RNA-Seq Analysis Identifies Transcription Factors Involved in Anthocyanin Biosynthesis of ‘Red Zaosu’ Pear Peel and Functional Study of PpPIF8
Abstract
:1. Introduction
2. Results
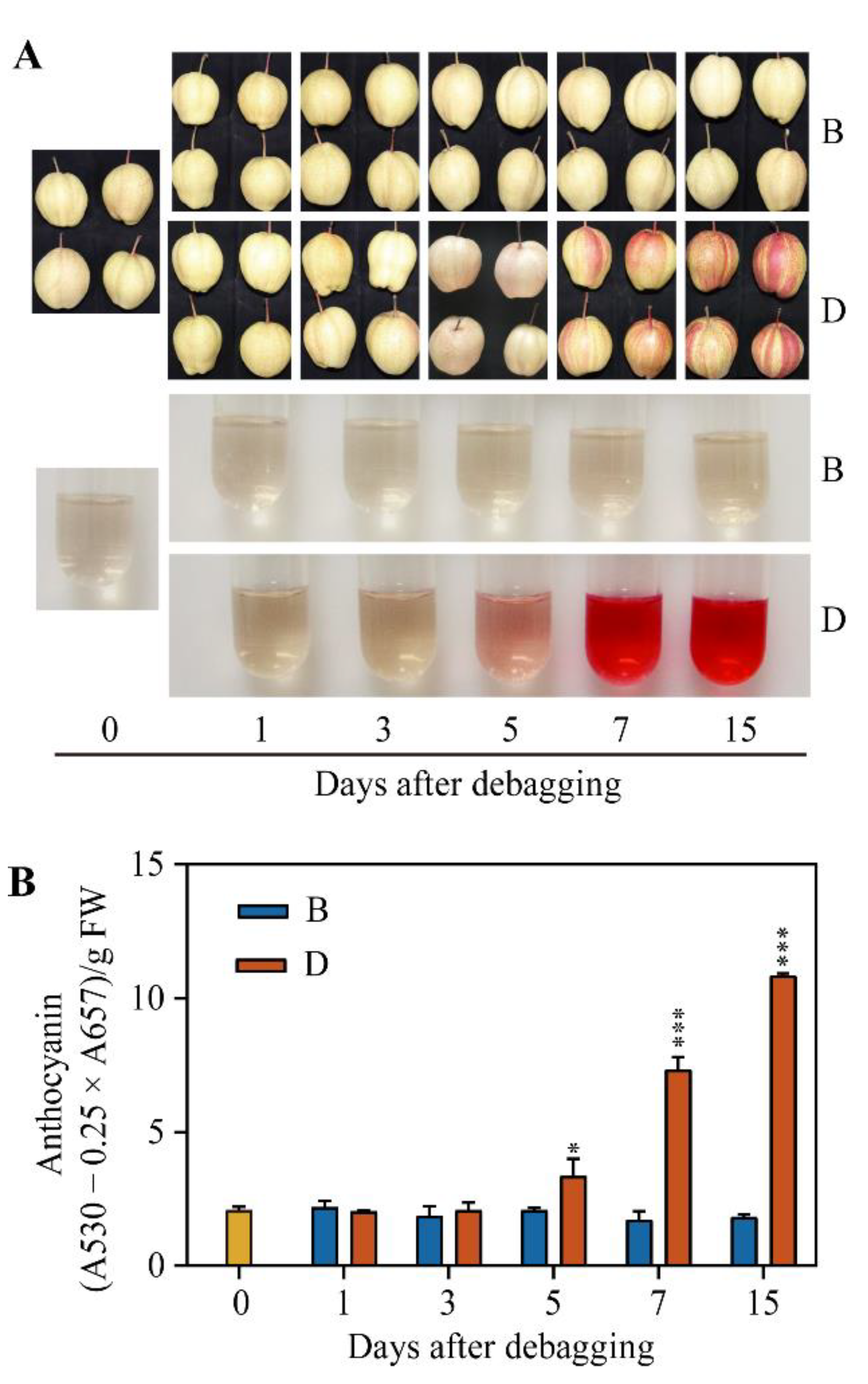
2.1. Anthocyanin Accumulation in the Peel of ‘Red Zaosu’ Pear after Debagging
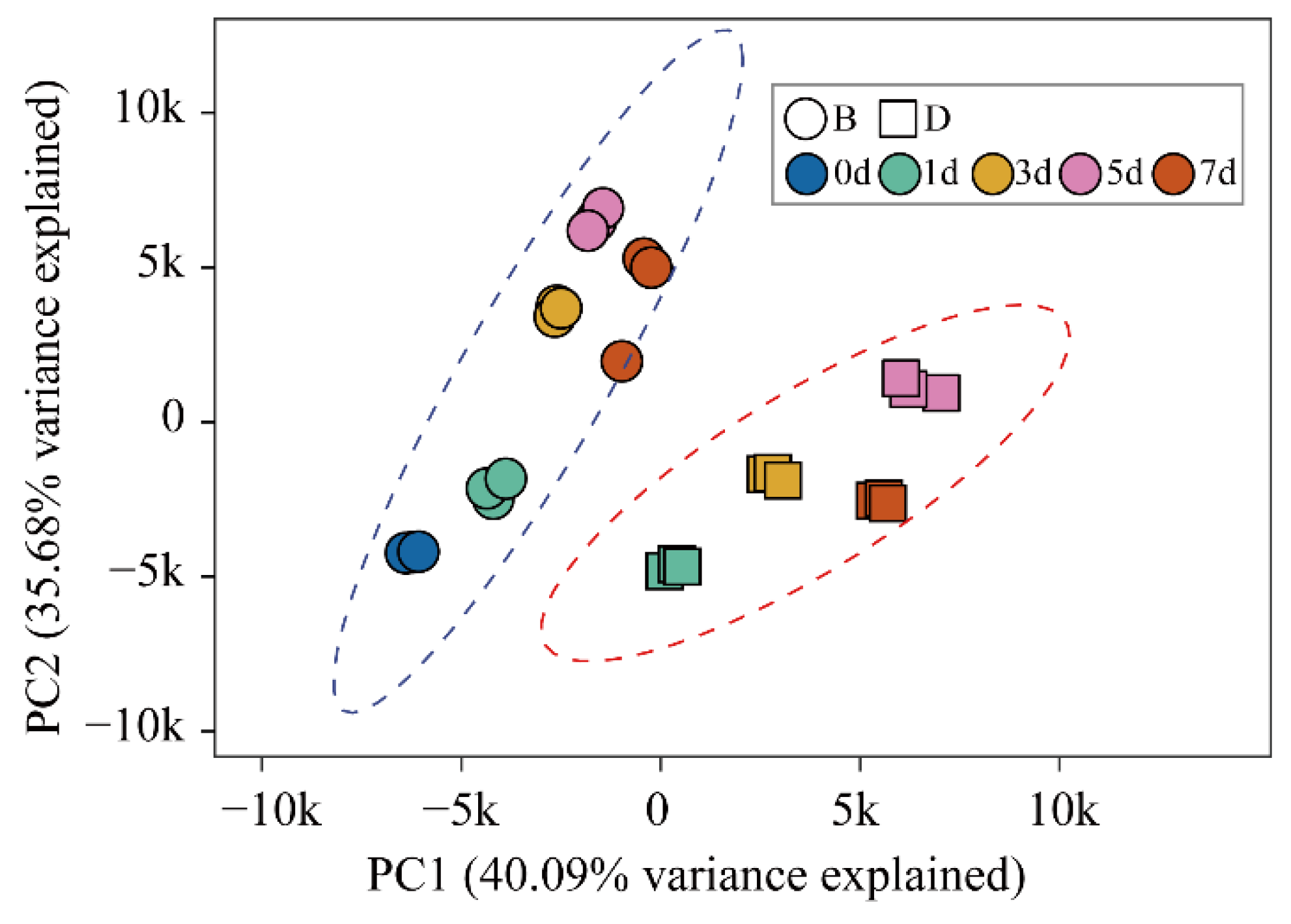
2.2. Overview of RNA Sequencing
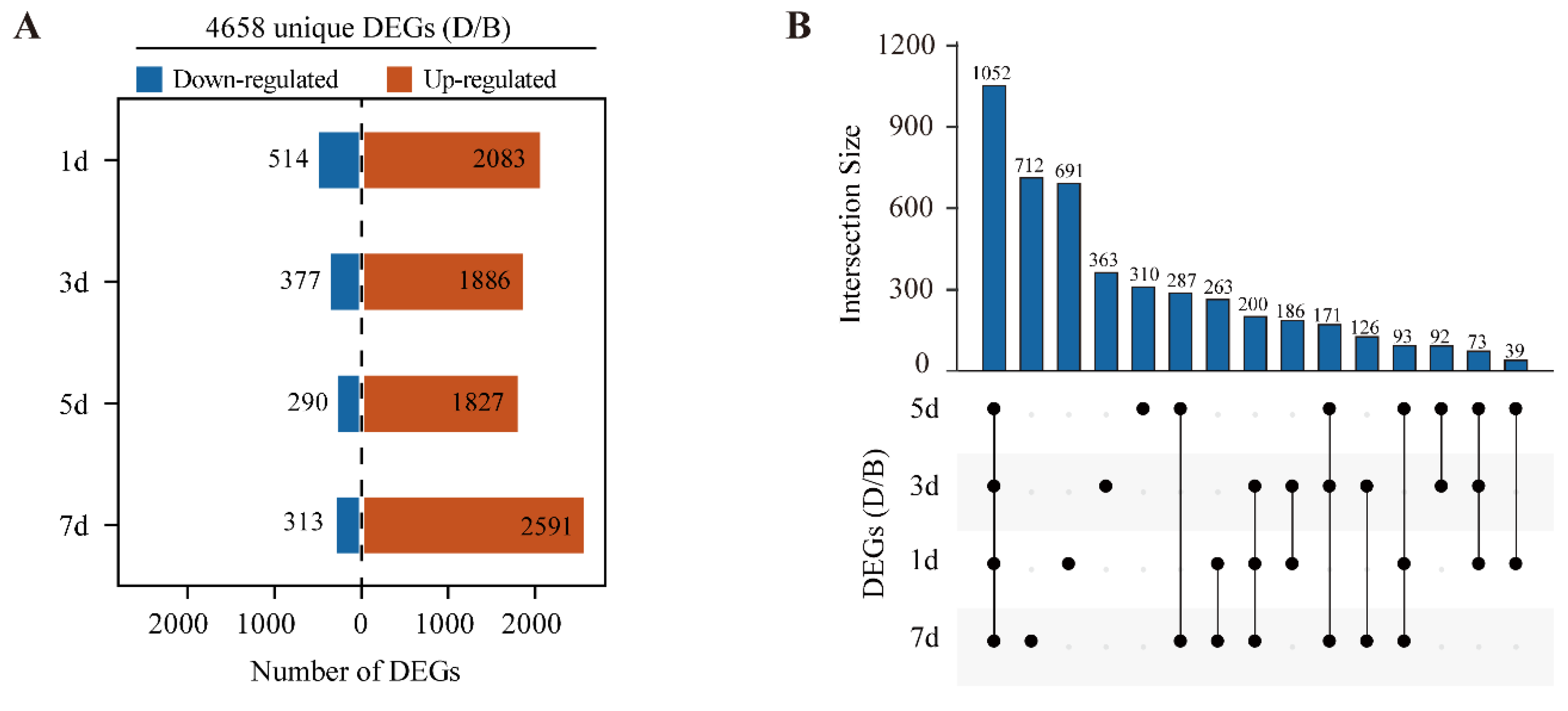
2.3. Differential Gene Expression Analysis
2.4. Weighted Gene Co-Expression Network Analysis (WGCNA)
2.4.1. Module Construct and Module–Trait Correlation
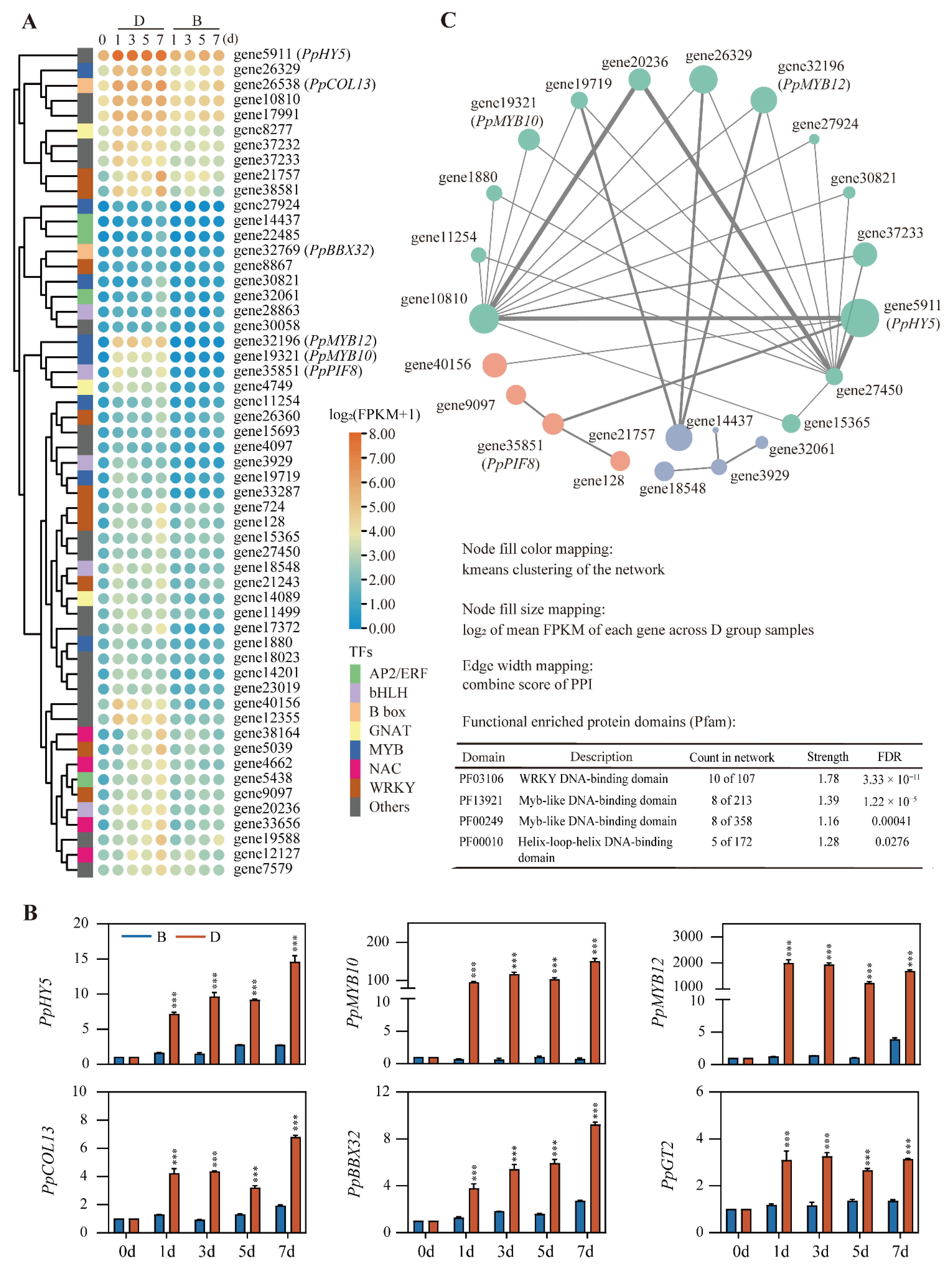
2.4.2. Analysis of Transcription Factors
2.4.3. Identifying Transcription Factors Regulating Gene Expression in “Blue” and “Turquoise” Modules
2.5. Functional Study of PpPIF8 in the Regulation of Anthocyanin Biosynthesis
2.5.1. Genome-Wide Identification of PIFs in Pear
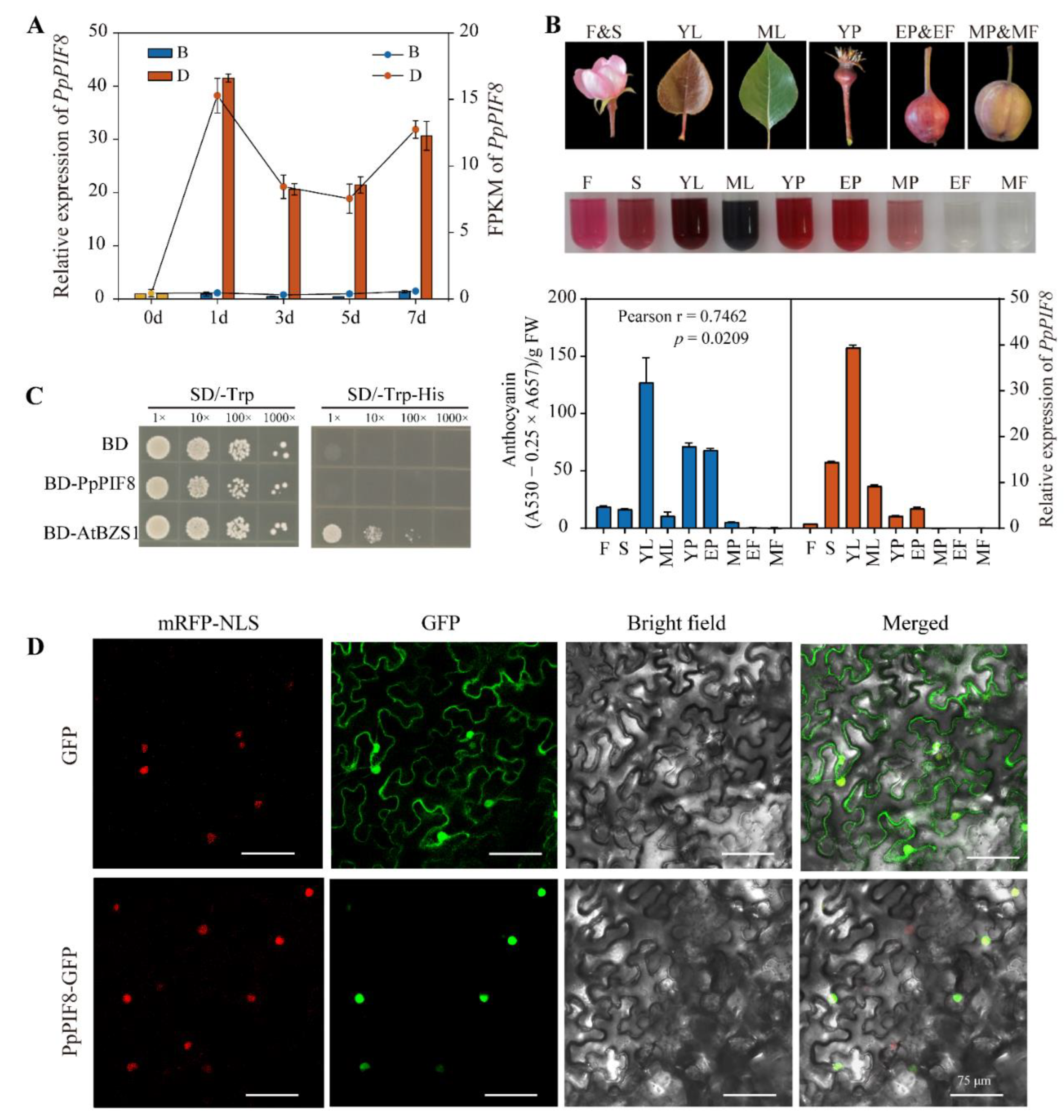
2.5.2. Verification of PpPIF8 Expression
2.5.3. Tissue-Specific Expression Analysis of PpPIF8
2.5.4. Transcriptional Activity and Subcellular Localization Analysis of PpPIF8
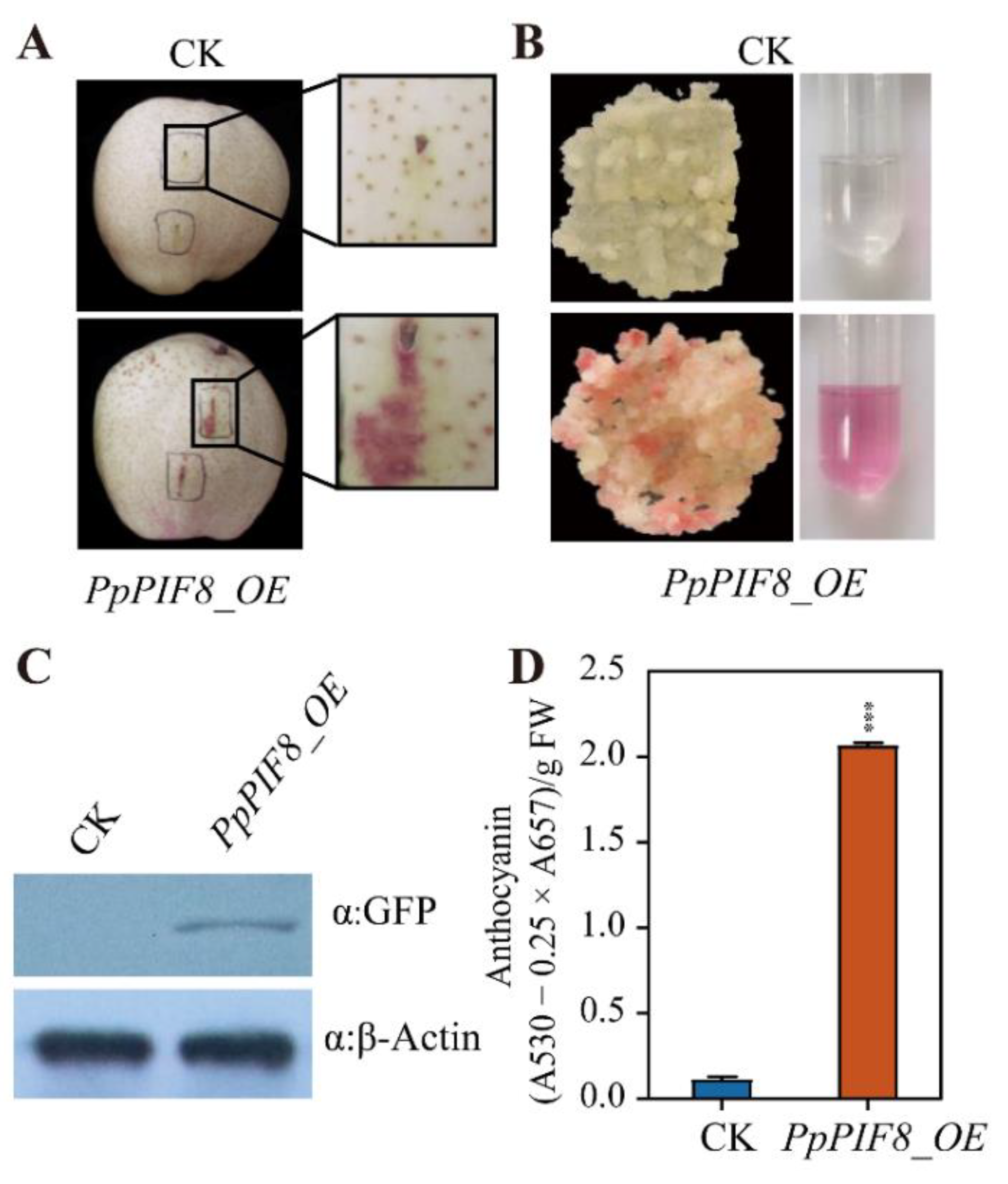
2.5.5. Overexpression of PpPIF8 Promotes Anthocyanin Accumulation in Pear
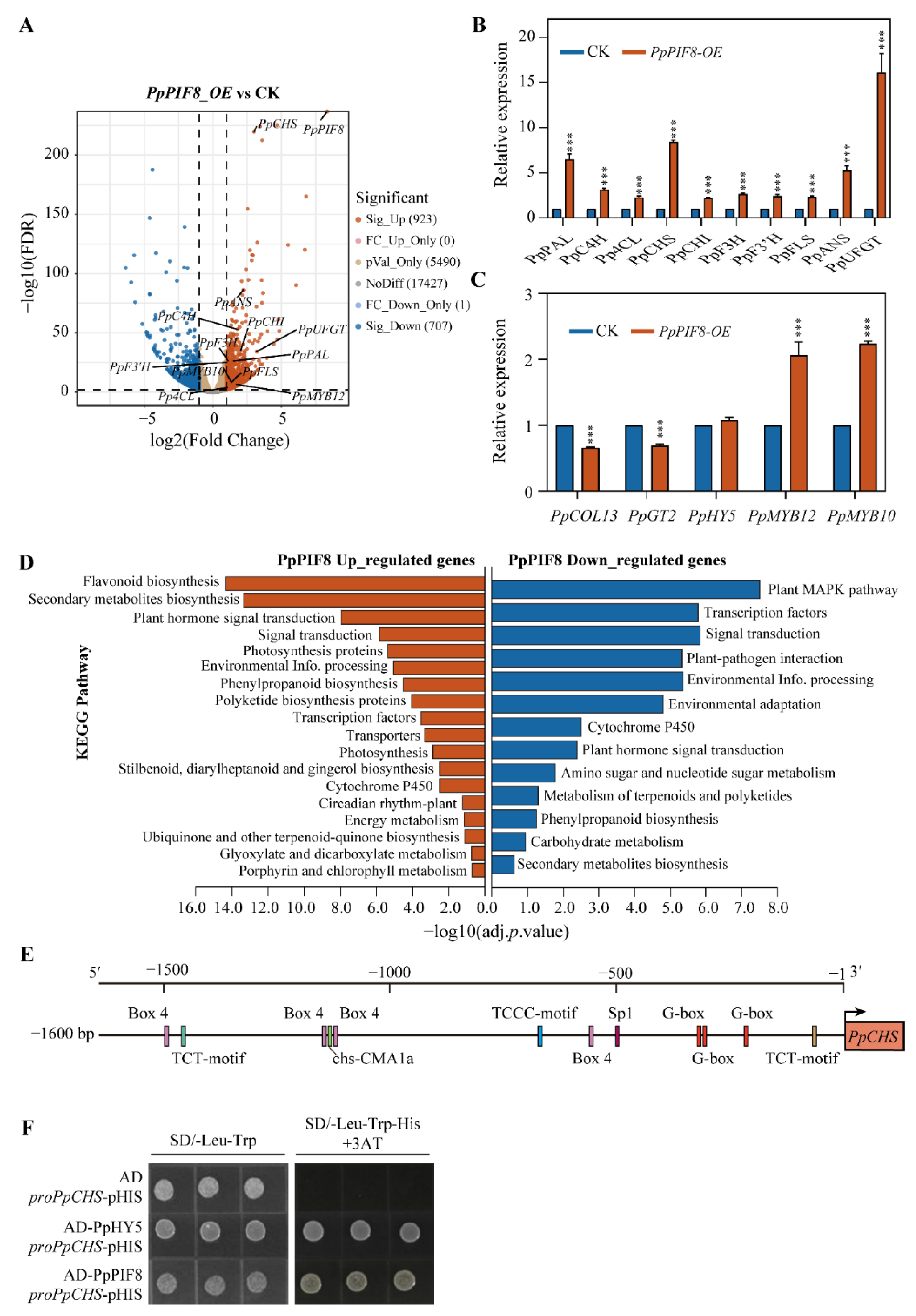
2.5.6. RNA-Seq Analysis of PpPIF8_OE Transgenic Pear Calli
3. Discussion
3.1. Light Is a Key Factor for Anthocyanin Biosynthesis in Pear Peel
3.2. Comprehensive RNA-Sequencing and Gene Co-Expression Network Analysis Identify Potential TFs Involved in Anthocyanin Biosynthesis
3.3. Identification of PIFs in Pear Genome
3.4. Tissue-Specific Expression, Subcellular Localization, and Transcriptional Activity of PpPIF8
3.5. PpPIF8 Positively Regulates Anthocyanin Biosynthesis
3.6. Molecular Mechanisms of PpPIF8 in Regulation of Anthocyanin Biosynthesis
4. Materials and Methods
4.1. Plant Materials and Treatments
4.2. Induction and Transformation of Pear Calli
