Tiller Angle Control 1 Is Essential for the Dynamic Changes in Plant Architecture in Rice
Abstract
:1. Introduction
2. Results
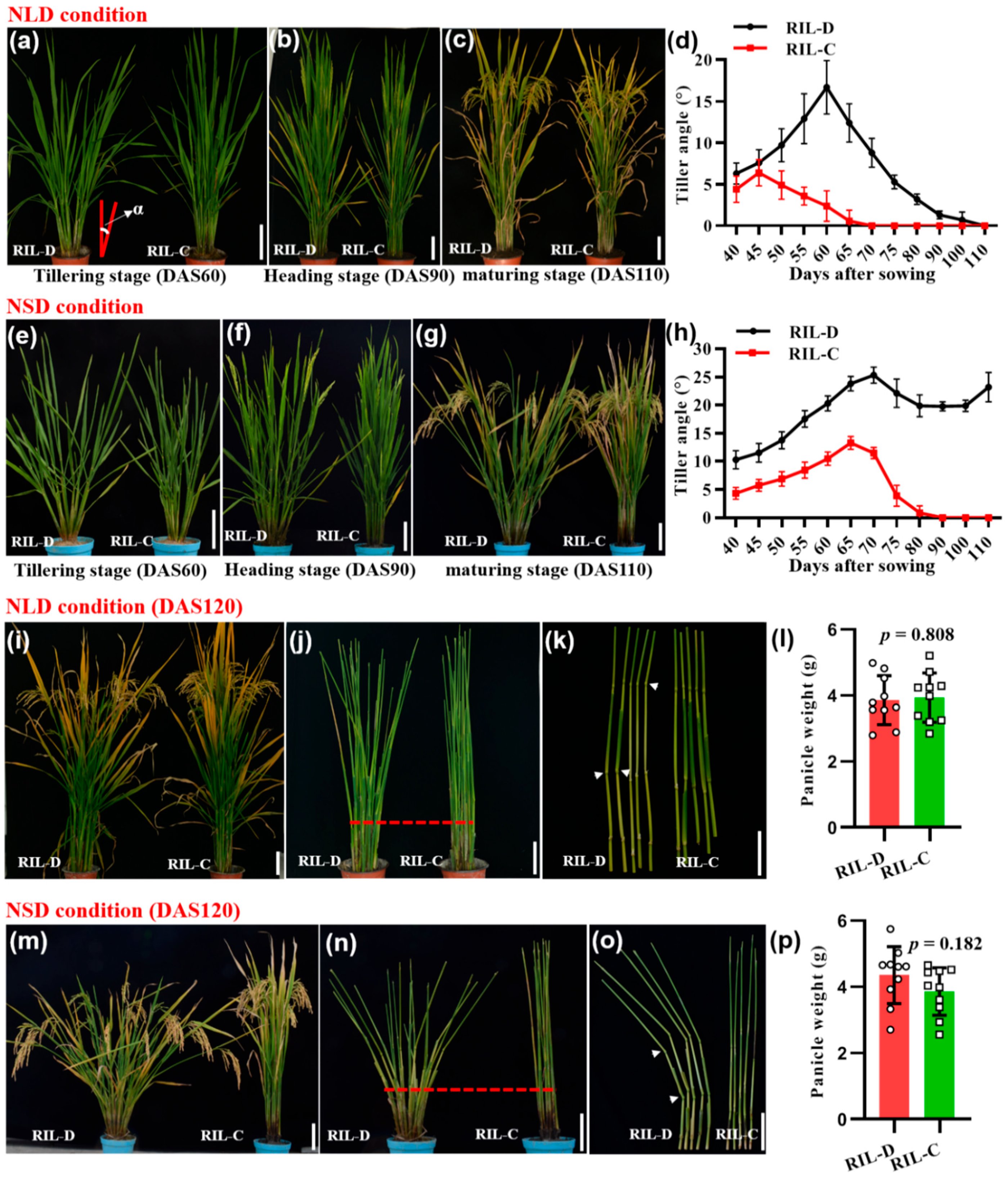
2.1. RIL-D Shows the Dynamic Plant Architecture under Natural Long-Day Conditions
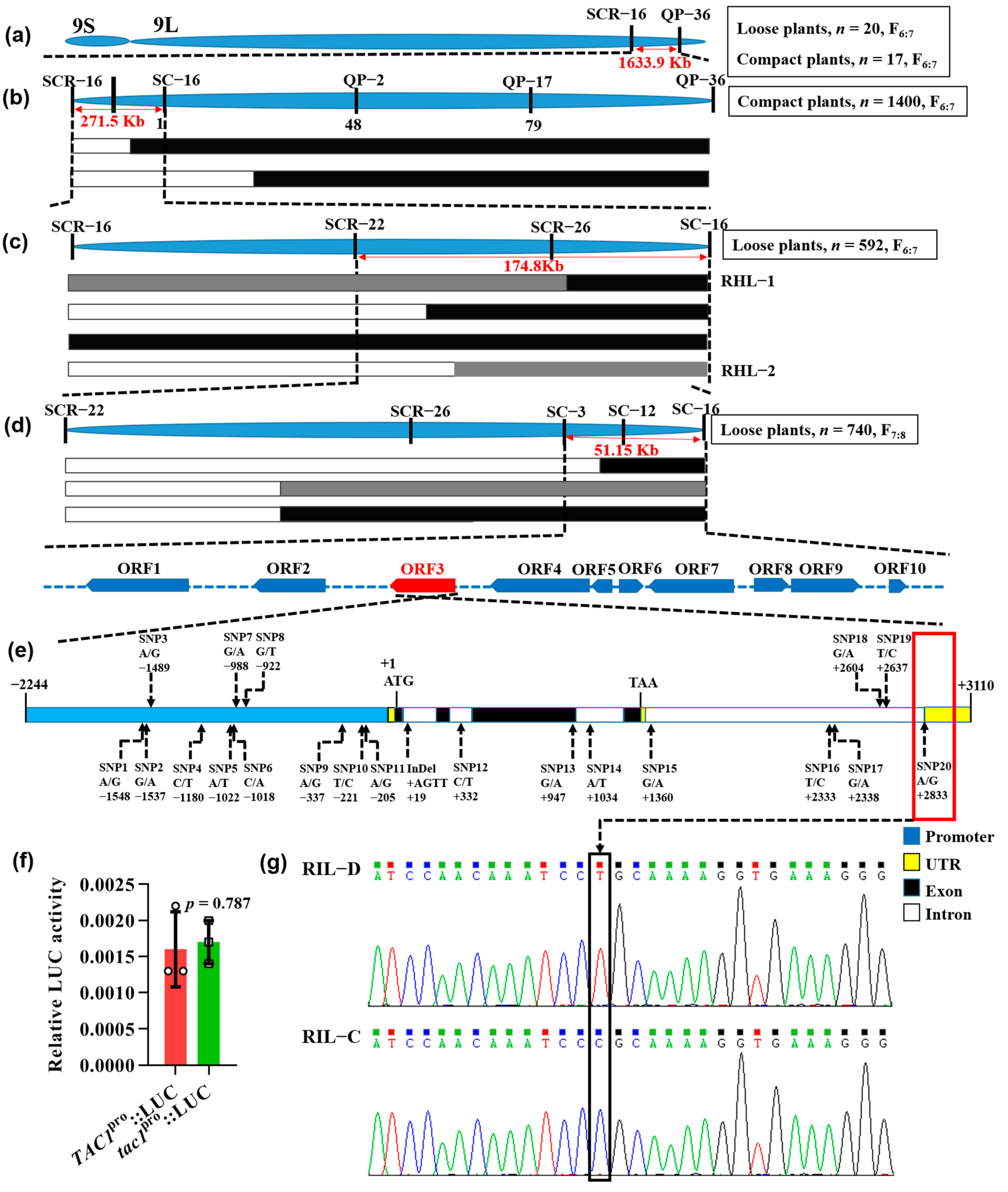
2.2. Genetic Analysis and Fine Mapping of the Candidate Gene
2.3. TAC1 Is the Target Gene Responsible for the Dynamic Plant Architecture of RIL-D
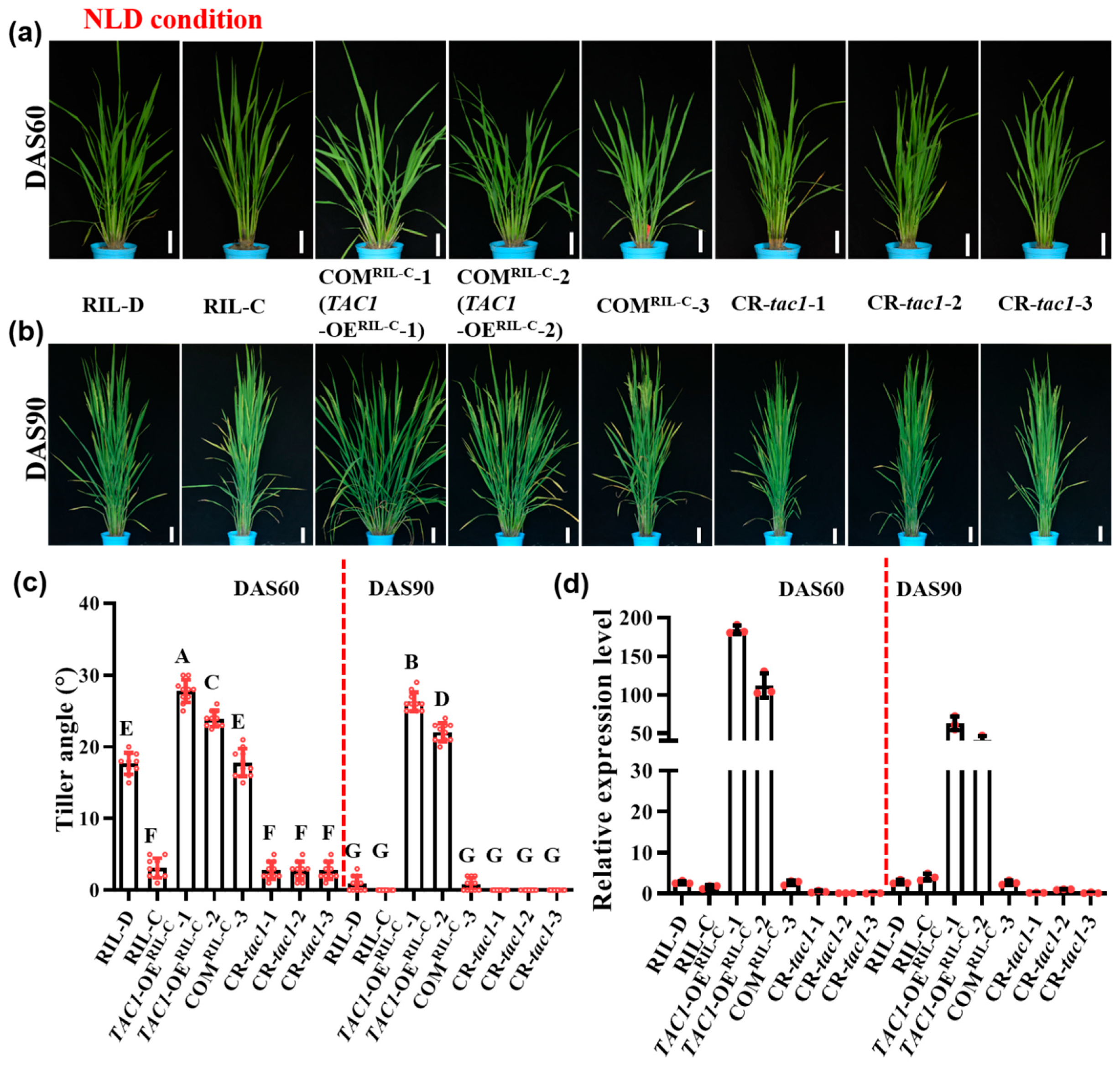
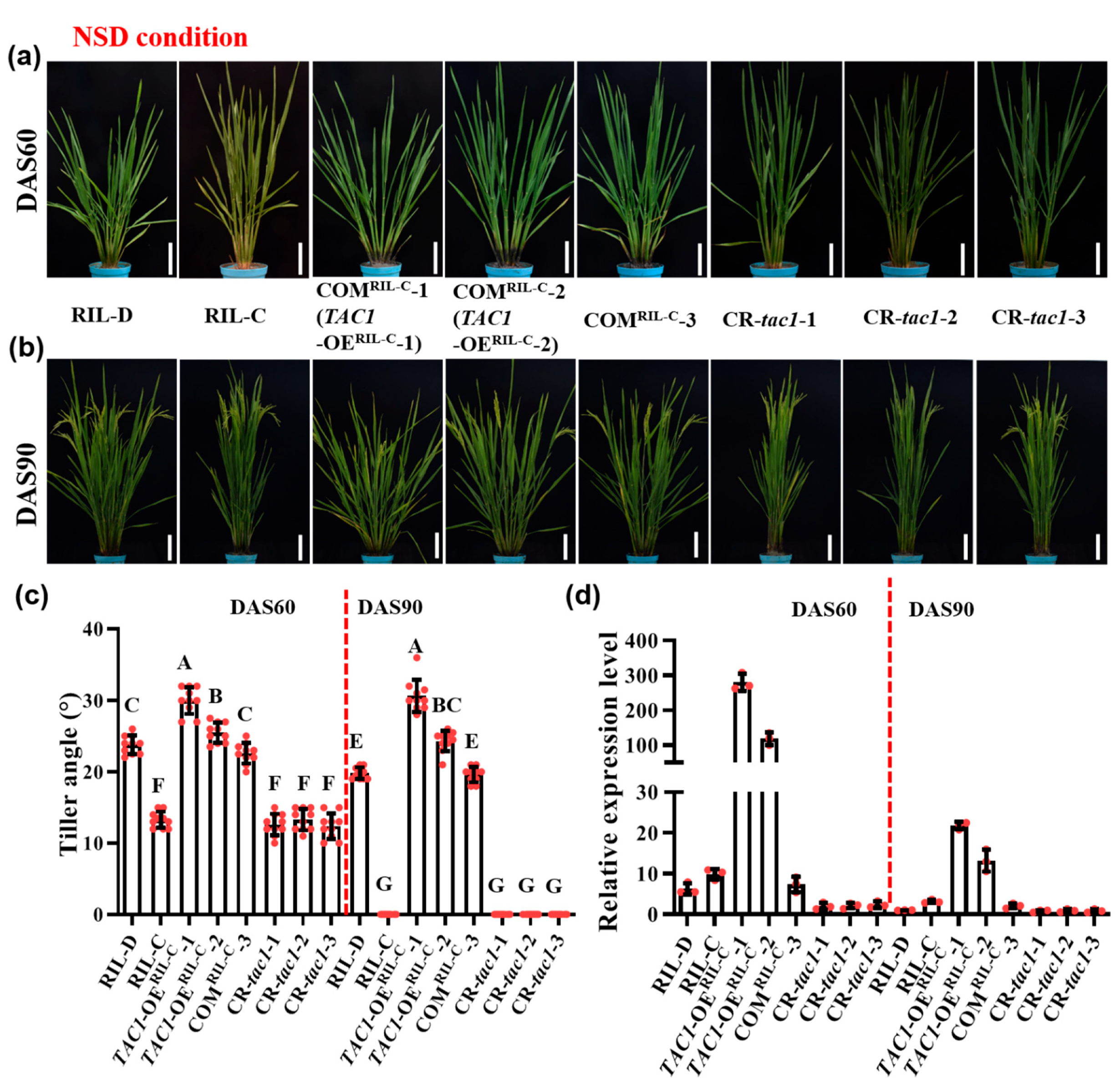
2.4. Transgenic Lines with High TAC1 Expression Displayed Looser Plant Architecture
2.5. Complementary Lines in the Nipponbare (NPB) Background Show Different Degrees of Looseness in Plant Architecture
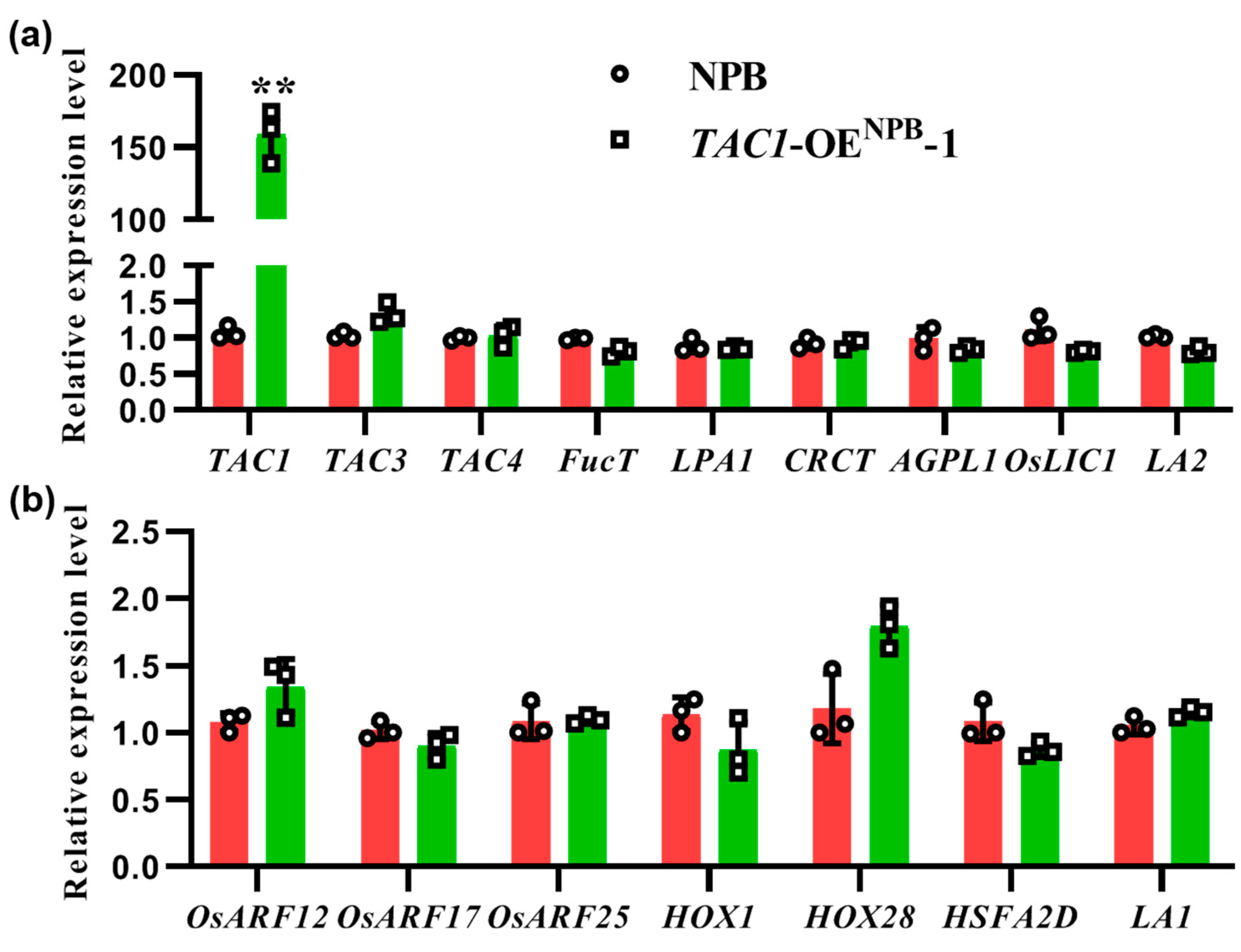
2.6. The Expression of Tiller-Angle-Related Genes Did Not Change between NPB and the TAC1-OENPB-1 Line
3. Discussion
3.1. TAC1 Is Responsible for the Dynamic Changes in Plant Architecture in Rice
3.2. TAC1 Positively Regulates Loose Plant Architecture in Rice
3.3. TAC1 Modulates Different Plant Architecture under NLD and NSD Conditions, Which May Be Related to Light Signals
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Measurements of Rice Tiller Angle
4.3. Rice 8K Chip Assay
4.4. Map-Based Cloning of TAC1
4.5. Generation of Constructs and Rice Transformation
4.6. RNA Extraction, cDNA Preparation, and Real-Time Quantitative PCR (qRT-PCR)
4.7. Rice Protoplast Preparation and Transient Transformation
4.8. Promoter Activity Assay in the Rice Protoplasts
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Reinhardt, D.; Kuhlemeier, C. Plant architecture. EMBO Rep. 2002, 3, 846–851. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, J. Molecular basis of plant architecture. Annu. Rev. Plant Biol. 2008, 59, 253–279. [Google Scholar] [CrossRef] [PubMed]
