Plants—Microorganisms-Based Bioremediation for Heavy Metal Cleanup: Recent Developments, Phytoremediation Techniques, Regulation Mechanisms, and Molecular Responses
Abstract
1. Introduction
2. Heavy Metals: An Alarming Threat to Soil and Environment
| Chemical Element (Contaminants) | Maximum Permissible Level in Irrigation Water (µg/mL) | Maximum Permissible Level in the Soil (µg/g) | Maximum Permissible Level in Vegetables (µg/g) |
|---|---|---|---|
| As | 0.001 | 20 | - |
| Cd | 0.0003 | 0.8 | 0.10 |
| Co | 0.05 | 50 | 50 |
| Cr | 0.55 | 100 | - |
| Cu | 0.017 | 36 | - |
| Fe | 0.02 | 50,000 | 425 |
| Mn | 0.04 | 2000 | 500 |
| Ni | 0.002 | 35 | 67 |
| Pb | 0.001 | 85 | 0.30 |
| Se | 0.02 | 10 | - |
| Zn | 0.20 | 50 | 100 |
3. Current Bioremediation Technologies: Status, Pitfalls, and Drawbacks
3.1. Phytoremediation
3.1.1. Different Phytoremediation Technologies Involving Contaminants’ Removal
Phytoextraction
Phytostabilization
Phytovolatilization
Phytotransformation
Phytofiltration
3.1.2. Plant Mechanisms for Metal Detoxification
3.1.3. Phytoremediation: Drawbacks and Future Application
3.2. Plant-Associated Microbe’s Affair for Environmental Clean-Up
| Microbial Species | PGP Features | Plant | HMs | Main Results | Reference |
|---|---|---|---|---|---|
| Proteus sp., Pseudomonas sp., E. meliloti, Glomus sp., Sclerocystis sp., Acaulospora sp. | IAA, Biofilm, P solubilization, K solubilization | M. sativa | Cu, Zn, Pb |
| [31] |
| Streptomyces sp. | IAA, ACCD, Zeatin, GA, P solubilization | Zea mays | As, Cr |
| [37] |
| Proteus sp., Pseudomonas sp., E. meliloti | IAA, Biofilm, P solubilization, K solubilization | M. sativa | Cu, Zn, Pb |
| [38] |
| Aspergillus niger, Penicillium chrysosporium | ACCD, IAA, Gibberellins P solubilization, siderophores | V. faba | Cd, Pb |
| [41] |
| Glomus mosseae, Sinorhizobium meliloti | n/s | M. sativa | Cd |
| [42] |
| Streptomyces pactum | n/s | Sorghum bicolor | Zn, Pb, Cd, Cu |
| [80] |
| Bacillus sp. | IAA, hydrolytic and ligninolytic enzymes, Siderophores | Phragmites communis | Fe, Cu, Zn, Cd, Mn, Ni, Pb, As |
| [96] |
| Acinetobacter lwoffii | IAA, ACCD, EPS, siderophore, P solubilization | Vigna radiata | As |
| [104] |
| Aliinostoc sp. | phosphatase production, N fixation | Oryza sativa | Cd |
| [105] |
| Arthrobacter sp., Bacillus altitudinis, Bacillus megatherium, Sphingomonas sp. | ACCD, IAA, Siderophore, P solubilization | Brassica napus | Cd |
| [106] |
| Arthrobacter sp., Microbacterium oxydans | IAA, ACCD, siderophores | Noccaea caerulescens and Arabidopsis | Ni, Cu, Co, Mn, Fe |
| [107] |
| Bacillus cereus, Pseudomonas moraviensis | n/s | Triticum aestivum | Cd, Co, Cr, Cu, Mn, Ni, Pb |
| [108] |
| B. cereus | IAA, siderophores | Zea mays | Cd, Cu, Ni, Pb, Zn |
| [109] |
| Bacillus licheniformis, Micrococcus luteus, Pseudomonas fluorescens | P solubilization, N fixation, siderophores | Vitis vinifera | As |
| [110] |
| Bacillus megaterium | IAA, arginine decarboxylase, siderophores | Brassica campestris and Brassica rapa | Cd |
| [111] |
| Bacillus safensis Kocuria rosea | n/s | Helianthus annuus | Cd, Fe, Zn |
| [112] |
| Bacillus sp., Klebsiella sp., Leifsonia sp., Enterobacter sp. | P solubilization, IAA and EPS production | Z. mays | Cd |
| [113] |
| Bacillus sp., Pseudomonas sp., G. mosseae | IAA, HCN, siderophores, P solubilization | Eucalyptus camaldulensis | Cd |
| [114] |
| B. safensis, P. fluorescens | ACCD, IAA Siderophore | H. annuus | Zn, Pb |
| [115] |
| Brevibacterium casei | NH3, HCN, IAA, ACCD | Sinapis alba | Cd, Zn, Cu |
| [116] |
| Chaetomium cupreum | Siderophore | Miscanthus sinensis | Al, Cu, Fe, Pb, Zn |
| [117] |
| Chlorella vulgaris, Pseudomonas putida | n/s | O. sativa | As |
| [118] |
| Debaryomyces hansenii | IAA, P and Zn solubilization, Siderophores | O. sativa | As |
| [119] |
| Funneliformis mosseae | n/s | Glycine max | Cu, Pb, Zn |
| [120] |
| F. mosseae, Diversispora spurcum | n/s | Cynodon dactylon | Pb, Zn, Cd |
| [121] |
| Glomus aggregatum, G. intraradices, Glomus elunicatum, Glomus versiforme | n/s | M. sativa | Cd |
| [122] |
| G. versiforme | n/s | Solanum nigrum | Cd |
| [123] |
| Klebsiella oxytoca | P solubilization | H. annuus | Co, Pb, Zn |
| [124] |
| Klebsiella sp. | IAA, EPS, NH3 P solubilization, | V. radiata | Cd, Cu, Pb |
| [125] |
| Kocuria flava, Bacillus vietnamensis | IAA, EPS siderophores | O. sativa | As |
| [126] |
| Oscillatoria sp. | n/s | Portulaca oleracea | Cr, Fe, Al, Zn |
| [127] |
| Oscillatoria sp., Leptolyngbya sp. | n/s | Lactuca sativa and Raphanus sativus | Fe, As, Pb, Cr, Ni |
| [128] |
| Paecilomyces formosus, Penicillium funiculosum | IAA, gibberellins, P solubilization | G. max | Al, Ni, and Cd |
| [129] |
| Pantoea agglomerans, Bacillus aryabhattai | ACCD, N fixation, P solubilization, siderophores | Spartina densiflora | As, Cu, Pb, Zn, Cd |
| [130] |
| Pantoea stewartii, Microbacterium arborescens, Enterobacter sp. | IAA, ACCD, siderophores | Leptochloa fusca | Cr, Cu, Fe, Ni, Pb, Ba, Cd, Co |
| [131] |
| Penicillium janthinellum | IAA adsorption | C. dactylon | Cd |
| [132] |
| Piriformospora indica | n/s | Artemisia annua | As |
| [133] |
| P. indica | n/s | Cenchrus purpureus | Cd |
| [134] |
| Planomicrobium chinense, B. cereus, P. fluorescens | P solubilization | Z. mays | Ni, Cd, Pb, Co, Cu, Fe, Zn |
| [135] |
| P. aeruginosa | IAA, HCN, NH3, ACCD siderophore, P solubilization | T. aestivum | Cu, Cr, Cd |
| [136] |
| P. aeruginosa, Actinomyces sp., Azotobacter sp., Azospirillum brasilense., Bacillus subtilis | n/s | Eichhornia crassipes | As |
| [137] |
| P. fluorescens | IAA, ACCD, siderophore | Sedum alfredii | Cd |
| [138] |
| P. fluorescens | n/s | Pisum sativum | Pb |
| [139] |
| Pseudomonas libanensis, Claroideoglomus claroideum | ACCD, IAA, P solubilization Siderophores | H. annuus | Ni |
| [140] |
| Pseudomonas sp. | IAA, EPS, HCN P solubilization, N fixation, siderophores | M. sativa | Cr |
| [141] |
| Pseudomonas sp., Serratia sp. | Organic acids, ACCD, IAA, Acetoin, P solubilization, N fixation | Helianthus tuberosus | Cd and Zn |
| [142] |
| Pseudomonas sp., Glomus sp. | n/s | Centaurea cyanus | Pb |
| [143] |
