Structural and Pharmacological Network Analysis of miRNAs Involved in Acute Ischemic Stroke: A Systematic Review
Abstract
:1. Introduction
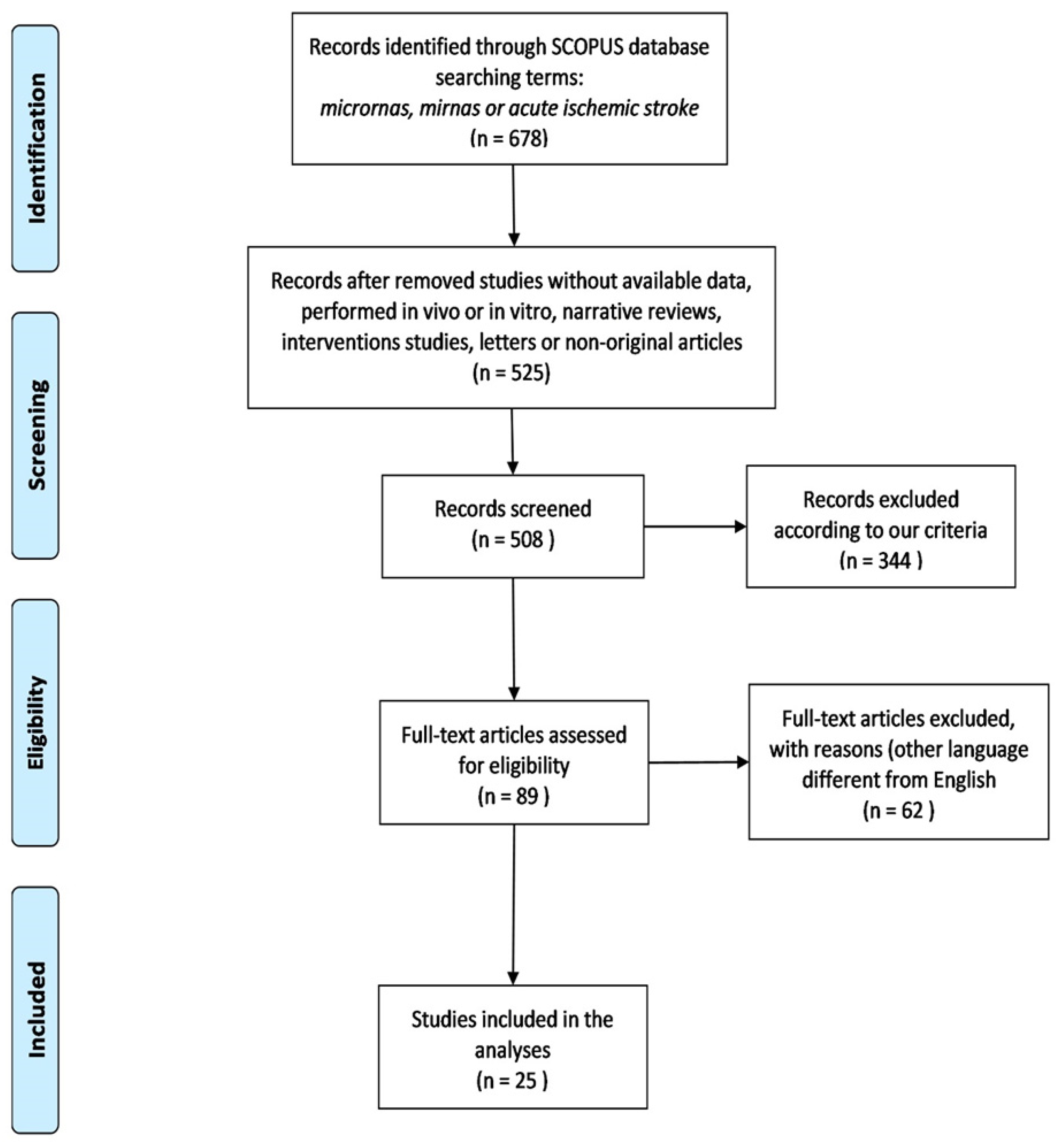
2. Materials and Methods
2.1. Study Strategy and Selection
2.1.1. Inclusion Criteria
- Studies were reported or published between 2015 and 2021.
