Smith MD, Roberts CT. Circular RNAs in Pregnancy and the Placenta. International Journal of Molecular Sciences. 2022; 23(9):4551. https://doi.org/10.3390/ijms23094551
Chicago/Turabian StyleArthurs, Anya L., Tanja Jankovic-Karasoulos, Melanie D. Smith, and Claire T. Roberts. 2022. "Circular RNAs in Pregnancy and the Placenta" International Journal of Molecular Sciences 23, no. 9: 4551. https://doi.org/10.3390/ijms23094551
APA StyleArthurs, A. L., Jankovic-Karasoulos, T., Smith, M. D., & Roberts, C. T. (2022). Circular RNAs in Pregnancy and the Placenta. International Journal of Molecular Sciences, 23(9), 4551. https://doi.org/10.3390/ijms23094551







