Adenosine Receptor A2B Negatively Regulates Cell Migration in Ovarian Carcinoma Cells
Abstract
:1. Introduction
2. Results
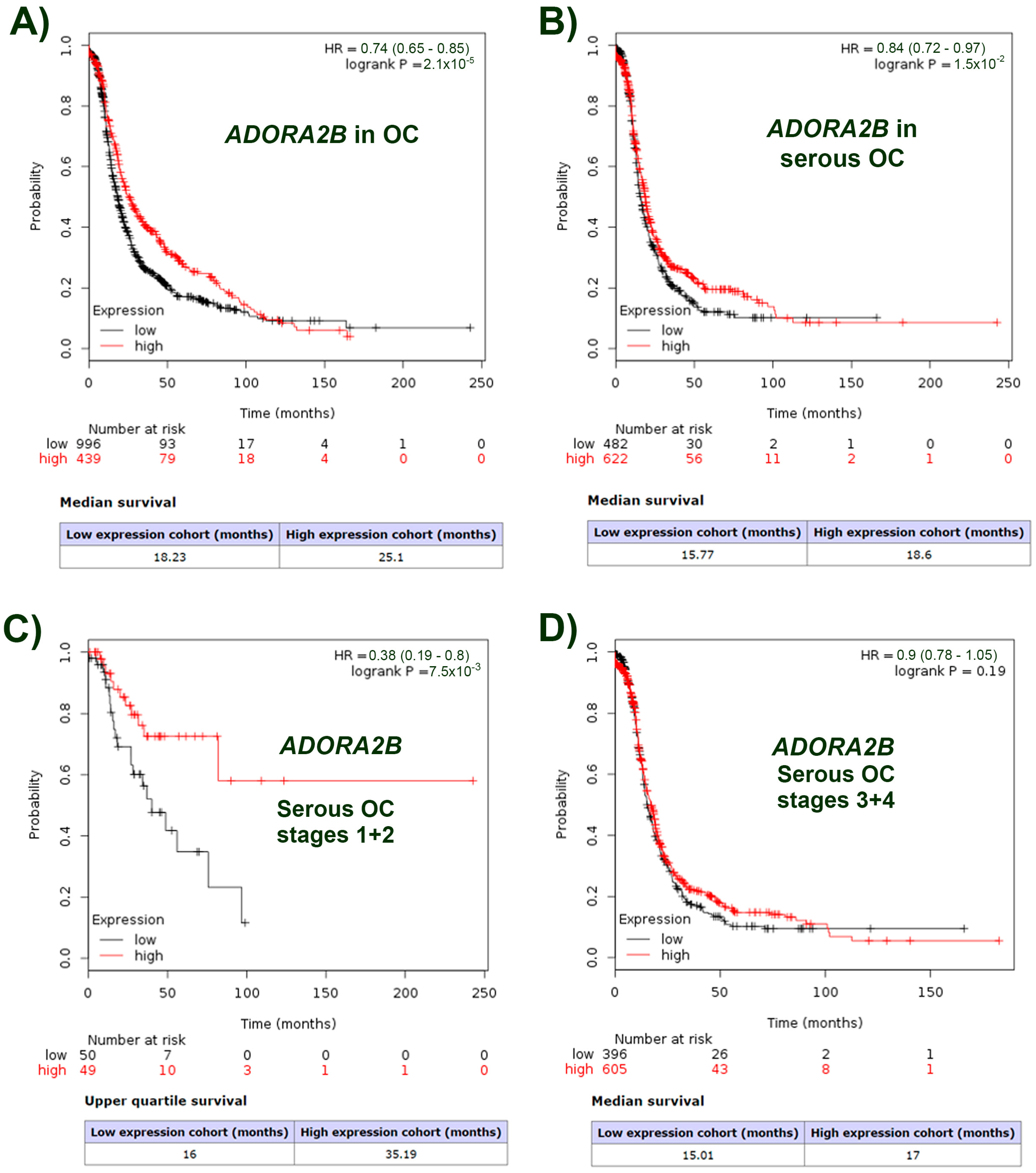
2.1. ADORA2B Expression Is Related to the Surveillance Probability of Ovarian Carcinoma Patients and Is Specific to the Tumor Type
2.2. SKOV-3 Cell Line Expresses a Functional A2BR
2.3. A2BR Stimulation Inhibited Cell Migration without Modifying Cell Proliferation
2.4. ADORA2B Enhances the Expression of the Epithelial Marker E-Cadherin in SKOV-3 Cells
2.5. Gene Expression Mediated by A2BR Activation in SKOV-3 Cells
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Reverse Transcription (RT) and Polymerase Chain Reaction (PCR)
4.3. Biotinylation of Plasma Membrane Proteins
4.4. Western Blot
4.5. Cell Viability
4.6. Lentiviral Infection
4.7. Immunofluorescence
4.8. Actin Cytoskeleton Labeling
4.9. Wound Closure Assay
4.10. Cancer Database Analysis
4.11. cDNA Microarray Analysis
4.12. Statistics Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Allard, B.; Longhi, M.S.; Robson, S.C.; Stagg, J. The Ectonucleotidases CD39 and CD73: Novel Checkpoint Inhibitor Targets. Immunol. Rev. 2017, 276, 121–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pellegatti, P.; Raffaghello, L.; Bianchi, G.; Piccardi, F.; Pistoia, V.; Di Virgilio, F. Increased Level of Extracellular ATP at Tumor Sites: In Vivo Imaging with Plasma Membrane Luciferase. PLoS ONE 2008, 3, e2599. [Google Scholar] [CrossRef] [PubMed]
- de Leve, S.; Wirsdörfer, F.; Jendrossek, V. Targeting the Immunomodulatory CD73/Adenosine System to Improve the Therapeutic Gain of Radiotherapy. Front. Immunol. 2019, 10, 698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antonioli, L.; Fornai, M.; Pellegrini, C.; D’Antongiovanni, V.; Turiello, R.; Morello, S.; Haskó, G.; Blandizzi, C. Adenosine Signaling in the Tumor Microenvironment. In Tumor Microenvironment: Signaling Pathways—Part B; Advances in Experimental Medicine and Biology; Birbrair, A., Ed.; Springer International Publishing: Cham, Switzerland, 2021; pp. 145–167. ISBN 978-3-030-47189-7. [Google Scholar]
- Deaglio, S.; Dwyer, K.M.; Gao, W.; Friedman, D.; Usheva, A.; Erat, A.; Chen, J.-F.; Enjyoji, K.; Linden, J.; Oukka, M.; et al. Adenosine Generation Catalyzed by CD39 and CD73 Expressed on Regulatory T Cells Mediates Immune Suppression. J. Exp. Med. 2007, 204, 1257–1265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kjaergaard, J.; Hatfield, S.; Jones, G.; Ohta, A.; Sitkovsky, M. A2A Adenosine Receptor Gene Deletion or Synthetic A2A Antagonist Liberate Tumor-Reactive CD8+ T Cells from Tumor-Induced Immunosuppression. J. Immunol. 