- Basu, U.; Parida, S.K. Restructuring plant types for developing tailor-made crops. Plant Biotechnol. J. 2021. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Gao, H.; Liang, Y.; Li, J.; Wang, Y. Molecular basis underlying rice tiller angle: Current progress and future perspectives. Mol. Plant 2022, 15, 125–137. [Google Scholar] [CrossRef]
- Yu, B.; Lin, Z.; Li, H.; Li, X.; Li, J.; Wang, Y.; Zhang, X.; Zhu, Z.; Zhai, W.; Wang, X.; et al. TAC1, a major quantitative trait locus controlling tiller angle in rice. Plant J. 2007, 52, 891–898. [Google Scholar] [CrossRef]
- Jiang, J.; Tan, L.; Zhu, Z.; Fu, Y.; Liu, F.; Cai, H.; Sun, C. Molecular evolution of the TAC1 gene from rice (Oryza sativa L.). J. Genet. Genom. 2012, 39, 551–560. [Google Scholar] [CrossRef]
- Ku, L.; Wei, X.; Zhang, S.; Zhang, J.; Guo, S.; Chen, Y. Cloning and characterization of a putative TAC1 ortholog associated with leaf angle in maize (Zea mays L.). PLoS ONE 2011, 6, e20621. [Google Scholar] [CrossRef]
- Dardick, C.; Callahan, A.; Horn, R.; Ruiz, K.B.; Zhebentyayeva, T.; Hollender, C.; Whitaker, M.; Abbott, A.; Scorza, R. PpeTAC1 promotes the horizontal growth of branches in peach trees and is a member of a functionally conserved gene family found in diverse plants species. Plant J. 2013, 75, 618–630. [Google Scholar] [CrossRef]
- Hollender, C.A.; Pascal, T.; Tabb, A.; Hadiarto, T.; Srinivasan, C.; Wang, W.; Liu, Z.; Scorza, R.; Dardick, C. Loss of a highly conserved sterile alpha motif domain gene (WEEP) results in pendulous branch growth in peach trees. Proc. Natl. Acad. Sci. USA 2018, 115, E4690–E4699. [Google Scholar] [CrossRef] [Green Version]
- Hill, J.L., Jr.; Hollender, C.A. Branching out: New insights into the genetic regulation of shoot architecture in trees. Curr. Opin. Plant Biol. 2019, 47, 73–80. [Google Scholar] [CrossRef]