- Studies discussed miRNAs differentially expressed in AIS.
- Studies presented both cases and control groups.
- Studies performed in human samples, such as whole blood, serum, plasma, exosomes, or blood cells; studies based on the ethnicity of study participants were not excluded.
- Only studies that validated AIS diagnosis by neuroimaging, such as computed tomography (CT) or magnetic resonance imaging (MRI), were included.
- Studies were conducted within 24 h of AIS symptoms.
2.1.2. Exclusion Criteria
- Studies published in languages excluding English.
- Narrative reviews, intervention studies, letters to editors, and non-original articles.
- Unpublished data, incomplete datasets, or preprints.
- Studies without available data.
- Studies without controls.
- Studies that used duplicated data.
- Studies performed in vivo.
- Studies performed in vitro, even when these were human-derived.
- Studies performed with already published databases.
2.2. Data Extraction
Data Collection
2.3. Network Structural Analysis
2.4. Network Pharmacology Analysis
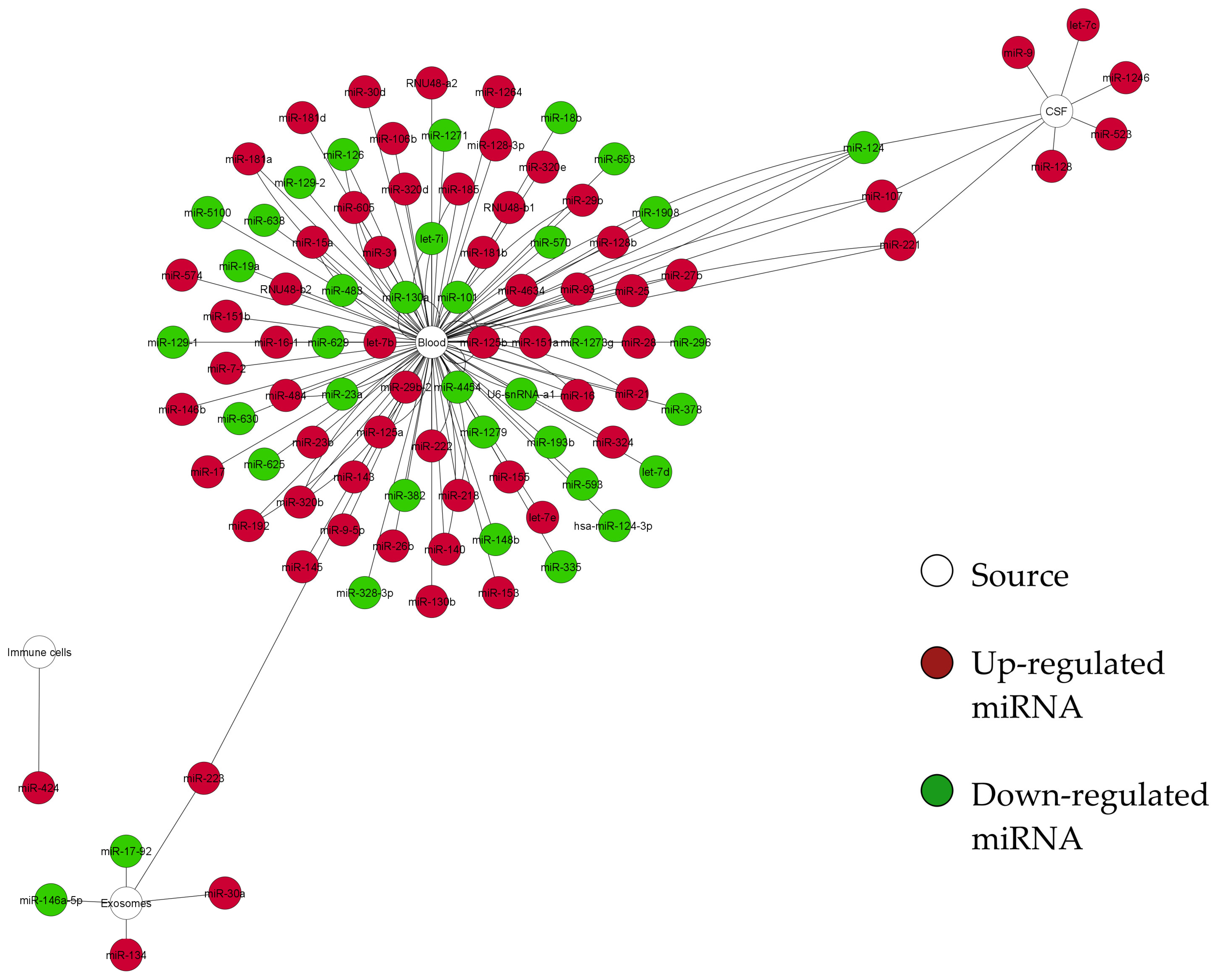
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Benjamin, E.J.; Muntner, P.; Alonso, A.; Bittencourt, M.S.; Callaway, C.W.; Carson, A.P.; Chamberlain, A.M.; Chang, A.R.; Cheng, S.; Das, S.R.; et al. Heart disease and stroke statistics-2019 update: A report from the american heart association. Circulation 2019, 139, e56–e528. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.P.; Liu, P.; Xu, C.M.; Li, G.W.; Gao, L.; Luo, Y.M. Unique MicroRNAs signature of lymphocyte of Yang and Yin syndromes in acute ischemic stroke patients. Chin. J. Integr. Med. 2019, 25, 590–597. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; De Silva, T.M.; Chen, J.; Faraci, F.M. Cerebral vascular disease and neurovascular injury in ischemic stroke. Circ. Res. 2017, 120, 449–471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fonarow, G.C.; Zhao, X.; Smith, E.E.; Saver, J.L.; Reeves, M.J.; Bhatt, D.L.; Xian, Y.; Hernandez, A.F.; Peterson, E.D.; Schwamm, L.H. Door-to-needle times for tissue plasminogen activator administration and clinical outcomes in acute ischemic stroke before and after a quality improvement initiative. JAMA 2014, 311, 1632–1640. [Google Scholar] [CrossRef]