2018, 201, 782–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aghaei, M.; Karami-Tehrani, F.; Panjehpour, M.; Salami, S.; Fallahian, F. Adenosine Induces Cell-Cycle Arrest and Apoptosis in Androgen-Dependent and -Independent Prostate Cancer Cell Lines, LNcap-FGC-10, DU-145, and PC3. Prostate 2012, 72, 361–375. [Google Scholar] [CrossRef]
- Shirali, S.; Aghaei, M.; Shabani, M.; Fathi, M.; Sohrabi, M.; Moeinifard, M. Adenosine Induces Cell Cycle Arrest and Apoptosis via CyclinD1/Cdk4 and Bcl-2/Bax Pathways in Human Ovarian Cancer Cell Line OVCAR-3. Tumor Biol. 2013, 34, 1085–1095. [Google Scholar] [CrossRef]
- Yang, D.; Song, J.; Wu, L.; Ma, Y.; Song, C.; Dovat, S.; Nishizaki, T.; Liu, J. Induction of Senescence by Adenosine Suppressing the Growth of Lung Cancer Cells. Biochem. Biophys. Res. Commun. 2013, 440, 62–67. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, J.; Zhang, Q.; Chen, P.; Song, J.; Yu, S.; Liu, H.; Liu, F.; Song, C.; Yang, D.; et al. Adenosine Induces Apoptosis in Human Liver Cancer Cells through ROS Production and Mitochondrial Dysfunction. Biochem. Biophys. Res. Commun. 2014, 448, 8–14. [Google Scholar] [CrossRef]
- Campos-Contreras, A.d.R.; Díaz-Muñoz, M.; Vázquez-Cuevas, F.G. Purinergic Signaling in the Hallmarks of Cancer. Cells 2020, 9, 1612. [Google Scholar] [CrossRef]
- Fredholm, B.B.; IJzerman, A.P.; Jacobson, K.A.; Linden, J.; Müller, C.E. International Union of Basic and Clinical Pharmacology. LXXXI. Nomenclature and Classification of Adenosine Receptors—An Update. Pharmacol. Rev. 2011, 63, 1–34. [Google Scholar] [CrossRef] [PubMed]
- Feoktistov, I.; Goldstein, A.E.; Biaggioni, I. Role of P38 Mitogen-Activated Protein Kinase and Extracellular Signal-Regulated Protein Kinase Kinase in Adenosine A2B Receptor-Mediated Interleukin-8 Production in Human Mast Cells. Mol. Pharmacol. 1999, 55, 726–734. [Google Scholar] [PubMed]
- Gao, Z.; Chen, T.; Weber, M.J.; Linden, J. A2B Adenosine and P2Y2 Receptors Stimulate Mitogen-Activated Protein Kinase in Human Embryonic Kidney-293 Cells: Cross-Talk between Cyclic Amp and Protein Kinase C Pathways. J. Biol. Chem. 1999, 274, 5972–5980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiang, H.; Liu, Z.; Wang, D.; Chen, Y.; Yang, Y.; Dou, K. Adenosine A2b Receptor Is Highly Expressed in Human Hepatocellular Carcinoma. Hepatol. Res. 2006, 36, 56–60. [Google Scholar] [CrossRef]