4.3. Anthocyanin Extraction and Measurement
4.4. RNA Extraction and RT-qPCR
4.5. RNA-Seq Analysis and WGCNA
4.6. Identification of the PpPIF Genes and Phylogenetic Tree Construction
4.7. Subcellular Localization Analysis
4.8. Transient Overexpression Assay in ‘Red Zaosu’ Fruit
4.9. Transactivation Activity Assay
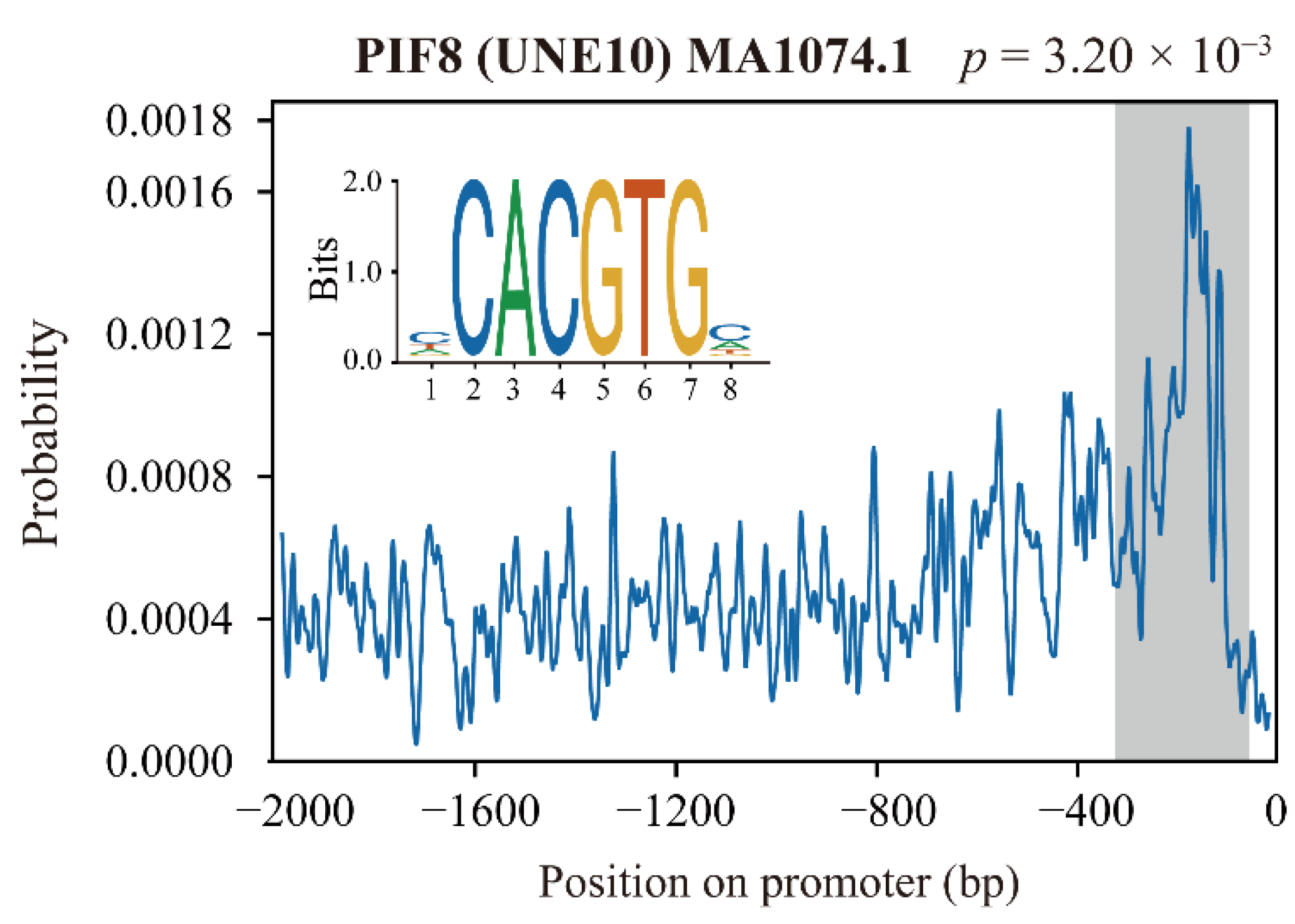
4.10. Yeast One-Hybrid Assay
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, H.; Liu, Z.; Wu, Y.; Zheng, L.; Zhang, G. Regulatory mechanisms of anthocyanin biosynthesis in apple and pear. Int. J. Mol. Sci. 2021, 22, 8441. [Google Scholar] [CrossRef]
- Zhang, Y.; Butelli, E.; Martin, C. Engineering anthocyanin biosynthesis in plants. Curr. Opin. Plant Biol. 2014, 19, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.-W.; Gong, C.-C.; Song, H.-F.; Cui, Y.-Y. Effects of anthocyanins on the prevention and treatment of cancer. Br. J. Pharmacol. 2017, 174, 1226–1243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaakola, L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gangappa, S.N.; Botto, J.F. The multifaceted roles of HY5 in plant growth and development. Mol. Plant 2016, 9, 1353–1365. [Google Scholar] [CrossRef] [Green Version]
- An, J.P.; Qu, F.J.; Yao, J.F.; Wang, X.N.; You, C.X.; Wang, X.F.; Hao, Y.J. The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Hortic. Res. 2017, 4, 17023. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Xu, P.; Chen, G.; Wu, J.; Liu, Z.; Lian, H. FvbHLH9 functions as a positive regulator of anthocyanin biosynthesis by forming a HY5-bHLH9 transcription complex in strawberry fruits. Plant Cell Physiol. 2020, 61, 826–837. [Google Scholar] [CrossRef]
- Shin, D.H.; Choi, M.; Kim, K.; Bang, G.; Cho, M.; Choi, S.B.; Choi, G.; Park, Y.I. HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Lett. 2013, 587, 1543–1547. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Wang, P.; Li, X.; Wang, Y.; Tian, S.; Qin, G. The transcription factor SlHY5 regulates the ripening of tomato fruit at both the transcriptional and translational levels. Hortic. Res. 2021, 8, 83. [Google Scholar] [CrossRef]