- O’Carroll, C.B.; Aguilar, M.I. Management of postthrombolysis hemorrhagic and orolingual angioedema complications. Neurohospitalist 2015, 5, 133–141. [Google Scholar] [CrossRef] [Green Version]
- Gomolka, R.S.; Chrzan, R.M.; Urbanik, A.; Kazmierski, R.; Grzanka, A.D.; Nowinski, W.L. Quantification of image contrast of infarcts on computed tomography scans. Neuroradiol. J. 2017, 30, 15–22. [Google Scholar] [CrossRef]
- Gurav, S.K.; Zirpe, K.G.; Wadia, R.S.; Pathak, M.K.; Deshmukh, A.M.; Sonawane, R.V.; Goli, N. Problems and limitations in thrombolysis of acute stroke patients at a tertiary care center. Indian J. Crit. Care Med. 2015, 19, 265–269. [Google Scholar] [CrossRef]
- Beermann, J.; Piccoli, M.T.; Viereck, J.; Thum, T. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol. Rev. 2016, 96, 1297–1325. [Google Scholar] [CrossRef] [Green Version]
- Maitiseyiti, A.; Ci, H.; Fang, Q.; Guan, S.; Shawuti, A.; Wang, H.; Ge, X. Identification of novel long noncoding RNAs and Their Role in Abdominal Aortic Aneurysm. BioMed Res. Int. 2020, 2020, 3502518. [Google Scholar] [CrossRef]
- Schulte, C.; Barwari, T.; Joshi, A.; Zeller, T.; Mayr, M. Noncoding RNAs versus protein biomarkers in cardiovascular disease. Trends Mol. Med. 2020, 26, 583–596. [Google Scholar] [CrossRef] [Green Version]
- Wijerathne, H.; Witek, M.A.; Baird, A.E.; Soper, S.A. Liquid biopsy markers for stroke diagnosis. Expert Rev. Mol. Diagn. 2020, 20, 771–788. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Morris-Blanco, K.C.; Lopez, M.S.; Yang, T.; Zhao, H.; Vemuganti, R.; Luo, Y. Impact of microRNAs on ischemic stroke: From pre- to post-disease. Prog. Neurobiol. 2018, 163–164, 59–78. [Google Scholar] [CrossRef]
- Tiedt, S.; Dichgans, M.J.S. Role of non-coding RNAs in stroke. Stroke 2018, 49, 3098–3106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ouyang, Y.-B.; Giffard, R.G. MicroRNAs affect BCL-2 family proteins in the setting of cerebral ischemia. Neurochem. Int. 2014, 77, 2–8. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Zhang, J. Identification of miRNA-21 and miRNA-24 in plasma as potential early stage markers of acute cerebral infarction. Mol. Med. Rep. 2014, 10, 971–976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhiraj, D.K.; Chrysanthou, E.; Mallucci, G.R.; Bushell, M. miRNAs-19b, -29b-2* and -339-5p show an early and sustained up-regulation in ischemic models of stroke. PLoS ONE 2013, 8, e83717. [Google Scholar] [CrossRef]