| Pseudomonas sp., Azotobacter sp., Paenibacillus sp., Streptomyces sp. Glomus sp. | P solubilization Siderophores, IAA production | Pennisetum glaucum S. bicolor | Fe |
| [144] |
| Rhizoglomus intraradices, Glomus etunicatum | n/s | T. aestivum | As |
| [145] |
| Rhizophagus fasciculatus, Rhizophagus intraradices, F. mosseae G. aggregatum | n/s | Z. mays | Cd, Cr, Ni, Pb, Fe, Zn, Cr, Mn |
| [146] |
| R. intraradices, G. versiforme | n/s | Z. mays | Cd |
| [147] |
| R. irregularis | n/s | M. sativa | Zn, Cd |
| [63] |
| Rhodobacter sphaeroides | IAA production | T. aestivum | Cd, Zn |
| [148] |
| Serratia sp. | IAA production, P solubilization, ACCD | H. annuus | Cu, Zn, Ni, Pb, As |
| [149] |
| Simplicillium chinense | n/s | P. communis | Pb and Cd |
| [150] |
| S. meliloti, P. fluorescence, P. indica | IAA, HCN, ACCD P solubilization siderophores | M. sativa | Cd |
| [151] |
| Spirulina platensis | n/s | Z. mays | Cd |
| [152] |
| S. pactum | n/s | Lolium perenne | Pb |
| [153] |
| S. pactum, Bacillus sp. | n/s | B. juncea | Cd, Pb, Cu, Zn |
| [154] |
| Talaromyces pinophilus | Gibberellic acid | T. aestivum | Cd, Ni, Cu, Zn |
| [155] |
| Trametes hirsuta | n/s | T. aestivum | Pb |
| [156] |
| Trichoderma asperellum | n/s | Suaeda salsa | Pb |
| [157] |
| Variovorax paradoxus, Rhizobium leguminosarum Glomus sp. | n/s | P. sativum and B. juncea | Cd, Zn, Fe, Mn |
| [158] |
3.3. Metaorganism as a Strategy to Improve Phytoremediation
3.4. Microbial Remediation: Heavy-Metal Remediation by Microorganisms
3.4.1. Different Microbial Remediation Techniques Involving Removal and Containment of Contaminants
Bio-Stimulation
Bio-Augmentation
Engineered Microbial Remediation
3.4.2. Mechanism of Heavy Metal Remediation by Microorganisms
| Group | Bioremediation | Metal | Metal Concentration (mg/L) | Remediation Efficiency (%) | Mechanism | Reference |
|---|---|---|---|---|---|---|
| Bacteria | P. aeruginosa | As, Cd, Zn | 182, 20, 983 | 53, 90, 80 | Biosurfactant production | [12] |
| R. opacus | Pb, Cd, Ni, Co, Cr | 100, 100, 250, 200, 100 | n/s | Adsorption in exopolysaccharides | [17] | |
| R. rhodochrous | Pb, Cd, Ni, Co, Cr | 100, 250, 250, 150, 150 | n/s | Adsorption in exopolysaccharides | [17] | |
| B. cereeus | Cr, Fe, Mn, Ni, Cu, Cd, Zn | 100, 100, 50, 50, 30, 30, 50 | 82, 92, 97, 43, 25, 31, 36 | Reduction (for Cr) | [18] | |
| S. ginsengisoli | As | 500 | 98 | Precipitation | [184] | |
| B. subtilis | Hg, Cd | 500 | 30, 76 | Biosrption | [185] | |
| B. cereus | Pb, Cd, Cr | 100 | 69, 54, 43 | Biosurfactant production | [210] | |
| Bacillus sp. | Pb, Hg, Mn, Cd | 1000 | 76, 98, 90, 100 | Biosurfactant production | [209] | |
| C. freundii | Al, Cd, Cu, Fe, Pb, Mn, Zn | n/s | 87, 40, 19, 34, 57, 25, 49 | Biosurfactant production | [212] | |
| D. desulfuricans | Cd, Ni, Cr | 100 | 100, 98, 74 | Sulfate-reduction | [215] | |
| Ensifer adhaerens | Cr, Cu, Cd, Ni, Zn, Pb | 150 | 80, 81, 80, 82, 80, 80 | Bioaccumulation and biosorption | [218] | |
| Acinetobacter sp. | Cr | 16 | 87 | Reduction | [238] | |
| Alcaligenes faecalis | Cd | 100 | 70 | Adsorption and/or precipitation | [239] | |
| Bacillus pumilus | Pb | 100 | 88 | Adsorption and/or precipitation | [239] | |
| Brevibacterium iodinium | Pb | 100 | 87 | Adsorption and/or precipitation | [239] | |
| P. aeruginosa | Cd, Pb | 100 | 76 | Adsorption and/or precipitation | [239] | |
| B. cereus | Pb | 100 | 79.26 | Bioaccumulation and biosorption | [240] | |
| Bacillus circulans | Cr | 1110 | 71 | Reduction | [241] | |
| Bacillus firmus | Pb, Cu, Zn | 1000 | 98, 75, 62 | Adsorption in exopolysaccharides | [242] | |
| B. licheniformis | Hg | 100 | 73 | - | [243] | |
| B. subtilis | Cr | 570 | 100 | Reduction | [244] | |
| P. aeruginosa | Cr | 570 | 100 | Reduction | [244] | |
| Bacillus xiamenensis | Pb | 100 | 99.19 | Bioaccumulation and biosorption | [245] | |
| Cellulosimicrobium sp. | Cr | 300 | 63 | Reduction | [246] | |
| D. desulfuricans | Cr, Cu, Ni | 200 | 56, 79, 90 | - | [247] | |
| Micrococcus sp. | Cr, Ni | 100, 50 | 90, 55 | Biosrption | [248] | |
| P. aeruginosa | Cd, Pb | 435, 905 | 92, 88 | Biosurfactant production | [249] | |
| Sporosarcina saromensis | Cr | 100 | 100 | Reduction | [250] | |
| Streptomyces sp. | Zn | 65.38, 32.69 | 36 or 43 | Bioaccumulation or biosorption | [251] | |
| Yeast | S. cerevisiae | Pb, Cd | 25, 80 | 71, 77 | Biosorption | [10] |
| S. cerevisiae | Hg, Cd | 500 | 19, 70 | Biosorption | [185] | |
| C. bombicola | Cr, Pb, Zn, Cu, Cd | 70 | 23, 10, 7, 5, 16 | Biosurfactant production | [211] | |
| C. tropicalis | Cd | 100 | 78 | Biosorption | [223] | |
| Candida parapsilosis | Hg | 100 | 80 | - | [243] | |
| S. cerevisiae | Cr | 570 | 96 | Reduction | [244] | |
| Candida sphaerica | Fe, Zn, Pb | 1877, 1470, 3038 | 89, 87, 70 | Biosurfactant production | [252] | |
| Cryptococcus sp. | Zn | 100 | 85 | Biosurfactant production | [253] | |
| Rhodotorula mucilaginosa | Cr | 200 | 27 | Reduction | [254] | |
| Fungi | A. niger | Cd, Cr | 0.6, 0.4 | 79, 48 | Bioaccumulation | [11] |
| Aspergillus fumigatus | Cd, Cr | 0.6, 0.4 | 76, 35 | Bioaccumulation | [11] | |
| Penicillium rubens | Cd, Cr | 0.6, 0.4 | 75, 35 | Bioaccumulation | [11] | |
| Lecythophora sp. | As | 10 | 32 | Reduction and volatilization | [19] | |
| S. chinense | Cd, Pb | 400, 2000 | 88, 58 | Biosorption | [150] | |
| Aspergillus sp. | Cr, Ni | 100, 50 | 92, 9 | Biosorption | [248] | |
| Aspergillus flavus | Cd, Cu, Fe, Mn, Pb, Zn | 1000 | 87, 83, 96, 92, 87, 70 | Biosorption | [255] | |
| Aspergillus gracilis | Cd, Cu, Fe, Mn, Pb, Zn | 1000 | 66, 57, 90, 77, 82, 7 | Biosorption | [255] | |
| Aspergillus penicillioides | Cd, Cu, Fe, Mn, Pb, Zn | 1000 | 53, 32, 90, 69, 77, 84 | Biosorption | [255] | |
| Aspergillus restrictus | Cd, Cu, Fe, Mn, Pb, Zn | 1000 | 61,77, 64, 72, 44, 87 | Biosorption | [255] | |
| Sterigmatomyces halophilus | Cd, Cu, Fe, Mn, Pb, Zn | 1000 | 95, 90, 77, 89, 57, 93 | Biosorption | [255] | |
| A. niger | Ni | 30 | 70.3 | Biosorption | [256] | |
| Aspergillus versicolor | Cr, Ni, Cu | 50 | 100, 30, 29 | Bioaccumulation | [257] | |
| Phanerochaete chrysosporium | Cd, Ni | 25, 16 | 96, 89 | Bioaccumulation | [258] | |