- Ma, D.-F.; Kondo, T.; Nakazawa, T.; Niu, D.-F.; Mochizuki, K.; Kawasaki, T.; Yamane, T.; Katoh, R. Hypoxia-Inducible Adenosine A2B Receptor Modulates Proliferation of Colon Carcinoma Cells. Hum. Pathol. 2010, 41, 1550–1557. [Google Scholar] [CrossRef]
- Zhou, Y.; Chu, X.; Deng, F.; Tong, L.; Tong, G.; Yi, Y.; Liu, J.; Tang, J.; Tang, Y.; Xia, Y.; et al. The Adenosine A2b Receptor Promotes Tumor Progression of Bladder Urothelial Carcinoma by Enhancing MAPK Signaling Pathway. Oncotarget 2017, 8, 48755–48768. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Gallardo, M.; González-Ramírez, R.; Sandoval, A.; Felix, R.; Monjaraz, E. Adenosine Stimulate Proliferation and Migration in Triple Negative Breast Cancer Cells. PLoS ONE 2016, 11, e0167445. [Google Scholar] [CrossRef] [Green Version]
- Klymenko, Y.; Kim, O.; Stack, M.S. Complex Determinants of Epithelial: Mesenchymal Phenotypic Plasticity in Ovarian Cancer. Cancers 2017, 9, 104. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Ramírez, A.S.; Díaz-Muñoz, M.; Butanda-Ochoa, A.; Vázquez-Cuevas, F.G. Nucleotides and Nucleoside Signaling in the Regulation of the Epithelium to Mesenchymal Transition (EMT). Purinergic Signal. 2017, 13, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Yi, Y.; Zhou, Y.; Chu, X.; Zheng, X.; Fei, D.; Lei, J.; Qi, H.; Dai, Y. Blockade of Adenosine A2b Receptor Reduces Tumor Growth and Migration in Renal Cell Carcinoma. J. Cancer 2020, 11, 421–431. [Google Scholar] [CrossRef] [Green Version]
- Gao, Z.W.; Wang, H.P.; Dong, K.; Lin, F.; Wang, X.; Zhang, H.Z. Adenosine Inhibits Migration, Invasion and Induces Apoptosis of Human Cervical Cancer Cells. Neoplasma 2016, 63, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Giacomelli, C.; Daniele, S.; Romei, C.; Tavanti, L.; Neri, T.; Piano, I.; Celi, A.; Martini, C.; Trincavelli, M.L. The A2BAdenosine Receptor Modulates the Epithelial- Mesenchymal Transition through the Balance of CAMP/PKA and MAPK/ERK Pathway Activation in Human Epithelial Lung Cells. Front. Pharmacol. 2018, 9, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez-Ramírez, A.S.; Díaz-Muñoz, M.; Battastini, A.M.; Campos-Contreras, A.; Olvera, A.; Bergamin, L.; Glaser, T.; Jacintho Moritz, C.E.; Ulrich, H.; Vázquez-Cuevas, F.G. Cellular Migration Ability Is Modulated by Extracellular Purines in Ovarian Carcinoma SKOV-3 Cells. J. Cell. Biochem. 2017, 118, 4468–4478. [Google Scholar] [CrossRef] [PubMed]
- Balázs, G. KM-Plot. Available online: http://www.kmplot.com/analysis (accessed on 19 November 2021).