- Spinetti, G.; Fortunato, O.; Caporali, A.; Shantikumar, S.; Marchetti, M.; Meloni, M.; Descamps, B.; Floris, I.; Sangalli, E.; Vono, R.; et al. MicroRNA-15a and microRNA-16 impair human circulating proangiogenic cell functions and are increased in the proangiogenic cells and serum of patients with critical limb ischemia. Circ. Res. 2013, 112, 335–346. [Google Scholar] [CrossRef] [Green Version]
- Sonawane, A.R.; Weiss, S.T.; Glass, K.; Sharma, A. Network medicine in the age of biomedical big data. Front. Genet. 2019, 10, 294. [Google Scholar] [CrossRef] [Green Version]
- Vocaturo, E.; Veltri, P. On the use of networks in biomedicine. Procedia Comput. Sci. 2017, 110, 498–503. [Google Scholar] [CrossRef]
- Bejleri, J.; Jirström, E.; Donovan, P.; Williams, D.J.; Pfeiffer, S. Diagnostic and prognostic circulating MicroRNA in acute stroke: A systematic and bioinformatic analysis of current evidence. J. Stroke 2021, 23, 162–182. [Google Scholar] [CrossRef]
- Xu, W.; Gao, L.; Zheng, J.; Li, T.; Shao, A.; Reis, C.; Chen, S.; Zhang, J. The roles of MicroRNAs in stroke: Possible therapeutic targets. Cell Transplant. 2018, 27, 1778–1788. [Google Scholar] [CrossRef] [Green Version]
- Dewdney, B.; Trollope, A.; Moxon, J.; Thomas Manapurathe, D.; Biros, E.; Golledge, J. Circulating MicroRNAs as biomarkers for acute ischemic stroke: A systematic review. J. Stroke Cerebrovasc. Dis. 2018, 27, 522–530. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Huang, L.; Liang, L.; Chen, L.; Zou, C.; Li, Z.; Li, R.; Jian, C.; Zou, D. Identification of an miRNA regulatory network and candidate markers for ischemic stroke related to diabetes. Int. J. Gen. Med. 2021, 14, 3213–3223. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Zhang, X.; Peng, S.; Sun, J.; Chen, X.; Deng, Y.; Yi, L. Identification of novel biomarkers in ischemic stroke: A genome-wide integrated analysis. BMC Med. Genet. 2020, 21, 66. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.W.; Ko, M.T.; Lin, C.Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8 (Suppl. 4), S11. [Google Scholar] [CrossRef] [Green Version]