- Li, P.; Wang, Y.; Qian, Q.; Fu, Z.; Wang, M.; Zeng, D.; Li, B.; Wang, X.; Li, J. LAZY1 controls rice shoot gravitropism through regulating polar auxin transport. Cell Res. 2007, 17, 402–410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Liang, Y.; Yuan, Y.; Wang, L.; Meng, X.; Xiong, G.; Zhou, J.; Cai, Y.; Han, N.; Hua, L.; et al. OsBRXL4 regulates shoot gravitropism and rice tiller angle through affecting LAZY1 nuclear localization. Mol. Plant 2019, 12, 1143–1156. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, M.; Furutani, M.; Nishimura, T.; Nakamura, M.; Fushita, T.; Iijima, K.; Baba, K.; Tanaka, H.; Toyota, M.; Tasaka, M.; et al. The arabidopsis LAZY1 family plays a key role in gravity signaling within statocytes and in branch angle control of roots and shoots. Plant Cell 2017, 29, 1984–1999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, M.; Nishimura, T.; Morita, M.T. Bridging the gap between amyloplasts and directional auxin transport in plant gravitropism. Curr. Opin. Plant Biol. 2019, 52, 54–60. [Google Scholar] [CrossRef]
- Zhang, J.; Ku, L.X.; Han, Z.P.; Guo, S.L.; Liu, H.J.; Zhang, Z.Z.; Cao, L.R.; Cui, X.J.; Chen, Y.H. The ZmCLA4 gene in the qLA4-1 QTL controls leaf angle in maize (Zea mays L.). J. Exp. Bot. 2014, 65, 5063–5076. [Google Scholar] [CrossRef] [Green Version]
- Hollender, C.A.; Hill, J.L., Jr.; Waite, J.; Dardick, C. Opposing influences of TAC1 and LAZY1 on lateral shoot orientation in arabidopsis. Sci. Rep. 2020, 10, 6051. [Google Scholar] [CrossRef] [Green Version]
- Zhang, N.; Yu, H.; Yu, H.; Cai, Y.; Huang, L.; Xu, C.; Xiong, G.; Meng, X.; Wang, J.; Chen, H.; et al. A core regulatory pathway controlling rice tiller angle mediated by the LAZY1-dependent asymmetric distribution of auxin. Plant Cell 2018, 30, 1461–1475. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Li, S.; Fan, X.; Song, S.; Zhou, X.; Weng, X.; Xiao, J.; Li, X.; Xiong, L.; You, A.; et al. OsHOX1 and OsHOX28 redundantly shape rice tiller angle by reducing HSFA2D expression and auxin content. Plant Physiol. 2020, 184, 1424–1437. [Google Scholar] [CrossRef]