- Gyorffy, B.; Lánczky, A.; Szállási, Z. Implementing an Online Tool for Genome-Wide Validation of Survival-Associated Biomarkers in Ovarian-Cancer Using Microarray Data from 1287 Patients. Endocr. Relat. Cancer 2012, 19, 197–208. [Google Scholar] [CrossRef] [Green Version]
- Sureechatchaiyan, P.; Hamacher, A.; Brockmann, N.; Stork, B.; Kassack, M.U. Adenosine Enhances Cisplatin Sensitivity in Human Ovarian Cancer Cells. Purinergic Signal. 2018, 14, 395–408. [Google Scholar] [CrossRef]
- Schulte, G.; Fredholm, B.B. Human Adenosine A1, A2A, A2B, and A3 Receptors Expressed in Chinese Hamster Ovary Cells All Mediate the Phosphorylation of Extracellular-Regulated Kinase 1/2. Mol. Pharmacol. 2000, 58, 477–482. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Ramírez, A.S.; Garay, E.; García-Carrancá, A.; Vázquez-Cuevas, F.G. The P2RY2 Receptor Induces Carcinoma Cell Migration and EMT Through Cross-Talk with Epidermal Growth Factor Receptor. J. Cell. Biochem. 2016, 117, 1016–1026. [Google Scholar] [CrossRef]
- Ohta, A.; Gorelik, E.; Prasad, S.J.; Ronchese, F.; Lukashev, D.; Wong, M.K.K.; Huang, X.; Caldwell, S.; Liu, K.; Smith, P.; et al. A2A Adenosine Receptor Protects Tumors from Antitumor T Cells. Proc. Natl. Acad. Sci. USA 2006, 103, 13132–13137. [Google Scholar] [CrossRef] [Green Version]
- Wilkat, M.; Bast, H.; Drees, R.; Dünser, J.; Mahr, A.; Azoitei, N.; Marienfeld, R.; Frank, F.; Brhel, M.; Ushmorov, A.; et al. Adenosine Receptor 2B Activity Promotes Autonomous Growth, Migration as Well as Vascularization of Head and Neck Squamous Cell Carcinoma Cells. Int. J. Cancer 2020, 147, 202–217. [Google Scholar] [CrossRef]
- Ou, Y.; Chan, G.; Zuo, J.; Rattner, J.B.; van der Hoorn, F.A. Purinergic A2b Receptor Activation by Extracellular Cues Affects Positioning of the Centrosome and Nucleus and Causes Reduced Cell Migration. J. Biol. Chem. 2016, 291, 15388–15403. [Google Scholar] [CrossRef] [Green Version]
- Vasiukov, G.; Menshikh, A.; Owens, P.; Novitskaya, T.; Hurley, P.; Blackwell, T.; Feoktistov, I.; Novitskiy, S.V. Adenosine/TGFβ Axis in Regulation of Mammary Fibroblast Functions. PLoS ONE 2021, 16, e0252424. [Google Scholar] [CrossRef] [PubMed]
- Yousefi, M.; Dehghani, S.; Nosrati, R.; Ghanei, M.; Salmaninejad, A.; Rajaie, S.; Hasanzadeh, M.; Pasdar, A. Current Insights into the Metastasis of Epithelial Ovarian Cancer—Hopes and Hurdles. Cell Oncol. 2020, 43, 515–538. [Google Scholar] [CrossRef] [PubMed]