- Maere, S.; Heymans, K.; Kuiper, M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005, 21, 3448–3449. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.J.; Xia, F.; Zou, J. Fast and covariate-adaptive method amplifies detection power in large-scale multiple hypothesis testing. Nat. Commun. 2019, 10, 3433. [Google Scholar] [CrossRef] [Green Version]
- Hassler, M.R.; Klisaroska, A.; Kollmann, K.; Steiner, I.; Bilban, M.; Schiefer, A.I.; Sexl, V.; Egger, G. Antineoplastic activity of the DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine in anaplastic large cell lymphoma. Biochimie 2012, 94, 2297–2307. [Google Scholar] [CrossRef] [Green Version]
- Yusefi, M.; Shameli, K.; Jahangirian, H.; Teow, S.Y.; Umakoshi, H.; Saleh, B.; Rafiee-Moghaddam, R.; Webster, T.J. The potential anticancer activity of 5-fluorouracil loaded in cellulose fibers isolated from rice straw. Int. J. Nanomed. 2020, 15, 5417–5432. [Google Scholar] [CrossRef]
- Li, X.; Chu, S.; Lin, M.; Gao, Y.; Liu, Y.; Yang, S.; Zhou, X.; Zhang, Y.; Hu, Y.; Wang, H. MC Anticancer property of ginsenoside Rh2 from ginseng. Eur. J. Med. Chem. 2020, 203, 112627. [Google Scholar] [CrossRef]
- Rager, J.E.; Smeester, L.; Jaspers, I.; Sexton, K.G.; Fry, R.C. Epigenetic changes induced by air toxics: Formaldehyde exposure alters miRNA expression profiles in human lung cells. Environ. Health Perspect. 2011, 119, 494–500. [Google Scholar] [CrossRef] [PubMed]
- Ghaffari, S.H.; Bashash, D.; Dizaji, M.Z.; Ghavamzadeh, A.; Alimoghaddam, K. Alteration in miRNA gene expression pattern in acute promyelocytic leukemia cell induced by arsenic trioxide: A possible mechanism to explain arsenic multi-target action. Tumour Biol. 2012, 33, 157–172. [Google Scholar] [CrossRef] [PubMed]
- Beljanski, V.; Trichostatin, A. xPharm: The Comprehensive Pharmacology Reference; Enna, S.J., Bylund, D.B., Eds.; Elsevier: New York, NY, USA, 2009; pp. 1–4. [Google Scholar] [CrossRef]
- Wei, K.; He, H.; Li, H.; Wang, L.; Ruan, L.; Pang, D.; Cheng, H. Gallotannin 1,2,6-tri-O-galloyl-β-d-glucopyranose: Its availability and changing patterns in tea (Camellia sinensis). Food Chem. 2019, 296, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Kalani, M.Y.S.; Alsop, E.; Meechoovet, B.; Beecroft, T.; Agrawal, K.; Whitsett, T.G.; Huentelman, M.J.; Spetzler, R.F.; Nakaji, P.; Kim, S.; et al. Extracellular microRNAs in blood differentiate between ischaemic and haemorrhagic stroke subtypes. J. Extracell. Vesicles 2020, 9, 1713540. [Google Scholar] [CrossRef] [Green Version]