- Jin, J.; Huang, W.; Gao, J.P.; Yang, J.; Shi, M.; Zhu, M.Z.; Luo, D.; Lin, H.X. Genetic control of rice plant architecture under domestication. Nat. Genet. 2008, 40, 1365–1369. [Google Scholar] [CrossRef]
- Tan, L.; Li, X.; Liu, F.; Sun, X.; Li, C.; Zhu, Z.; Fu, Y.; Cai, H.; Wang, X.; Xie, D.; et al. Control of a key transition from prostrate to erect growth in rice domestication. Nat. Genet. 2008, 40, 1360–1364. [Google Scholar] [CrossRef]
- Zhang, W.; Tan, L.; Sun, H.; Zhao, X.; Liu, F.; Cai, H.; Fu, Y.; Sun, X.; Gu, P.; Zhu, Z.; et al. Natural variations at TIG1 encoding a TCP transcription factor contribute to plant architecture domestication in rice. Mol. Plant 2019, 12, 1075–1089. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Wang, W.; Zhang, N.; Cai, Y.; Liang, Y.; Meng, X.; Yuan, Y.; Li, J.; Wu, D.; Wang, Y. LAZY2 controls rice tiller angle through regulating starch biosynthesis in gravity-sensing cells. New Phytol. 2021, 231, 1073–1087. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Li, Y.; Zhang, H.; Liu, W.; Dong, Z.; Liu, L.; Liu, S.; Sheng, X.; Min, J.; Huang, R.; et al. The chloroplast-localized protein LTA1 regulates tiller angle and yield of rice. Crop J. 2021. [Google Scholar] [CrossRef]
- Li, H.; Sun, H.; Jiang, J.; Sun, X.; Tan, L.; Sun, C. TAC4 controls tiller angle by regulating the endogenous auxin content and distribution in rice. Plant Biotechnol. J. 2021, 19, 64–73. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Wang, Y.; Xue, D.; Wang, J.; Yan, M.; Liu, G.; Dong, G.; Zeng, D.; Lu, Z.; Zhu, X.; et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 2010, 42, 541–544. [Google Scholar] [CrossRef] [PubMed]
- Miura, K.; Ikeda, M.; Matsubara, A.; Song, X.J.; Ito, M.; Asano, K.; Matsuoka, M.; Kitano, H.; Ashikari, M. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 2010, 42, 545–549. [Google Scholar] [CrossRef] [PubMed]
- Springer, N. Shaping a better rice plant. Nat. Genet. 2010, 42, 475–476. [Google Scholar] [CrossRef]