- Lee, J.; He, K.; Stolc, V.; Lee, H.; Figueroa, P.; Gao, Y.; Tongprasit, W.; Zhao, H.; Lee, I.; Deng, X.W. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 2007, 19, 731–749. [Google Scholar] [CrossRef] [Green Version]
- Bai, S.; Tao, R.; Yin, L.; Ni, J.; Yang, Q.; Yan, X.; Yang, F.; Guo, X.; Li, H.; Teng, Y. Two B-box proteins, PpBBX18 and PpBBX21, antagonistically regulate anthocyanin biosynthesis via competitive association with Pyrus pyrifolia ELONGATED HYPOCOTYL 5 in the peel of pear fruit. Plant J. 2019, 100, 1208–1223. [Google Scholar] [CrossRef] [PubMed]
- Bursch, K.; Toledo-Ortiz, G.; Pireyre, M.; Lohr, M.; Braatz, C.; Johansson, H. Identification of BBX proteins as rate-limiting cofactors of HY5. Nat. Plants 2020, 6, 921–928. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Chu, L.; Zhang, Y.; Bian, Y.; Xiao, J.; Xu, D. HY5: A pivotal regulator of light-dependent development in higher plants. Front Plant Sci. 2021, 12, 800989. [Google Scholar] [CrossRef] [PubMed]
- Ou, C.; Zhang, X.; Wang, F.; Zhang, L.; Zhang, Y.; Fang, M.; Wang, J.; Wang, J.; Jiang, S.; Zhang, Z. A 14 nucleotide deletion mutation in the coding region of the PpBBX24 gene is associated with the red skin of “Zaosu Red” pear (Pyrus pyrifolia White Pear Group): A deletion in the PpBBX24 gene is associated with the red skin of pear. Hortic. Res. 2020, 7, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Tao, R.; Yu, W.; Gao, Y.; Ni, J.; Yin, L.; Zhang, X.; Li, H.; Wang, D.; Bai, S.; Teng, Y. Light-induced basic/helix-loop-helix64 enhances anthocyanin biosynthesis and undergoes CONSTITUTIVELY PHOTOMORPHOGENIC1-mediated degradation in pear. Plant Physiol. 2020, 184, 1684–1701. [Google Scholar] [CrossRef] [PubMed]