- Khoshnam, S.E.; Winlow, W.; Farbood, Y.; Moghaddam, H.F.; Farzaneh, M. Emerging roles of microRNAs in ischemic stroke: As possible therapeutic agents. J. Stroke 2017, 19, 166–187. [Google Scholar] [CrossRef]
- Martinez, B.; Peplow, P.V. MicroRNAs as diagnostic markers and therapeutic targets for traumatic brain injury. Neural Regen. Res. 2017, 12, 1749–1761. [Google Scholar] [CrossRef]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as biomarkers in disease: Latest findings regarding their role in diagnosis and prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef] [Green Version]
- Bulygin, K.V.; Beeraka, N.M.; Saitgareeva, A.R.; Nikolenko, V.N.; Gareev, I.; Beylerli, O.; Akhmadeeva, L.R.; Mikhaleva, L.M.; Torres Solis, L.F.; Solís Herrera, A.; et al. Can miRNAs be considered as diagnostic and therapeutic molecules in ischemic stroke pathogenesis?-Current status. Int. J. Mol. Sci. 2020, 21, 6728. [Google Scholar] [CrossRef]
- Bruno, D.C.F.; Donatti, A.; Martin, M.; Almeida, V.S.; Geraldis, J.C.; Oliveira, F.S.; Dogini, D.B.; Lopes-Cendes, I. Circulating nucleic acids in the plasma and serum as potential biomarkers in neurological disorders. Braz. J. Med. Biol. Res. 2020, 53, e9881. [Google Scholar] [CrossRef]
- He, X.W.; Shi, Y.H.; Liu, Y.S.; Li, G.F.; Zhao, R.; Hu, Y.; Lin, C.C.; Zhuang, M.T.; Su, J.J.; Liu, J.R. Increased plasma levels of miR-124-3p, miR-125b-5p and miR-192-5p are associated with outcomes in acute ischaemic stroke patients receiving thrombolysis. Atherosclerosis 2019, 289, 36–43. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, Y.; Graves, D.T. FOXO transcription factors: Their clinical significance and regulation. BioMed Res. Int. 2014, 2014, 925350. [Google Scholar] [CrossRef] [Green Version]
- Zhan, L.; Li, D.; Liang, D.; Wu, B.; Zhu, P.; Wang, Y.; Sun, W.; Xu, E. Activation of Akt/FoxO and inactivation of MEK/ERK pathways contribute to induction of neuroprotection against transient global cerebral ischemia by delayed hypoxic postconditioning in adult rats. Neuropharmacology 2012, 63, 873–882. [Google Scholar] [CrossRef]