| Cyanobacteria | N. linckia | Cr, Fe, Ni, Zn, Cu | 2.5, 2, 0.5, 0.5, 0.5 | n/s | Bioaccumulation | [13] |
| Chlorella pyrenoidosa | Cu | 5 | 83 | Biosorption | [53] | |
| Anabaena variabilis | Pb | 15 | 71.4 | Biosorption and/or bioaccumulation | [259] | |
| Nostoc muscorum | Pb | 15 | 97.8 | Biosorption and/or bioaccumulation | [259] | |
| Botryocossuss sp. | Cr | 5 | 94 | Reduction and biosorption | [260] | |
| C. pyrenoidosa | Cd | 1.5 | 45.45 | Biosorption and bioaccumulation | [261] | |
| Scenedesmus acutus | Cd | 1.5 | 57.14 | Biosorption and bioaccumulation | [261] | |
| Scenedesmus sp. | Cr | 10 | 93 | Biosorption | [262] | |
| Spirogyra sp. | Cr, Cu, Fe, Mn, Se, Zn | 5 | 98, 90, 100, 100, 98, 81 | Biosorption and/or bioaccumulation | [263] | |
| S. platensis | Cr, Fe, Ni, Zn | 10, 5, 2, 2 | n/s | Bioaccumulation | [264] | |
| Spirulina sp. | Cr, Cu, Fe, Mn, Se, Zn | 5 | 98, 81, 99, 100, 99, 79 | Biosorption and/or bioaccumulation | [263] |
3.4.3. Soil Microbiota Evolution under HMs Contamination and Phytoremediation Approach
3.4.4. Microbial Remediation: Pitfalls, Drawbacks, and Future Application
3.5. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xu, Y.; Dai, S.; Meng, K.; Wang, Y.; Ren, W.; Zhao, L.; Christie, P.; Teng, Y. Occurrence and risk assessment of potentially toxic elements and typical organic pollutants in contaminated rural soils. Sci. Total Environ. 2018, 630, 618–629. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.J.; Huang, X.P.; Xiang, L.; Wang, Y.Z.; Li, Y.W.; Li, H.; Cai, Q.Y.; Mo, C.H.; Wong, M.H. Source, migration and toxicology of microplastics in soil. Environ. Int. 2020, 137, 105263. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, S.; Priya, T.A.K.; Khoo, K.S.; Hoang, T.K.A.; Ng, H.S.; Munawaroh, H.S.H.; Karaman, C.; Orooji, Y.; Show, P.L. A Critical review on various remediation approaches for heavy metal contaminants removal from contaminated soils. Chemosphere 2021, 287, 132369. [Google Scholar] [CrossRef] [PubMed]
- Emenike, C.U.; Jayanthi, B.; Agamuthu, P.; Fauziah, S.H. Biotransformation and removal of heavy metals: A review of phytoremediation and microbial remediation assessment on contaminated soil. Environ. Rev. 2018, 26, 156–168. [Google Scholar] [CrossRef]
- Tang, J.; Zhang, J.; Ren, L.; Zhou, Y.; Gao, J.; Luo, L.; Yang, Y.; Peng, Q.; Huang, H.; Chen, A. Diagnosis of soil contamination using microbiological indices: A review on heavy metal pollution. J. Environ. Manag. 2019, 242, 121–130. [Google Scholar] [CrossRef]
- El Alaoui, A.; Raklami, A.; Bechtaoui, N.; El Gharmali, A.; Ouhammou, A.; Imziln, B.; Achouak, W.; Pajuelo, E.; Oufdou, K. Use of native plants and their associated bacteria rhizobiomes to remediate-restore Draa Sfar and Kettara mining sites, Morocco. Environ. Monit. Assess. 2021, 193, 232. [Google Scholar] [CrossRef]
- Ojuederie, O.B.; Babalola, O.O. Microbial and plant-assisted bioremediation of heavy metal polluted environments: A review. Int. J. Environ. Res. Public Health 2017, 14, 1504. [Google Scholar] [CrossRef]
- Liu, S.; Yang, B.; Liang, Y.; Xiao, Y.; Fang, J. Prospect of phytoremediation combined with other approaches for remediation of heavy metal-polluted soils. Environ. Sci. Pollut. Res. 2020, 27, 16069–16085. [Google Scholar] [CrossRef]
- Yin, K.; Wang, Q.; Lv, M.; Chen, L. Microorganism remediation strategies towards heavy metals. Chem. Eng. J. 2019, 360, 1553–1563. [Google Scholar] [CrossRef]
- Hadiani, M.R.; Darani, K.K.; Rahimifard, N.; Younesi, H. Biosorption of low concentration levels of lead (II) and cadmium (II) from aqueous solution by Saccharomyces cerevisiae: Response surface methodology. Biocatal. Agric. Biotechnol. 2018, 15, 25–34. [Google Scholar] [CrossRef]
- Khan, I.; Aftab, M.; Shakir, S.U.; Ali, M.; Qayyum, S.; Rehman, M.U.; Haleem, K.S.; Touseef, I. Mycoremediation of heavy metal (Cd and Cr)–polluted soil through indigenous metallotolerant fungal isolates. Environ. Monit. Assess. 2019, 191, 585. [Google Scholar] [CrossRef] [PubMed]
- Lopes, C.S.C.; Teixeira, D.B.; Braz, B.F.; Santelli, R.E.; de Castilho, L.V.A.; Gomez, J.G.C.; Castro, R.P.V.; Seldin, L.; Freire, D.M.G. Application of rhamnolipid surfactant for remediation of toxic metals of long- and short-term contamination sites. Int. J. Environ. Sci. Technol. 2021, 18, 575–588. [Google Scholar] [CrossRef]
- Cepoi, L.; Zinicovscaia, I.; Valuta, A.; Codreanu, L.; Rudi, L.; Chiriac, T.; Yushin, N.; Grozdov, D.; Peshkova, A. Bioremediation capacity of edaphic cyanobacteria Nostoc linckia for chromium in association with other heavy-metals-contaminated soils. Environments 2022, 9, 1. [Google Scholar] [CrossRef]
- Guarino, F.; Miranda, A.; Castiglione, S.; Cicatelli, A. Arsenic phytovolatilization and epigenetic modifications in Arundo donax L. assisted by a PGPR consortium. Chemosphere 2020, 251, 126310. [Google Scholar] [CrossRef] [PubMed]
- Yanitch, A.; Kadri, H.; Frenette-Dussault, C.; Joly, S.; Pitre, F.E.; Labrecque, M. A four-year phytoremediation trial to decontaminate soil polluted by wood preservatives: Phytoextraction of Arsenic, Chromium, Copper, Dioxins and Furans. Int. J. Phytoremed. 2020, 22, 1505–1514. [Google Scholar] [CrossRef] [PubMed]
- Gavrilescu, M. Enhancing phytoremediation of soils polluted with heavy metals. Curr. Opin. Biotechnol. 2022, 74, 21–31. [Google Scholar] [CrossRef]
- Dobrowolski, R.; Szcześ, A.; Czemierska, M.; Jarosz-Wikołazka, A. Studies of Cadmium(II), Lead(II), Nickel(II), Cobalt(II) and Chromium(VI) sorption on extracellular polymeric substances produced by Rhodococcus opacus and Rhodococcus rhodochrous. Bioresour. Technol. 2017, 225, 113–120. [Google Scholar] [CrossRef]
- Nayak, A.K.; Panda, S.S.; Basu, A.; Dhal, N.K. Enhancement of toxic Cr (VI), Fe, and other heavy metals phytoremediation by the synergistic combination of native Bacillus cereus Strain and Vetiveria zizanioides L. Int. J. Phytoremed. 2018, 20, 682–691. [Google Scholar] [CrossRef]
- Chang, J.; Si, G.; Dong, J.; Yang, Q.; Shi, Y.; Chen, Y.; Zhou, K.; Chen, J. Transcriptomic analyses reveal the pathways associated with the volatilization and resistance of Mercury(II) in the fungus Lecythophora sp. DC-F1. Sci. Total Environ. 2021, 752, 142172. [Google Scholar] [CrossRef]