- Cui, D.; Zhao, S.; Xu, H.; Allan, A.C.; Zhang, X.; Fan, L.; Chen, L.; Su, J.; Shu, Q.; Li, K. The interaction of MYB, bHLH and WD40 transcription factors in red pear (Pyrus pyrifolia) peel. Plant Mol. Biol. 2021, 106, 407–417. [Google Scholar] [CrossRef]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef]
- Feng, S.; Wang, Y.; Yang, S.; Xu, Y.; Chen, X. Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10. Planta 2010, 232, 245–255. [Google Scholar] [CrossRef]
- Jin, W.; Wang, H.; Li, M.; Wang, J.; Yang, Y.; Zhang, X.; Yan, G.; Zhang, H.; Liu, J.; Zhang, K. The R2R3 MYB transcription factor PavMYB10.1 involves in anthocyanin biosynthesis and determines fruit skin colour in sweet cherry (Prunus avium L.). Plant Biotechnol. J. 2016, 14, 2120–2133. [Google Scholar] [CrossRef] [Green Version]
- Espley, R.V.; Hellens, R.P.; Putterill, J.; Stevenson, D.E.; Kutty-Amma, S.; Allan, A.C. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 2007, 49, 414–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, Z.Z.; Lin-Wang, K.; Zhou, D.R.; Lin, Y.J.; Jiang, C.C.; Pan, S.L.; Espley, R.V.; Andre, C.M.; Ye, X.F. Activation of PsMYB10.2 transcription causes anthocyanin accumulation in flesh of the red-fleshed mutant of ‘Sanyueli’ (Prunus salicina Lindl.). Front Plant Sci. 2021, 12, 1167. [Google Scholar] [CrossRef] [PubMed]
- Zhai, R.; Zhao, Y.; Wu, M.; Yang, J.; Li, X.; Liu, H.; Wu, T.; Liang, F.; Yang, C.; Wang, Z.; et al. The MYB transcription factor PbMYB12b positively regulates flavonol biosynthesis in pear fruit. BMC Plant Biol. 2019, 19, 85. [Google Scholar] [CrossRef] [PubMed]
- Yao, G.; Ming, M.; Allan, A.C.; Gu, C.; Li, L.; Wu, X.; Wang, R.; Chang, Y.; Qi, K.; Zhang, S.; et al. Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. Plant J. 2017, 92, 437–451. [Google Scholar] [CrossRef] [Green Version]
- Castillon, A.; Shen, H.; Huq, E. Phytochrome interacting factors: Central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 2007, 12, 514–521. [Google Scholar] [CrossRef]