- Clayton, A.; Al-Taei, S.; Webber, J.; Mason, M.D.; Tabi, Z. Cancer Exosomes Express CD39 and CD73, Which Suppress T Cells through Adenosine Production. J. Immunol. 2011, 187, 676–683. [Google Scholar] [CrossRef] [PubMed]
- Nooshabadi, V.T.; Arab, S. Targeting Tumor-Derived Exosomes Expressing CD73: New Opportunities in the Pathogenesis and Treatment of Cancer. Curr. Mol. Med. 2021, 21, 476–483. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, R.; Ray Chaudhuri, S.; Roy, S.S. FGF9-induced ovarian cancer cell invasion involves VEGF-A/VEGFR2 augmentation by virtue of ETS1 upregulation and metabolic reprogramming. J. Cell. Biochem. 2018, 119, 8174–8189. [Google Scholar] [CrossRef] [PubMed]
- Yu, A.-S.; Zhao, L. Effects of the GSK-3β Inhibitor (2Z,3E)-6-Bromoindirubin-3′-Oxime upon Ovarian Cancer Cells. Tumour Biol. 2016, 37, 4857–4864. [Google Scholar] [CrossRef]
- Hao, P.; Li, H.; Wu, A.; Zhang, J.; Wang, C.; Xian, X.; Ren, Q.; Hao, N.; Wang, Y.; Yue, F.; et al. Lipocalin2 Promotes Cell Proliferation and Migration in Ovarian Cancer through Activation of the ERK/GSK3β/β-Catenin Signaling Pathway. Life Sci. 2020, 262, 118492. [Google Scholar] [CrossRef]
- Cheon, D.-J.; Li, A.J.; Beach, J.A.; Walts, A.E.; Tran, H.; Lester, J.; Karlan, B.Y.; Orsulic, S. ADAM12 Is a Prognostic Factor Associated with an Aggressive Molecular Subtype of High-Grade Serous Ovarian Carcinoma. Carcinogenesis 2015, 36, 739–747. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Zhang, Y.; Bai, J.; Xue, Y.; Peng, Q. MMP1 and MMP9 Are Potential Prognostic Biomarkers and Targets for Uveal Melanoma. BMC Cancer 2021, 21, 1068. [Google Scholar] [CrossRef]
- Li, J.; Zhang, S.; Wu, L.; Pei, M.; Jiang, Y. Berberine Inhibited Metastasis through MiR-145/MMP16 Axis in Vitro. J. Ovarian Res. 2021, 14, 4. [Google Scholar] [CrossRef]
- Luo, X.; Zhang, X.; Peng, J.; Chen, Y.; Zhao, W.; Jiang, X.; Su, L.; Xie, M.; Lin, B. MiR-371b-5p Promotes Cell Proliferation, Migration and Invasion in Non-Small Cell Lung Cancer via SCAI. Biosci. Rep. 2020, 40, BSR20200163. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Tsai, Y.-H.; Tseng, S.-H. Regulation of ZMYND8 to Treat Cancer. Molecules 2021, 26, 1083. [Google Scholar] [CrossRef] [PubMed]
- Basu, M.; Sengupta, I.; Khan, M.W.; Srivastava, D.K.; Chakrabarti, P.; Roy, S.; Das, C. Dual Histone Reader ZMYND8 Inhibits Cancer Cell Invasion by Positively Regulating Epithelial Genes. Biochem. J. 2017, 474, 1919–1934. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Liu, J.; Chen, X.; Li, Y.; Li, Z.; Shen, C.; Chen, K.; Zhang, X. Low Expression of ZMYND8 Correlates with Aggressive Features and Poor Prognosis in Nasopharyngeal Carcinoma. CMAR 2019, 11, 7835–7843. [Google Scholar] [CrossRef] [Green Version]