- Ashraf, S.; Ali, Q.; Zahir, Z.A.; Ashraf, S.; Asghar, H.N. Phytoremediation: Environmentally sustainable way for reclamation of heavy metal polluted soils. Ecotoxicol. Environ. Saf. 2019, 174, 714–727. [Google Scholar] [CrossRef]
- Gustin, M.S.; Hou, D.; Tack, F.M.G. The term “heavy metal(s)”: History, current debate, and future use. Sci. Total Environ. 2021, 789, 147951. [Google Scholar] [CrossRef] [PubMed]
- Pratush, A.; Kumar, A.; Hu, Z. Adverse Effect of Heavy Metals (As, Pb, Hg, and Cr) on health and their bioremediation strategies: A review. Int. Microbiol. 2018, 21, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Asad, S.A.; Farooq, M.; Afzal, A.; West, H. Integrated phytobial heavy metal remediation strategies for a sustainable clean environment—A review. Chemosphere 2019, 217, 925–941. [Google Scholar] [CrossRef] [PubMed]
- García-García, J.D.; Sánchez-Thomas, R.; Moreno-Sánchez, R. Bio-recovery of non-essential heavy metals by intra- and extracellular mechanisms in free-living microorganisms. Biotechnol. Adv. 2016, 34, 859–873. [Google Scholar] [CrossRef]
- Zwolak, A.; Sarzyńska, M.; Szpyrka, E.; Stawarczyk, K. Sources of soil pollution by heavy metals and their accumulation in vegetables: A review. Water. Air. Soil Pollut. 2019, 230, 164. [Google Scholar] [CrossRef]
- Dhaliwal, S.S.; Singh, J.; Taneja, P.K.; Mandal, A. Remediation techniques for removal of heavy metals from the soil contaminated through different sources: A review. Environ. Sci. Pollut. Res. 2020, 27, 1319–1333. [Google Scholar] [CrossRef]
- Kandziora-Ciupa, M.; Nadgórska-Socha, A.; Barczyk, G. The influence of heavy metals on biological soil quality assessments in the Vaccinium myrtillus L. rhizosphere under different field conditions. Ecotoxicology 2021, 30, 292–310. [Google Scholar] [CrossRef]
- Sharma, R.; Singh, N.S.; Singh, D.K. Impact of heavy metal contamination and seasonal variations on enzyme’s activity of Yamuna river soil in Delhi and NCR. Appl. Water Sci. 2020, 10, 83. [Google Scholar] [CrossRef]
- Kumar, S.; Prasad, S.; Yadav, K.K.; Shrivastava, M.; Gupta, N.; Nagar, S.; Bach, Q.V.; Kamyab, H.; Khan, S.A.; Yadav, S.; et al. Hazardous heavy metals contamination of vegetables and food chain: Role of sustainable remediation approaches—A review. Environ. Res. 2019, 179, 108792. [Google Scholar] [CrossRef]
- Mensah, A.K.; Marschner, B.; Antoniadis, V.; Stemn, E.; Shaheen, S.M.; Rinklebe, J. Human health risk via soil ingestion of potentially toxic elements and remediation potential of native plants near an abandoned mine spoil in Ghana. Sci. Total Environ. 2021, 798, 149272. [Google Scholar] [CrossRef]
- Raklami, A.; Tahiri, A.; Bechtaoui, N.; Abdelhay, E.G.; Pajuelo, E.; Baslam, M.; Meddich, A.; Oufdou, K. Restoring the plant productivity of heavy metal-contaminated soil using phosphate sludge, marble waste, and beneficial microorganisms. J. Environ. Sci. 2020, 99, 210–221. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Quan, Q.; Gan, Y.; Dong, J.; Fang, J.; Wang, L.; Liu, J. Effects of heavy metals on microbial communities in sediments and establishment of bioindicators based on microbial taxa and function for environmental monitoring and management. Sci. Total Environ. 2020, 749, 141555. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.T.; Wang, J.; Dong, H.; Chen, J.M.; Ge, Y. Relative importance of soil properties and heavy metals/metalloids to modulate microbial community and activity at a smelting site. J. Soils Sediments 2021, 21, 1–12. [Google Scholar] [CrossRef]
- Shuaib, M.; Azam, N.; Bahadur, S.; Romman, M.; Yu, Q.; Xuexiu, C. Variation and succession of microbial communities under the conditions of persistent heavy metal and their survival mechanism. Microb. Pathog. 2021, 150, 104713. [Google Scholar] [CrossRef] [PubMed]
- Jacob, J.M.; Karthik, C.; Saratale, R.G.; Kumar, S.S.; Prabakar, D.; Kadirvelu, K.; Pugazhendhi, A. Biological approaches to tackle heavy metal pollution: A survey of literature. J. Environ. Manag. 2018, 217, 56–70. [Google Scholar] [CrossRef] [PubMed]
- Aponte, H.; Meli, P.; Butler, B.; Paolini, J.; Matus, F.; Merino, C.; Cornejo, P.; Kuzyakov, Y. Meta-analysis of heavy metal effects on soil enzyme activities. Sci. Total Environ. 2020, 737, 139744. [Google Scholar] [CrossRef]
- AL-huqail, A.A.; El-bondkly, A.M.A. Improvement of Zea mays L. Growth parameters under chromium and arsenic stress by the heavy metal-resistant Streptomyces sp. NRC21696. Int. J. Environ. Sci. Technol. 2021, 18, 1–22. [Google Scholar] [CrossRef]
- Raklami, A.; Oufdou, K.; Tahiri, A.I.; Mateos-Naranjo, E.; Navarro-Torre, S.; Rodríguez-Llorente, I.D.; Meddich, A.; Redondo-Gómez, S.; Pajuelo, E. Safe cultivation of Medicago sativa in metal-polluted soils from semi-arid regions assisted by heat-and metallo-resistant PGPR. Microorganisms 2019, 7, 212. [Google Scholar] [CrossRef]
- Raklami, A.; Oubane, M.; Meddich, A.; Hafidi, M.; Marschner, B.; Heinze, S.; Oufdou, K. Phytotoxicity and genotoxicity as a new approach to assess heavy metals effect on Medicago sativa L.: Role of metallo-resistant rhizobacteria. Environ. Technol. Innov. 2021, 24, 101833. [Google Scholar] [CrossRef]
- Tauqeer, H.M.; Ali, S.; Rizwan, M.; Ali, Q.; Saeed, R.; Iftikhar, U.; Ahmad, R.; Farid, M.; Abbasi, G.H. Phytoremediation of heavy metals by Alternanthera bettzickiana: Growth and physiological response. Ecotoxicol. Environ. Saf. 2016, 126, 138–146. [Google Scholar] [CrossRef]
- El-Mahdy, O.M.; Mohamed, H.I.; Mogazy, A.M. Biosorption effect of Aspergillus niger and Penicillium chrysosporium for Cd- and Pb-contaminated soil and their physiological effects on Vicia faba L. Environ. Sci. Pollut. Res. 2021, 28, 67608–67631. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Fang, L.; Beiyuan, J.; Cui, Y.; Peng, Q.; Zhu, S.; Wang, M.; Zhang, X. Improvement of alfalfa resistance against Cd stress through rhizobia and arbuscular mycorrhiza fungi co-inoculation in cd-contaminated soil. Environ. Pollut. 2021, 277, 116758. [Google Scholar] [CrossRef] [PubMed]