- Leivar, P.; Monte, E. PIFs: Systems integrators in plant development. Plant Cell 2014, 26, 56–78. [Google Scholar] [CrossRef] [Green Version]
- Khanna, R.; Huq, E.; Kikis, E.A.; Al-Sady, B.; Lanzatella, C.; Quail, P.H. A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell 2004, 16, 3033–3044. [Google Scholar] [CrossRef] [Green Version]
- Toledo-Ortiz, G.; Huq, E.; Quail, P.H. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 2003, 15, 1749–1770. [Google Scholar] [CrossRef] [Green Version]
- Shin, J.; Park, E.; Choi, G. PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J. 2007, 49, 981–994. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, Y.; Wang, J.; Li, P.; Zhao, C.; Chen, Y.; Bi, Y. Phytochrome-interacting factors PIF4 and PIF5 negatively regulate anthocyanin biosynthesis under red light in Arabidopsis seedlings. Plant Sci. 2015, 238, 64–72. [Google Scholar] [CrossRef]
- Liu, H.; Shu, Q.; Lin-Wang, K.; Allan, A.C.; Espley, R.V.; Su, J.; Pei, M.; Wu, J. The PyPIF5-PymiR156a-PySPL9-PyMYB114/MYB10 module regulates light-induced anthocyanin biosynthesis in red pear. Mol. Hortic. 2021, 1, 14. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, X.W.; Liu, X.; Zheng, P.F.; Su, L.; Wang, G.L.; Wang, X.F.; Li, Y.Y.; You, C.X.; An, J.P. Phytochrome interacting factor MdPIF7 modulates anthocyanin biosynthesis and hypocotyl growth in apple. Plant Physiol. 2022, 188, 2342–2363. [Google Scholar] [CrossRef] [PubMed]
- Xue, H.; Wang, S.; Yao, J.-L.; Deng, C.H.; Wang, L.; Su, Y.; Zhang, H.; Zhou, H.; Sun, M.; Li, X.; et al. Chromosome level high-density integrated genetic maps improve the Pyrus bretschneideri ‘DangshanSuli’ v1.0 genome. BMC Genom. 2018, 19, 833. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, T.L.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994, 2, 28–36. [Google Scholar]