- Chen, Y.; Fan, X.; Song, W.; Zhang, Y.; Xu, G. Over-expression of OsPIN2 leads to increased tiller numbers, angle and shorter plant height through suppression of OsLAZY1. Plant Biotechnol. J. 2012, 10, 139–149. [Google Scholar] [CrossRef]
- Liu, J.M.; Park, S.J.; Huang, J.; Lee, E.J.; Xuan, Y.H.; Je, B.I.; Kumar, V.; Priatama, R.A.; Raj, K.V.; Kim, S.H.; et al. Loose Plant Architecture1 (LPA1) determines lamina joint bending by suppressing auxin signalling that interacts with C-22-hydroxylated and 6-deoxo brassinosteroids in rice. J. Exp. Bot. 2016, 67, 1883–1895. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Tang, D.; Li, M.; Wang, K.; Cheng, Z. Loose plant architecture1, an INDETERMINATE DOMAIN protein involved in shoot gravitropism, regulates plant architecture in rice. Plant Physiol. 2013, 161, 317–329. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.; Tan, L.; Zhu, Z.; Xiao, L.; Xie, D.; Sun, C. PAY1 improves plant architecture and enhances grain yield in rice. Plant J. 2015, 83, 528–536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, C.; Lin, X.; Zuo, Y.; Yu, Z.; Baerson, S.R.; Pan, Z.; Zeng, R.; Song, Y. Transcription factor OsbZIP49 controls tiller angle and plant architecture through the induction of indole-3-acetic acid-amido synthetases in rice. Plant J. 2021, 108, 1346–1364. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Smith, S.M.; Li, J. Genetic regulation of shoot architecture. Annu. Rev. Plant Biol. 2018, 69, 437–468. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wagner, D. Plant inflorescence architecture: The formation, activity, and fate of axillary meristems. Cold Spring Harb. Perspect. Biol. 2020, 12, a034652. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Zhao, H.; Xie, W.; Han, Z.; Li, G.; Yao, W.; Bai, X.; Hu, Y.; Guo, Z.; Lu, K.; et al. A novel tiller angle gene, TAC3, together with TAC1 and D2 largely determine the natural variation of tiller angle in rice cultivars. PLoS Genet. 2016, 12, e1006412. [Google Scholar] [CrossRef] [Green Version]