- Jia, Z.; Li, S.; Wang, L. Assessment of soil heavy metals for eco-environment and human health in a rapidly urbanization area of the upper Yangtze Basin. Sci. Rep. 2018, 8, 3256. [Google Scholar] [CrossRef] [PubMed]
- Edokpayi, J.N.; Enitan, A.M.; Mutileni, N.; Odiyo, J.O. Evaluation of water quality and human risk assessment due to heavy metals in groundwater around Muledane area of Vhembe district, Limpopo province. S. Afr. Chem. Cent. J. 2018, 12, 2. [Google Scholar] [CrossRef] [PubMed]
- Ravindra, K.; Mor, S. Distribution and health risk assessment of arsenic and selected heavy metals in groundwater of Chandigarh, India. Environ. Pollut. 2019, 250, 820–830. [Google Scholar] [CrossRef]
- Njoga, E.O.; Ezenduka, E.V.; Ogbodo, C.G.; Ogbonna, C.U.; Jaja, I.F.; Ofomatah, A.C.; Okpala, C.O.R. Detection, Distribution and health risk assessment of toxic heavy metals/metalloids, arsenic, cadmium, and lead in goat carcasses processed for human consumption in south-eastern Nigeria. Foods 2021, 10, 798. [Google Scholar] [CrossRef]
- WHO (World Health Organization). Evolution of Who Air Quality Guidelines; WHO: Geneva, Switzerland, 2017. [Google Scholar]
- Lai, H.Y.; Hseu, Z.Y.; Chen, T.C.; Chen, B.C.; Guo, H.Y.; Chen, Z.S. Health risk-based assessment and management of heavy metals-contaminated soil sites in Taiwan. Int. J. Environ. Res. Public Health 2010, 7, 3595–3614. [Google Scholar] [CrossRef]
- Li, C.; Zhou, K.; Qin, W.; Tian, C.; Qi, M.; Yan, X.; Han, W. A Review on heavy metals contamination in soil: Effects, sources, and remediation techniques. Soil Sediment Contam. 2019, 28, 380–394. [Google Scholar] [CrossRef]
- Xia, X.; Lin, S.; Zhao, J.; Zhang, W.; Lin, K.; Lu, Q.; Zhou, B. Toxic responses of microorganisms to nickel exposure in farmland soil in the presence of earthworm (Eisenia fetida). Chemosphere 2018, 192, 43–50. [Google Scholar] [CrossRef]
- Roychowdhury, A.; Datta, R.; Sarkar, D. Heavy metal pollution and remediation. In Green Chemistry; Elsevier Inc.: Amsterdam, The Netherlands, 2018; pp. 359–373. ISBN 9780128092705. [Google Scholar]
- Mosa, K.A.; Saadoun, I.; Kumar, K.; Helmy, M.; Dhankher, O.P. Potential biotechnological strategies for the cleanup of heavy metals and metalloids. Front. Plant Sci. 2016, 7, 303. [Google Scholar] [CrossRef]
- Moreira, V.R.; Lebron, Y.A.R.; Freire, S.J.; Santos, L.V.S.; Palladino, F.; Jacob, R.S. Biosorption of copper ions from aqueous solution using Chlorella pyrenoidosa: Optimization, equilibrium and kinetics studies. Microchem. J. 2019, 145, 119–129. [Google Scholar] [CrossRef]
- McGrath, S.P.; Zhao, F.J.; Lombi, E. Plant and rhizosphere processes involved in phytoremediation of metal-contaminated soils. Plant Soil 2001, 232, 207–214. [Google Scholar] [CrossRef]
- Ullah, A.; Heng, S.; Farooq, M.; Munis, H.; Fahad, S.; Yang, X. Phytoremediation of heavy metals assisted by plant growth promoting (PGP) bacteria: A review. Environ. Exp. Bot. 2015, 117, 28–40. [Google Scholar] [CrossRef]
- Oladoye, P.O.; Olowe, O.M.; Asemoloye, M.D. Phytoremediation technology and food security impacts of heavy metal contaminated soils: A review of literature. Chemosphere 2022, 288, 132555. [Google Scholar] [CrossRef]
- Antoniadis, V.; Levizou, E.; Shaheen, S.M.; Ok, Y.S.; Sebastian, A.; Baum, C.; Prasad, M.N.V.; Wenzel, W.W.; Rinklebe, J. Trace elements in the soil-plant interface: Phytoavailability, translocation, and phytoremediation—A review. Earth-Sci. Rev. 2017, 171, 621–645. [Google Scholar] [CrossRef]
- Raklami, A.; Meddich, A.; Pajuelo, E.; Marschner, B.; Heinze, S.; Oufdou, K. Combined application of marble waste and beneficial microorganisms: Toward a cost-effective approach for restoration of heavy metals contaminated sites. Environ. Sci. Pollut. Res. 2022, 1–15. [Google Scholar] [CrossRef] [PubMed]
- DalCorso, G.; Fasani, E.; Manara, A.; Visioli, G.; Furini, A. Heavy metal pollutions: State of the art and innovation in phytoremediation. Int. J. Mol. Sci. 2019, 20, 3412. [Google Scholar] [CrossRef] [PubMed]
- Ali, H.; Khan, E.; Sajad, M.A. Phytoremediation of heavy metals-concepts and applications. Chemosphere 2013, 91, 869–881. [Google Scholar] [CrossRef]
- Kavanagh, L.; Keohane, J.; Cabellos, G.G.; Lloyd, A.; Cleary, J. Induced plant accumulation of lithium. Geosciences 2018, 8, 56. [Google Scholar] [CrossRef]
- Mahar, A.; Wang, P.; Ali, A.; Awasthi, M.K.; Lahori, A.H.; Wang, Q.; Li, R.; Zhang, Z. Challenges and opportunities in the phytoremediation of heavy metals contaminated soils: A review. Ecotoxicol. Environ. Saf. 2016, 126, 111–121. [Google Scholar] [CrossRef]
- Raklami, A.; El Gharmali, A.; Rahou, Y.A.; Oufdou, K. Compost and mycorrhizae application as a technique to alleviate Cd and Zn stress in Medicago sativa. Int. J. Phytoremed. 2020, 23, 190–201. [Google Scholar] [CrossRef] [PubMed]
- Fagorzi, C.; Checcucci, A.; DiCenzo, G.; Debiec-Andrzejewska, K.; Dziewit, L.; Pini, F.; Mengoni, A. Harnessing rhizobia to improve heavy-metal phytoremediation by legumes. Genes 2018, 9, 542. [Google Scholar] [CrossRef] [PubMed]
- Nirola, R.; Megharaj, M.; Palanisami, T.; Aryal, R.; Venkateswarlu, K.; Naidu, R. Evaluation of metal uptake factors of native trees colonizing an abandoned copper mine—A quest for phytostabilization. J. Sustain. Min. 2015, 14, 115–123. [Google Scholar] [CrossRef]
- Jebara, S.H.; Saadani, O.; Fatnassi, I.C.; Chiboub, M.; Abdelkrim, S.; Jebara, M. Inoculation of Lens culinaris with Pb-resistant bacteria shows potential for phytostabilization. Environ. Sci. Pollut. Res. 2015, 22, 2537–2545. [Google Scholar] [CrossRef]
- Saadani, O.; Fatnassi, I.C.; Chiboub, M.; Abdelkrim, S.; Barhoumi, F.; Jebara, M.; Jebara, S.H. Ecotoxicology and environmental safety in situ phytostabilisation capacity of three legumes and their associated plant growth promoting bacteria (PGPBs) in mine tailings of northern Tunisia. Ecotoxicol. Environ. Saf. 2016, 130, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Oleńska, E.; Imperato, V.; Małek, W.; Włostowski, T.; Wójcik, M.; Swiecicka, I.; Vangronsveld, J.; Thijs, S. Trifolium repens-Associated bacteria as a potential tool to facilitate phytostabilization of zinc and lead polluted waste heaps. Plants 2020, 9, 1002. [Google Scholar] [CrossRef]