- Mandai, K.; Nakanishi, H.; Satoh, A.; Takahashi, K.; Satoh, K.; Nishioka, H.; Mizoguchi, A.; Takai, Y. Ponsin/SH3P12: An l-Afadin- and Vinculin-Binding Protein Localized at Cell-Cell and Cell-Matrix Adherens Junctions. J. Cell Biol. 1999, 144, 1001–1017. [Google Scholar] [CrossRef]
- Takahashi, K.; Nakanishi, H.; Miyahara, M.; Mandai, K.; Satoh, K.; Satoh, A.; Nishioka, H.; Aoki, J.; Nomoto, A.; Mizoguchi, A.; et al. Nectin/PRR: An Immunoglobulin-like Cell Adhesion Molecule Recruited to Cadherin-Based Adherens Junctions through Interaction with Afadin, a PDZ Domain-Containing Protein. J. Cell Biol. 1999, 145, 539–549. [Google Scholar] [CrossRef]
- Takai, Y.; Irie, K.; Shimizu, K.; Sakisaka, T.; Ikeda, W. Nectins and Nectin-like Molecules: Roles in Cell Adhesion, Migration, and Polarization. Cancer Sci. 2003, 94, 655–667. [Google Scholar] [CrossRef]
- Oshima, T.; Sato, S.; Kato, J.; Ito, Y.; Watanabe, T.; Tsuji, I.; Hori, A.; Kurokawa, T.; Kokubo, T. Nectin-2 Is a Potential Target for Antibody Therapy of Breast and Ovarian Cancers. Mol. Cancer 2013, 12, 60. [Google Scholar] [CrossRef] [Green Version]
- Chomczynski, P.; Sacchi, N. Single-Step Method of RNA Isolation by Acid Guanidinium Thiocyanate-Phenol-Chloroform Extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef]
- Vázquez-Cuevas, F.G.; Cruz-Rico, A.; Garay, E.; García-Carrancá, A.; Pérez-Montiel, D.; Juárez, B.; Arellano, R.O. Differential Expression of the P2X7 Receptor in Ovarian Surface Epithelium during the Oestrous Cycle in the Mouse. Reprod. Fertil. Dev. 2013, 25, 971–984. [Google Scholar] [CrossRef]






| Down-Regulated Transcripts | |||
|---|---|---|---|
| Gene symbol | Gene name | Description | Z score |
| AKT3 | AKT serine/threonine kinase 3 | Peptidyl-serine phosphorylation | −3.08 |
| GSK3B | Glycogen synthase kinase 3 beta | −2.18 | |
| FGF9 | Fibroblast Growth Factor 9 | Regulation of cell migration | −2.46 |
| VCL | Vinculin | −3.68 | |
| ENNP2 | Ectonucleotide pyrophosphatase/phosphodiesterase 2 | −3.43 | |
| ADAM15 | ADAM Metallopeptidase Domain 15 | Proteolysis, extracellular matrix reorganization | −4.96 |
| ADAM12 | ADAM Metallopeptidase Domain 12 | −2.15 | |
| MMP2 | Matrix Metallopeptidase 2 | −2.69 | |
| MMP16 | Matrix Metallopeptidase 16 | −2.08 | |
| COL6A3 | Collagen, Type VI, Alpha 3 | −2.29 | |
| FGD1 | FYVE, RhoGEF and PH domain containing 1 | Actin cytoskeleton organization | −5.56 |