- Bai, S.; Sun, Y.; Qian, M.; Yang, F.; Ni, J.; Tao, R.; Li, L.; Shu, Q.; Zhang, D.; Teng, Y. Transcriptome analysis of bagging-treated red Chinese sand pear peels reveals light-responsive pathway functions in anthocyanin accumulation. Sci. Rep. 2017, 7, 63. [Google Scholar] [CrossRef] [Green Version]
- Takos, A.M.; Jaffé, F.W.; Jacob, S.R.; Bogs, J.; Robinson, S.P.; Walker, A.R. Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 2006, 142, 1216–1232. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Zhu, Z.Z.; Qu, D.; Wang, B.C.; Hao, N.N.; Yang, Y.Z.; Yang, H.J.; Zhao, Z.Y. MdBBX21, a B-box protein, positively regulates light-induced anthocyanin accumulation in apple peel. Front. Plant Sci. 2021, 12, 774446. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinf. 2008, 9, 559. [Google Scholar] [CrossRef] [Green Version]
- Oh, J.; Park, E.; Song, K.; Bae, G.; Choi, G. PHYTOCHROME INTERACTING FACTOR8 inhibits phytochrome A-mediated far-red light responses in Arabidopsis. Plant Cell 2020, 32, 186–205. [Google Scholar] [CrossRef]
- Zhang, K.; Zheng, T.; Zhu, X.; Jiu, S.; Liu, Z.; Guan, L.; Jia, H.; Fang, J. Genome-wide identification of PIFs in grapes (Vitis vinifera L.) and their transcriptional analysis under lighting/shading conditions. Genes 2018, 9, 451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, P.F.; Wang, X.; Yang, Y.Y.; You, C.X.; Zhang, Z.L.; Hao, Y.J. Identification of phytochrome-interacting factor family members and functional analysis of MdPIF4 in Malus domestica. Int. J. Mol. Sci. 2020, 21, 7350. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Wang, Y.; Li, Q.F.; Bjorn, L.O.; He, J.X.; Li, S.S. Arabidopsis STO/BBX24 negatively regulates UV-B signaling by interacting with COP1 and repressing HY5 transcriptional activity. Cell Res. 2012, 22, 1046–1057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, S.; Tao, R.; Tang, Y.; Yin, L.; Ma, Y.; Ni, J.; Yan, X.; Yang, Q.; Wu, Z.; Zeng, Y.; et al. BBX16, a B-box protein, positively regulates light-induced anthocyanin accumulation by activating MYB10 in red pear. Plant Biotechnol. J. 2019, 17, 1985–1997. [Google Scholar] [CrossRef] [Green Version]