- Yoo, K.Y.; Kwon, S.H.; Lee, C.H.; Yan, B.; Park, J.H.; Ahn, J.H.; Choi, J.H.; Ohk, T.G.; Cho, J.H.; Won, M.H. FoxO3a changes in pyramidal neurons and expresses in non-pyramidal neurons and astrocytes in the gerbil hippocampal CA1 region after transient cerebral ischemia. Neurochem. Res. 2012, 37, 588–595. [Google Scholar] [CrossRef]
- Zhou, H.; Wang, X.; Ma, L.; Deng, A.; Wang, S.; Chen, X. FoxO3 transcription factor promotes autophagy after transient cerebral ischemia/reperfusion. Int. J. Neurosci. 2019, 129, 738–745. [Google Scholar] [CrossRef]
- Deng, C.; Zhang, P.; Zhang, Y. Downregulation of FOxO4 promotes neuronal survival by mediating oxidative-stress–induced apoptosis after cerebral ischemia/reperfusion injury. Trop. J. Pharm. Res. 2021, 20, 23–28. [Google Scholar] [CrossRef]
- Garg, N.; Kumar, P.; Gadhave, K.; Giri, R. Chapter ten—The dark proteome of cancer: Intrinsic disorderedness and functionality of HIF-1α along with its interacting proteins. In Progress in Molecular Biology and Translational Science; Uversky, V.N., Ed.; Academic Press: Cambridge, MA, USA, 2019; Volume 166, pp. 371–403. [Google Scholar]
- Sneha, P.; Thirumal Kumar, D.; Lijo, J.; Megha, M.; Siva, R.; George Priya Doss, C. Probing the protein-protein interaction network of proteins causing maturity onset diabetes of the young. Adv. Protein Chem. Struct. Biol. 2018, 110, 167–202. [Google Scholar] [CrossRef]
- Van der Heide, L.P.; Smidt, M.P. Regulation of FoxO activity by CBP/p300-mediated acetylation. Trends Biochem. Sci. 2005, 30, 81–86. [Google Scholar] [CrossRef]


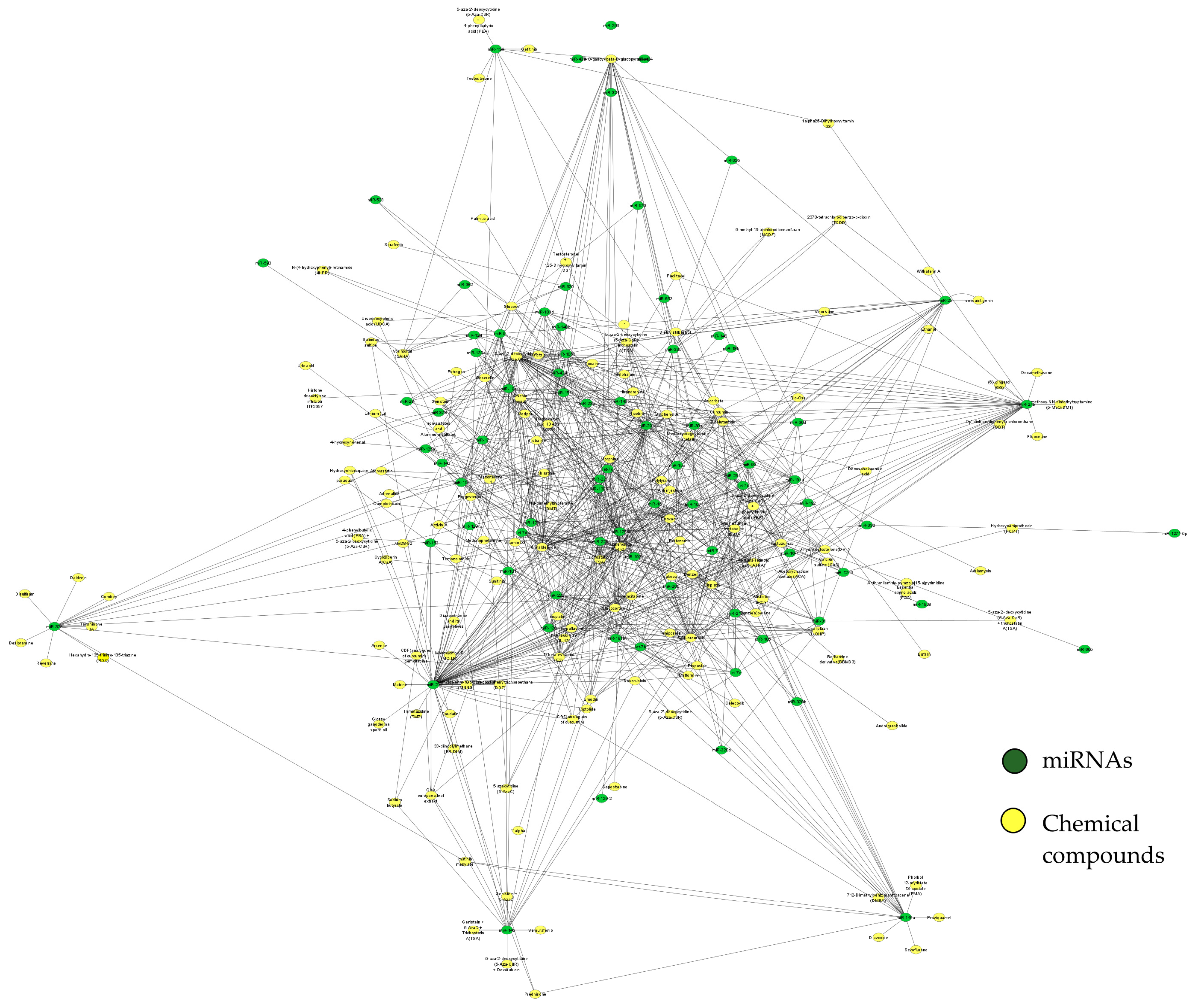
| Name | Score | Function | Type of Agent |
|---|---|---|---|
| 5-aza-2-deoxycytidine (5-Aza-CdR) | 55 | DNA methyltransferase inhibitor was able to reactivate genes silenced by DNA methylation and is a very potent epigenetic drug in several hematological malignancies [29]. | Chemotherapeutic drug |
| 5-Fluorouracil | 42 | In vitro studies have shown potential anticancer activity [30]. | Chemotherapeutic drug |