- Singh, J.; Sharma, D.; Kumar, G.; Sharma, N.R. Bioremediation: An eco-sustainable approach for restoration of contaminated sites. In Microbial Bioprospecting for Sustainable Development; Singh, J., Sharma, D., Kumar, G., Sharma, N.R., Eds.; Springer: Singapore, 2018; pp. 1–397. ISBN 978-981-13-0052-3. [Google Scholar]
- Saha, L.; Tiwari, J.; Bauddh, K.; Ma, Y. Recent developments in microbe–plant-based bioremediation for tackling heavy metal-polluted soils. Front. Microbiol. 2021, 12, 731723. [Google Scholar] [CrossRef]
- Sakakibara, M.; Watanabe, A.; Inoue, M.; Sano, S.; Kaise, T. Phytoextraction and phytovolatilization of arsenic from as-contaminated soils by Pteris vittata. In Proceedings of the Annual International Conference on Soils, Sediments Water and Energy, Amherst, MA, USA, 18–21 October 2010; Volume 12, pp. 267–272. [Google Scholar]
- Tyagi, B.; Kumar, N. Bioremediation: Principles and applications in environmental management. In Bioremediation for Environmental Sustainability; Elsevier: Amsterdam, The Netherlands, 2021; pp. 3–28. ISBN 9780128205242. [Google Scholar]
- Dolphen, R.; Thiravetyan, P. Phytodegradation of ethanolamines by Cyperus alternifolius: Effect of molecular size. Int. J. Phytoremed. 2015, 17, 686–692. [Google Scholar] [CrossRef]
- Chen, F.; Huber, C.; May, R.; Schröder, P. Metabolism of Oxybenzone in a Hairy Root Culture: Perspectives for phytoremediation of a widely used sunscreen agent. J. Hazard. Mater. 2016, 306, 230–236. [Google Scholar] [CrossRef]
- Rezania, S.; Taib, S.M.; Md Din, M.F.; Dahalan, F.A.; Kamyab, H. Comprehensive review on phytotechnology: Heavy metals removal by diverse aquatic plants species from wastewater. J. Hazard. Mater. 2016, 318, 587–599. [Google Scholar] [CrossRef]
- Hakeem, K.R.; Bhat, R.A.; Qadri, H. Phytoremediation of heavy metals: An eco-friendly and sustainable approach. In Bioremediation and Biotechnology: Sustainable Approaches to Pollution Degradation; Hakeem, K.R., Bhat, R.A., Qadri, H., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 1–327. ISBN 978-3-030-35690-3. [Google Scholar]
- Kumar, R.; Mishra, R.K.; Mishra, V.; Qidwai, A.; Pandey, A.; Shukla, S.K.; Pandey, M.; Pathak, A.; Dikshit, A. Detoxification and Tolerance of Heavy Metals in Plants; Elsevier Inc.: Amsterdam, The Netherlands, 2015; ISBN 9780128031582. [Google Scholar]
- Sytar, O.; Ghosh, S.; Malinska, H.; Zivcak, M.; Brestic, M. Physiological and molecular mechanisms of metal accumulation in hyperaccumulator plants. Physiol. Plant. 2021, 173, 148–166. [Google Scholar] [CrossRef] [PubMed]
- Gupta, D.K.; Corpas, F.J.; Palma, J.M. Role of phytochelatins in heavy metal stress and detoxification mechanisms in plants. In Heavy Metal Stress in Plants; Gupta, D.K., Corpas, F.J., Palma, J.M., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 1–242. ISBN 9783642384691. [Google Scholar]
- Ali, A.; Guo, D.; Mahar, A.; Ma, F.; Li, R.; Shen, F.; Wang, P.; Zhang, Z. Streptomyces pactum assisted phytoremediation in Zn/Pb smelter contaminated soil of feng county and its impact on enzymatic activities. Sci. Rep. 2017, 7, 46087. [Google Scholar] [CrossRef] [PubMed]
- Haydon, M.J.; Cobbett, C.S. Transporters of ligands for essential metal ions in plants: Research review. New Phytol. 2007, 174, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.Y.; Peng, Z.L.; Han, F.; Shan, X.Q.; Lin, J.M. Study of low-molecular weight organic acids in maize roots under the stress of cadmium using capillary zone electrophoresis. J. Sep. Sci. 2007, 30, 2742–2747. [Google Scholar] [CrossRef]
- Kanwal, S.; Bano, A.; Malik, R.N. Effects of arbuscular mycorrhizal fungi on metals uptake, physiological and biochemical response of Medicago sativa L. with increasing Zn and Cd concentrations in Soil. Am. J. Plant Sci. 2015, 6, 2906–2923. [Google Scholar] [CrossRef]
- Kushwaha, A.; Rani, R.; Kumar, S.; Gautam, A. Heavy metal detoxification and tolerance mechanisms in plants: Implications for phytoremediation. Environ. Rev. 2015, 24, 39–51. [Google Scholar] [CrossRef]
- Talukder, P. Role of Metallothionein and Phytochelatin in Combating Abiotic Stress Imparted by Copper-Induced Toxicity in Brinjal (Solanum melongena); Springer: Singapore, 2021; ISBN 9789811574085. [Google Scholar]
- Yan, A.; Wang, Y.; Tan, S.N.; Mohd Yusof, M.L.; Ghosh, S.; Chen, Z. Phytoremediation: A promising approach for revegetation of heavy metal-polluted land. Front. Plant Sci. 2020, 11, 359. [Google Scholar] [CrossRef]
- Jian, C.; Yang, Z.; Su, Y.; Han, F.X.; Monts, D.L. Phytoremediation of heavy metal/metalloid-contaminated soils. In Contaminated Soils: Environmental Impact, Disposal and Treatment; Steinberg, V., Ed.; Nova Science Publishers, Inc.: London, UK, 2011; p. 50. ISBN 9781607417910. [Google Scholar]
- Fernández, L.G.; Fernández-pascual, M.; Javier, F.; Mañero, G.; Antonio, J.; García, L. Phytoremediation of contaminated waters to improve water quality. In Phytoremediation; Ansari, A.A., Gill, S.S., Gill, R., Lanza, G., Newman, L., Eds.; Springer International Publishing: Cham, Switzerland, 2015; Volume 2, pp. 11–26. ISBN 9783319109695. [Google Scholar]
- Liu, W.; Wang, B.; Wang, Q.; Hou, J.; Wu, L.; Wood, J.L.; Luo, Y.; Franks, A.E. Characteristics of metal-tolerant plant growth-promoting yeast (Cryptococcus sp. NSE1) and its influence on Cd hyperaccumulator Sedum plumbizincicola. Environ. Sci. Pollut. Res. 2016, 23, 18621–18629. [Google Scholar] [CrossRef]
- Anli, M.; Baslam, M.; Tahiri, A.; Raklami, A.; Symanczik, S.; Boutasknit, A.; Ait-El-Mokhtar, M.; Ben-Laouane, R.; Toubali, S.; Ait Rahou, Y.; et al. Biofertilizers as strategies to improve photosynthetic apparatus, growth, and drought stress tolerance in the date palm. Front. Plant Sci. 2020, 11, 1560. [Google Scholar] [CrossRef]
- Ben-Laouane, R.; Ait-El-Mokhtar, M.; Anli, M.; Boutasknit, A.; Ait Rahou, Y.; Raklami, A.; Oufdou, K.; Wahbi, S.; Meddich, A. Green compost combined with mycorrhizae and rhizobia: A strategy for improving alfalfa growth and yield under field conditions. Gesunde Pflanz. 2021, 73, 193–207. [Google Scholar] [CrossRef]