- Jeong, S.W.; Das, P.K.; Jeoung, S.C.; Song, J.Y.; Lee, H.K.; Kim, Y.K.; Kim, W.J.; Park, Y.I.; Yoo, S.D.; Choi, S.B.; et al. Ethylene suppression of sugar-induced anthocyanin pigmentation in Arabidopsis. Plant Physiol. 2010, 154, 1514–1531. [Google Scholar] [CrossRef] [Green Version]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Premathilake, A.T.; Gao, Y.; Yu, W.; Tao, R.; Teng, Y.; Bai, S. Ethylene-activated PpERF105 induces the expression of the repressor-type R2R3-MYB gene PpMYB140 to inhibit anthocyanin biosynthesis in red pear fruit. Plant J. 2020, 105, 167–181. [Google Scholar] [CrossRef] [PubMed]



| Type of TF | No. | Description |
|---|---|---|
| AP2-ERF | 35 | Ethylene responsive transcription factor |
| AUX/IAA | 6 | AUX/IAA transcriptional regulator |
| bHLH | 18 | Basic helix–loop–helix protein |
| bZIP | 7 | Basic-leucine zipper protein |
| B-box | 8 | B-box type zinc finger family protein |
| C2H2 | 12 | C2H2-type zinc finger family protein |
| C3H | 3 | zinc finger (CCCH-type) family protein |
| GNAT | 7 | Acyl-CoA N-acyltransferases protein |
| GRAS | 5 | GRAS family transcription factor |
| HB | 14 | Homeobox-leucine zipper protein |
| LOB | 5 | LOB domain-containing protein |
| MYB | 40 | Myb domain protein |
| NAC | 15 | NAC domain-containing protein |
| SWI/SNF | 3 | SWIB/MDM2 domain superfamily |
| TAZ | 3 | BTB and TAZ domain protein |
| Tify | 3 | Jasmonate-zim-domain protein |
| Trihelix | 7 | Trihelix transcription factor |
| WRKY | 24 | WRKY family transcription factor |
| Other TFs | 48 | |
| Total TFs | 263 |
| Gene Name | Gene ID | Amino Acid Length (aa) | MW (KD) | pI | Instability Index | Best Hits |