| Ginsenoside Rh2 | 38 | Major bioactive ginsenosides from Panax ginseng with anti-proliferation, anti-invasion, anti-metastasis, induction of cell cycle arrest, promotion of differentiation, and reversal of multi-drug resistance activities against multiple tumor cells, and also alleviates the side effects of chemotherapy or radiotherapy [31]. | Chemotherapeutic drug |
| Formaldehyde | 38 | Formaldehyde alters miRNA patterns that regulate gene expression, potentially leading to the initiation of various diseases [32]. | Carcinogenic compound |
| Arsenic trioxide | 32 | Alters the miRNA gene expression pattern in acute promyelocytic leukemia cells [33]. | Toxicant |
| Trichostatin A (TSA) | 29 | A fungistatic antibiotic was obtained from Streptomyces platensis. It causes an accumulation of acetylated histones in a variety of mammalian tumor cell lines [34]. | Antibiotic drug |
| 1,2,6-Tri-O-galloyl-beta-D-glucopyranose | 29 | The natural compound from Camellia sinensis plays multiple roles against multidrug-resistant bacteria and other diseases [35]. | Antibiotic drug |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barrera-Vázquez, O.S.; Gomez-Verjan, J.C.; Ramírez-Aldana, R.; Torre, P.G.-d.; Rivero-Segura, N.A. Structural and Pharmacological Network Analysis of miRNAs Involved in Acute Ischemic Stroke: A Systematic Review. Int. J. Mol. Sci. 2022, 23, 4663. https://doi.org/10.3390/ijms23094663
Barrera-Vázquez OS, Gomez-Verjan JC, Ramírez-Aldana R, Torre PG-d, Rivero-Segura NA. Structural and Pharmacological Network Analysis of miRNAs Involved in Acute Ischemic Stroke: A Systematic Review. International Journal of Molecular Sciences. 2022; 23(9):4663. https://doi.org/10.3390/ijms23094663
Chicago/Turabian StyleBarrera-Vázquez, Oscar Salvador, Juan Carlos Gomez-Verjan, Ricardo Ramírez-Aldana, Paola García-dela Torre, and Nadia Alejandra Rivero-Segura. 2022. "Structural and Pharmacological Network Analysis of miRNAs Involved in Acute Ischemic Stroke: A Systematic Review" International Journal of Molecular Sciences 23, no. 9: 4663. https://doi.org/10.3390/ijms23094663
APA StyleBarrera-Vázquez, O. S., Gomez-Verjan, J. C., Ramírez-Aldana, R., Torre, P. G.-d., & Rivero-Segura, N. A. (2022). Structural and Pharmacological Network Analysis of miRNAs Involved in Acute Ischemic Stroke: A Systematic Review. International Journal of Molecular Sciences, 23(9), 4663. https://doi.org/10.3390/ijms23094663