| CDC42EP2 | CDC42 effector protein 2 | −2.40 | |
| LIMK1 | LIM domain kinase 1 | −2.08 | |
| LIMK2 | LIM domain kinase 2 | −2.10 | |
| GNAI2 | Guanine Nucleotide-Binding Protein G(I) Subunit Alpha-2 | G protein-coupled receptor signaling pathway | −2.50 |
| PDE3B | Cyclic GMP-Inhibited Phosphodiesterase B | −4.49 | |
| PDE9A | Phosphodiesterase 9A | −2.99 | |
| Up-Regulated Transcripts | |||
| Gene symbol | Gene name | Description | Z score |
| GNAS | Adenylate Cyclase-Stimulating G Alpha Protein | G protein-coupled receptor signaling pathway | 2.44 |
| ADCYAP1R1 | ADCYAP receptor type I | 2.00 | |
| ARHGEF7 | Rho Guanine Nucleotide Exchange Factor 7 | 2.20 | |
| PFN1 | Profilin 1 | Actin cytoskeleton organization | 6.10 |
| CDC42BPA | CDC42 Binding Protein Kinase Alpha | 2.31 | |
| ADORA3 | Adenosine A3 Receptor | Negative regulation of cell migration | 3.44 |
| C9orf126 | Suppressor Of Cancer Cell Invasion | 2.51 | |
| MCTP1 | Multiple C2 And Transmembrane Domain Containing 1 | 2.26 | |
| PRKCBP1 | Protein Kinase C Beta | 2.05 | |
| ACTG1 | Actin Gamma 1 | Cytoskeleton organization | 2.11 |
| ANK1 | Ankyrin 1 | 3.13 | |
| AJAP1 | Adherents Junctions Associated Protein 1 | Cell adhesion | 3.21 |
| PVRL2 | Nectin Cell Adhesion Molecule 2 | 2.52 | |
| DSG1 | Desmoglein 1 | 2.32 | |
| CXADR | CXADR Ig-Like Cell Adhesion Molecule | 2.03 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Campos-Contreras, A.d.R.; González-Gallardo, A.; Díaz-Muñoz, M.; Vázquez-Cuevas, F.G. Adenosine Receptor A2B Negatively Regulates Cell Migration in Ovarian Carcinoma Cells. Int. J. Mol. Sci. 2022, 23, 4585. https://doi.org/10.3390/ijms23094585
Campos-Contreras AdR, González-Gallardo A, Díaz-Muñoz M, Vázquez-Cuevas FG. Adenosine Receptor A2B Negatively Regulates Cell Migration in Ovarian Carcinoma Cells. International Journal of Molecular Sciences. 2022; 23(9):4585. https://doi.org/10.3390/ijms23094585
Chicago/Turabian StyleCampos-Contreras, Anaí del Rocío, Adriana González-Gallardo, Mauricio Díaz-Muñoz, and Francisco G. Vázquez-Cuevas. 2022. "Adenosine Receptor A2B Negatively Regulates Cell Migration in Ovarian Carcinoma Cells" International Journal of Molecular Sciences 23, no. 9: 4585. https://doi.org/10.3390/ijms23094585
APA StyleCampos-Contreras, A. d. R., González-Gallardo, A., Díaz-Muñoz, M., & Vázquez-Cuevas, F. G. (2022). Adenosine Receptor A2B Negatively Regulates Cell Migration in Ovarian Carcinoma Cells. International Journal of Molecular Sciences, 23(9), 4585. https://doi.org/10.3390/ijms23094585