- Harmoko, R.; Yoo, J.Y.; Ko, K.S.; Ramasamy, N.K.; Hwang, B.Y.; Lee, E.J.; Kim, H.S.; Lee, K.J.; Oh, D.B.; Kim, D.Y.; et al. N-glycan containing a core α1,3-fucose residue is required for basipetal auxin transport and gravitropic response in rice (Oryza sativa). New Phytol. 2016, 212, 108–122. [Google Scholar] [CrossRef] [Green Version]
- Morita, R.; Sugino, M.; Hatanaka, T.; Misoo, S.; Fukayama, H. CO2-responsive CONSTANS, CONSTANS-like, and time of chlorophyll a/b binding protein Expression1 protein is a positive regulator of starch synthesis in vegetative organs of rice. Plant Physiol. 2015, 167, 1321–1331. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Xu, Y.; Zhang, C.; Ma, Q.; Joo, S.H.; Kim, S.K.; Xu, Z.; Chong, K. OsLIC, a novel CCCH-type zinc finger protein with transcription activation, mediates rice architecture via brassinosteroids signaling. PLoS ONE 2008, 3, e3521. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Xu, Y.; Guo, S.; Zhu, J.; Huan, Q.; Liu, H.; Wang, L.; Luo, G.; Wang, X.; Chong, K. Dynamics of brassinosteroid response modulated by negative regulator LIC in rice. PLoS Genet. 2012, 8, e1002686. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Li, J.; Chen, Z.; Wei, Y.; Qi, Y.; Wu, C. OsmiR167a-targeted auxin response factors modulate tiller angle via fine-tuning auxin distribution in rice. Plant Biotechnol. J. 2020, 18, 2015–2026. [Google Scholar] [CrossRef] [Green Version]
- Kami, C.; Lorrain, S.; Hornitschek, P.; Fankhauser, C. Light-regulated plant growth and development. Curr. Top. Dev. Biol. 2010, 91, 29–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paik, I.; Huq, E. Plant photoreceptors: Multi-functional sensory proteins and their signaling networks. Semin. Cell Dev. Biol. 2019, 92, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Oakenfull, R.J.; Davis, S.J. Shining a light on the Arabidopsis circadian clock. Plant Cell Environ. 