- Navarro-Torre, S.; Mateos-Naranjo, E.; Caviedes, M.A.; Pajuelo, E.; Rodríguez-Llorente, I.D. Isolation of plant-growth-promoting and metal-resistant cultivable bacteria from Arthrocnemum macrostachyum in the odiel marshes with potential use in phytoremediation. Mar. Pollut. Bull. 2016, 110, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Raklami, A.; Bechtaoui, N.; Tahiri, A.I.; Anli, M.; Meddich, A.; Oufdou, K. Use of rhizobacteria and mycorrhizae consortium in the open field as a strategy for improving crop nutrition, productivity and soil fertility. Front. Microbiol. 2019, 10, 1106. [Google Scholar] [CrossRef] [PubMed]
- Nafis, A.; Raklami, A.; Bechtaoui, N.; El Khalloufi, F.; El Alaoui, A.; Glick, B.R.; Hafidi, M.; Kouisni, L.; Ouhdouch, Y.; Hassani, L. Actinobacteria from extreme niches in morocco and their plant growth-promoting potentials. Diversity 2019, 11, 139. [Google Scholar] [CrossRef]
- Manoj, S.R.; Karthik, C.; Kadirvelu, K.; Arulselvi, P.I.; Shanmugasundaram, T.; Bruno, B.; Rajkumar, M. Understanding the molecular mechanisms for the enhanced phytoremediation of heavy metals through plant growth promoting rhizobacteria: A review. J. Environ. Manag. 2020, 254, 109779. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Tripathi, S.; Chaturvedi, P.; Chaurasia, D.; Chandra, R. Newly isolated Bacillus sp. PS-6 assisted phytoremediation of heavy metals using Phragmites communis: Potential application in wastewater treatment. Bioresour. Technol. 2021, 320, 124353. [Google Scholar] [CrossRef] [PubMed]
- Coninx, L.; Martinova, V.; Rineau, F. Mycorrhiza-assisted phytoremediation. Adv. Bot. Res. 2017, 83, 127–188. [Google Scholar] [CrossRef]
- Riaz, M.; Kamran, M.; Fang, Y.; Wang, Q.; Cao, H.; Yang, G.; Deng, L.; Wang, Y.; Zhou, Y.; Anastopoulos, I.; et al. Arbuscular mycorrhizal fungi-induced mitigation of heavy metal phytotoxicity in metal contaminated soils: A critical review. J. Hazard. Mater. 2021, 402, 123919. [Google Scholar] [CrossRef]
- Liu, L.; Li, J.; Yue, F.; Yan, X.; Wang, F.; Bloszies, S.; Wang, Y. Effects of arbuscular mycorrhizal inoculation and biochar amendment on maize growth, cadmium uptake and soil cadmium speciation in cd-contaminated soil. Chemosphere 2018, 194, 495–503. [Google Scholar] [CrossRef]
- Janoušková, M.; Pavlíková, D.; Vosátka, M. Potential contribution of arbuscular mycorrhiza to cadmium immobilisation in soil. Chemosphere 2006, 65, 1959–1965. [Google Scholar] [CrossRef]
- Cabral, L.; Soares, C.R.F.S.; Giachini, A.J.; Siqueira, J.O. Arbuscular mycorrhizal fungi in phytoremediation of contaminated areas by trace elements: Mechanisms and major benefits of their applications. World J. Microbiol. Biotechnol. 2015, 31, 1655–1664. [Google Scholar] [CrossRef]
- Khalid, M.; Ur-rahman, S.; Hassani, D.; Hayat, K.; Zhou, P.; Hui, N. Advances in fungal-assisted phytoremediation of heavy metals: A review. Pedosphere 2021, 31, 475–495. [Google Scholar] [CrossRef]
- Handa, Y.; Nishide, H.; Takeda, N.; Suzuki, Y.; Kawaguchi, M.; Saito, K. RNA-Seq transcriptional profiling of an arbuscular mycorrhiza provides insights into regulated and coordinated gene expression in Lotus japonicus and Rhizophagus irregularis. Plant Cell Physiol. 2015, 56, 1490–1511. [Google Scholar] [CrossRef] [PubMed]
- Das, J.; Sarkar, P. Remediation of arsenic in mung bean (Vigna radiata) with growth enhancement by unique arsenic-resistant bacterium Acinetobacter lwoffii. Sci. Total Environ. 2018, 624, 1106–1118. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.; Chen, A.; Jiang, Y.; Sun, C.; Luo, S.; Shao, J. Application of a newly recorded diazotrophic cyanobacterium in acidified and Cd contaminated paddy soil: Promotes rice yield and decreases Cd accumulation. Sci. Total Environ. 2022, 814, 152630. [Google Scholar] [CrossRef]
- Pan, F.; Meng, Q.; Luo, S.; Shen, J.; Chen, B.; Khan, K.Y.; Japenga, J.; Ma, X.; Yang, X.; Feng, Y. Enhanced Cd extraction of oilseed rape (Brassica napus) by plant growth-promoting bacteria isolated from cd hyperaccumulator Sedum alfredii Hance. Int. J. Phytoremed. 2017, 19, 281–289. [Google Scholar] [CrossRef]
- Visioli, G.; Vamerali, T.; Mattarozzi, M.; Dramis, L.; Sanangelantoni, A.M. Combined endophytic inoculants enhance nickel phytoextraction from serpentine soil in the hyperaccumulator Noccaea caerulescens. Front. Plant Sci. 2015, 6, 638. [Google Scholar] [CrossRef]
- Hassan, T.U.; Bano, A.; Naz, I. Alleviation of heavy metals toxicity by the application of plant growth promoting rhizobacteria and effects on wheat grown in saline sodic field. Int. J. Phytoremed. 2017, 19, 522–529. [Google Scholar] [CrossRef]
- Bruno, L.B.; Anbuganesan, V.; Karthik, C.; Tripti; Kumar, A.; Banu, J.R.; Freitas, H.; Rajkumar, M. Enhanced phytoextraction of multi-metal contaminated soils under increased atmospheric temperature by bioaugmentation with plant growth promoting Bacillus cereus. J. Environ. Manag. 2021, 289, 112553. [Google Scholar] [CrossRef]
- Funes Pinter, I.; Salomon, M.V.; Berli, F.; Bottini, R.; Piccoli, P. Characterization of the As(III) tolerance conferred by plant growth promoting rhizobacteria to in vitro-grown grapevine. Appl. Soil Ecol. 2017, 109, 60–68. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, W.J.; He, L.Y.; Sheng, X.F. Increased biomass and quality and reduced heavy metal accumulation of edible tissues of vegetables in the presence of Cd-tolerant and immobilizing Bacillus megaterium H3. Ecotoxicol. Environ. Saf. 2018, 148, 269–274. [Google Scholar] [CrossRef]
- Mohammadzadeh, A.; Tavakoli, M.; Motesharezadeh, B.; Chaichi, M.R. Effects of plant growth-promoting bacteria on the phytoremediation of cadmium-contaminated soil by sunflower. Arch. Agron. Soil Sci. 2017, 63, 807–816. [Google Scholar] [CrossRef]
- Ahmad, I.; Akhtar, M.J.; Asghar, H.N.; Ghafoor, U.; Shahid, M. Differential effects of plant growth-promoting rhizobacteria on maize growth and cadmium uptake. J. Plant Growth Regul. 2016, 35, 303–315. [Google Scholar] [CrossRef]
- Motesharezadeh, B.; Kamal-poor, S.; Alikhani, H.; Zarei, M.; Azimi, S. Investigating the effects of plant growth promoting bacteria and Glomus mosseae on cadmium phytoremediation by Eucalyptus camaldulensis L. Pollution 2017, 3, 575–588. [Google Scholar] [CrossRef]