|---|---|---|---|---|---|---|
| PpPIF1 | gene2064 (LOC103961475) | 508 | 55.21 | 9.39 | 68.07 | AtPIF1 |
| PpPIF3 | gene16013 (LOC103955840) | 713 | 76.40 | 6.09 | 53.64 | AtPIF3 |
| PpPIF3a | gene15524 (LOC103955304) | 716 | 76.56 | 6.12 | 55.57 | AtPIF3 |
| PpPIF5 | gene8334 (LOC103947396) | 550 | 60.37 | 6.47 | 54.50 | AtPIF5 |
| PpPIF5a | gene2521 (LOC103965596) | 541 | 59.29 | 6.32 | 57.78 | AtPIF5 |
| PpPIF8 | gene35851 (LOC103935970) | 442 | 47.55 | 7.14 | 54.29 | AtPIF8 |
| PpPIF7a | gene31629 (LOC103931367) | 398 | 44.00 | 9.20 | 71.73 | AtPIF8 |
| PpPIF7b | gene3216 (LOC103931008) | 406 | 44.85 | 9.02 | 64.49 | AtPIF8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, Z.; Wei, C.; Cheng, Y.; Shang, Z.; Guo, X.; Guan, J. RNA-Seq Analysis Identifies Transcription Factors Involved in Anthocyanin Biosynthesis of ‘Red Zaosu’ Pear Peel and Functional Study of PpPIF8. Int. J. Mol. Sci. 2022, 23, 4798. https://doi.org/10.3390/ijms23094798
Ma Z, Wei C, Cheng Y, Shang Z, Guo X, Guan J. RNA-Seq Analysis Identifies Transcription Factors Involved in Anthocyanin Biosynthesis of ‘Red Zaosu’ Pear Peel and Functional Study of PpPIF8. International Journal of Molecular Sciences. 2022; 23(9):4798. https://doi.org/10.3390/ijms23094798
Chicago/Turabian StyleMa, Zhenyu, Chuangqi Wei, Yudou Cheng, Zhonglin Shang, Xiulin Guo, and Junfeng Guan. 2022. "RNA-Seq Analysis Identifies Transcription Factors Involved in Anthocyanin Biosynthesis of ‘Red Zaosu’ Pear Peel and Functional Study of PpPIF8" International Journal of Molecular Sciences 23, no. 9: 4798. https://doi.org/10.3390/ijms23094798
APA StyleMa, Z., Wei, C., Cheng, Y., Shang, Z., Guo, X., & Guan, J. (2022). RNA-Seq Analysis Identifies Transcription Factors Involved in Anthocyanin Biosynthesis of ‘Red Zaosu’ Pear Peel and Functional Study of PpPIF8. International Journal of Molecular Sciences, 23(9), 4798. https://doi.org/10.3390/ijms23094798