2017, 40, 2571–2585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Wit, M.; Galvao, V.C.; Fankhauser, C. Light-mediated hormonal regulation of plant growth and development. Annu. Rev. Plant Biol. 2016, 67, 513–537. [Google Scholar] [CrossRef] [PubMed]
- Lv, B.; Zhu, J.; Kong, X.; Ding, Z. Light participates in the auxin-dependent regulation of plant growth. J. Integr. Plant Biol. 2021, 63, 819–822. [Google Scholar] [CrossRef]
- Su, C.; Wang, Y.; Yu, Y.; He, Y.; Wang, L. Coordinative regulation of plants growth and development by light and circadian clock. aBIOTECH 2021, 2, 176–189. [Google Scholar] [CrossRef]
- Wei, X.; Xu, J.; Guo, H.; Jiang, L.; Chen, S.; Yu, C.; Zhou, Z.; Hu, P.; Zhai, H.; Wan, J. DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol. 2010, 153, 1747–1758. [Google Scholar] [CrossRef] [Green Version]
- Xue, W.; Xing, Y.; Weng, X.; Zhao, Y.; Tang, W.; Wang, L.; Zhou, H.; Yu, S.; Xu, C.; Li, X.; et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 2008, 40, 761–767. [Google Scholar] [CrossRef]
- Yan, W.H.; Wang, P.; Chen, H.X.; Zhou, H.J.; Li, Q.P.; Wang, C.R.; Ding, Z.H.; Zhang, Y.S.; Yu, S.B.; Xing, Y.Z.; et al. A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol. Plant 2011, 4, 319–330. [Google Scholar] [CrossRef]
- Waite, J.M.; Dardick, C. IGT/LAZY family genes are differentially influenced by light signals and collectively required for light-induced changes to branch angle. bioRxiv 2020. bioRxiv:2020.07.15.205625. Available online: https://www.biorxiv.org/content/10.1101/2020.07.15.205625v1 (accessed on 16 July 2020).
- Waite, J.M.; Dardick, C. TILLER ANGLE CONTROL 1 modulates plant architecture in response to photosynthetic signals. J. Exp. Bot. 2018, 69, 4935–4944. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.; Ren, Y.; Jian, Y.; Guo, Z.; Zhang, Y.; Xie, C.; Fu, J.; Wang, H.; Wang, G.; Xu, Y.; et al. Development of a maize 55 K SNP array with improved genome coverage for molecular breeding. Mol. Breed. 2017, 37, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Zhang, Y.; Sun, L.; Xu, P.; Tu, R.; Meng, S.; Wu, W.; Anis, G.B.; Hussain, K.; Riaz, A.; et al. WB1, a regulator of endosperm development in rice, is identified by a modified MutMap method. Int. J. Mol. Sci. 2018, 19, 2159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caldana, C.; Scheible, W.R.; Mueller-Roeber, B.; Ruzicic, S. A quantitative RT-PCR platform for high-throughput expression profiling of 2500 rice transcription factors. Plant Methods 2007, 3, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Tu, R.; Sun, L.; Wang, D.; Ruan, Z.; Zhang, Y.; Peng, Z.; Zhou, X.; Fu, J.; Liu, Q.; et al. Tiller Angle Control 1 Is Essential for the Dynamic Changes in Plant Architecture in Rice. Int. J. Mol. Sci. 2022, 23, 4997. https://doi.org/10.3390/ijms23094997
Wang H, Tu R, Sun L, Wang D, Ruan Z, Zhang Y, Peng Z, Zhou X, Fu J, Liu Q, et al. Tiller Angle Control 1 Is Essential for the Dynamic Changes in Plant Architecture in Rice. International Journal of Molecular Sciences. 2022; 23(9):4997. https://doi.org/10.3390/ijms23094997
Chicago/Turabian StyleWang, Hong, Ranran Tu, Lianping Sun, Dongfei Wang, Zheyan Ruan, Yue Zhang, Zequn Peng, Xingpeng Zhou, Junlin Fu, Qunen Liu, and et al. 2022. "Tiller Angle Control 1 Is Essential for the Dynamic Changes in Plant Architecture in Rice" International Journal of Molecular Sciences 23, no. 9: 4997. https://doi.org/10.3390/ijms23094997
APA StyleWang, H., Tu, R., Sun, L., Wang, D., Ruan, Z., Zhang, Y., Peng, Z., Zhou, X., Fu, J., Liu, Q., Wu, W., Zhan, X., Shen, X., Zhang, Y., Cao, L., & Cheng, S. (2022). Tiller Angle Control 1 Is Essential for the Dynamic Changes in Plant Architecture in Rice. International Journal of Molecular Sciences, 23(9), 4997. https://doi.org/10.3390/ijms23094997