- Mousavi, S.M.; Motesharezadeh, B.; Hosseini, H.M.; Alikhani, H.; Zolfaghari, A.A. Root-Induced Changes of Zn and Pb dynamics in the rhizosphere of sunflower with different plant growth promoting treatments in a heavily contaminated soil. Ecotoxicol. Environ. Saf. 2018, 147, 206–216. [Google Scholar] [CrossRef]
- Płociniczak, T.; Sinkkonen, A.; Romantschuk, M.; Sułowicz, S.; Piotrowska-Seget, Z. Rhizospheric bacterial strain Brevibacterium casei MH8a colonizes plant tissues and enhances Cd, Zn, Cu phytoextraction by white mustard. Front. Plant Sci. 2016, 7, 101. [Google Scholar] [CrossRef] [PubMed]
- Haruma, T.; Yamaji, K.; Ogawa, K.; Masuya, H.; Sekine, Y.; Kozai, N. Root-endophytic Chaetomium cupreum chemically enhances aluminium tolerance in Miscanthus sinensis via increasing the aluminium detoxicants, chlorogenic acid and oosporein. PLoS ONE 2019, 14, e0212644. [Google Scholar] [CrossRef] [PubMed]
- Awasthi, S.; Chauhan, R.; Dwivedi, S.; Srivastava, S.; Srivastava, S.; Tripathi, R.D. A Consortium of alga (Chlorella vulgaris) and bacterium (Pseudomonas putida) for amelioration of arsenic toxicity in rice: A promising and feasible approach. Environ. Exp. Bot. 2018, 150, 115–126. [Google Scholar] [CrossRef]
- Kaur, J.; Anand, V.; Srivastava, S.; Bist, V.; Tripathi, P.; Naseem, M.; Nand, S.; Anshu; Khare, P.; Srivastava, P.K.; et al. Yeast strain Debaryomyces hansenii for amelioration of arsenic stress in rice. Ecotoxicol. Environ. Saf. 2020, 195, 110480. [Google Scholar] [CrossRef]
- Adeyemi, N.O.; Atayese, M.O.; Sakariyawo, O.S.; Azeez, J.O.; Abayomi Sobowale, S.P.; Olubode, A.; Mudathir, R.; Adebayo, R.; Adeoye, S. Alleviation of heavy metal stress by arbuscular mycorrhizal symbiosis in Glycine max (L.) grown in copper, lead and zinc contaminated soils. Rhizosphere 2021, 18, 100325. [Google Scholar] [CrossRef]
- Zhan, F.; Li, B.; Jiang, M.; Li, T.; He, Y.; Li, Y.; Wang, Y. Effects of arbuscular mycorrhizal fungi on the growth and heavy metal accumulation of bermudagrass [Cynodon dactylon (L.) Pers.] grown in a lead–zinc mine wasteland. Int. J. Phytoremed. 2019, 21, 849–856. [Google Scholar] [CrossRef]
- Zhang, F.; Liu, M.; Li, Y.; Che, Y.; Xiao, Y. Effects of arbuscular mycorrhizal fungi, biochar and cadmium on the yield and element uptake of Medicago sativa. Sci. Total Environ. 2019, 655, 1150–1158. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Yuan, M.; Tan, S.; Yang, X.; Lan, Z.; Jiang, Q.; Ye, Z.; Jing, Y. Enhancement of arbuscular mycorrhizal fungus (Glomus versiforme) on the growth and Cd uptake by Cd-hyperaccumulator Solanum nigrum. Appl. Soil Ecol. 2015, 89, 44–49. [Google Scholar] [CrossRef]
- Arunakumara, K.K.I.U.; Walpola, B.C.; Yoon, M.H. Bioaugmentation-assisted phytoextraction of Co, Pb and Zn: An assessment with a phosphate-solubilizing bacterium isolated from metal-contaminated mines of Boryeong area in South Korea. Biotechnol. Agron. Soc. Environ. 2015, 19, 143–152. [Google Scholar]
- Chakraborty, S.; Das, S.; Banerjee, S.; Mukherjee, S.; Ganguli, A.; Mondal, S. Heavy metals bio-removal potential of the isolated Klebsiella Sp TIU20 strain which improves growth of economic crop plant (Vigna radiata L.) under heavy metals stress by exhibiting plant growth promoting and protecting traits. Biocatal. Agric. Biotechnol. 2021, 38, 102204. [Google Scholar] [CrossRef]
- Mallick, I.; Bhattacharyya, C.; Mukherji, S.; Dey, D.; Sarkar, S.C.; Mukhopadhyay, U.K.; Ghosh, A. Effective rhizoinoculation and biofilm formation by arsenic immobilizing halophilic plant growth promoting bacteria (PGPB) Isolated from mangrove rhizosphere: A step towards arsenic rhizoremediation. Sci. Total Environ. 2018, 610–611, 1239–1250. [Google Scholar] [CrossRef] [PubMed]
- Zanganeh, F.; Heidari, A.; Sepehr, A.; Rohani, A. Bioaugmentation and bioaugmentation–assisted phytoremediation of heavy metal contaminated soil by a synergistic effect of cyanobacteria inoculation, biochar, and purslane (Portulaca oleracea L.). Environ. Sci. Pollut. Res. 2022, 29, 6040–6059. [Google Scholar] [CrossRef]
- Atigh, Z.B.Q.; Heidari, A.; Sepehr, A.; Bahreini, M.; Mahbub, K.R. Bioremediation of heavy metal contaminated soils originated from iron ore mine by bio-augmentation with native cyanobacteria. Iran. J. Energy Environ. 2020, 11, 89–96. [Google Scholar] [CrossRef]
- Bilal, S.; Shahzad, R.; Imran, M.; Jan, R.; Kim, K.M.; Lee, I.J. Synergistic association of endophytic fungi enhances Glycine max L. resilience to combined abiotic stresses: Heavy metals, high temperature and drought stress. Ind. Crops Prod. 2020, 143, 111931. [Google Scholar] [CrossRef]
- Paredes-Páliz, K.; Rodríguez-Vázquez, R.; Duarte, B.; Caviedes, M.A.; Mateos-Naranjo, E.; Redondo-Gómez, S.; Caçador, M.I.; Rodríguez-Llorente, I.D.; Pajuelo, E. Investigating the mechanisms underlying phytoprotection by plant growth-promoting rhizobacteria in Spartina densiflora under metal stress. Plant Biol. 2018, 20, 497–506. [Google Scholar] [CrossRef]
- Ashraf, S.; Afzal, M.; Naveed, M.; Shahid, M.; Zahir, Z.A. Endophytic bacteria enhance remediation of tannery effluent in constructed wetlands vegetated with Leptochloa fusca. Int. J. Phytoremed. 2018, 20, 121–128. [Google Scholar] [CrossRef]
- Xie, Y.; Bu, H.; Feng, Q.; Wassie, M.; Amee, M.; Jiang, Y.; Bi, Y.; Hu, L.; Chen, L. Identification of Cd-resistant microorganisms from heavy metal-contaminated soil and its potential in promoting the growth and cd accumulation of bermudagrass. Environ. Res. 2021, 200, 111730. [Google Scholar] [CrossRef] [PubMed]
- Rahman, S.; Khalid, M.; Kayani, S.I.; Tang, K. The ameliorative effects of exogenous inoculation of Piriformospora indica on molecular, biochemical and physiological parameters of Artemisia annua L. under arsenic stress condition. Ecotoxicol. Environ. Saf. 2020, 206, 111202. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zheng, X.; Lin, L.; An, Q.; Jiao, Y.; Li, Q.; Li, Z.; Hong, Y.; Zhang, K.; Xie, C.; et al. Remediation of soils co-contaminated with cadmium and dichlorodiphenyltrichloroethane by king grass associated with Piriformospora indica: Insights into the regulation of root excretion and reshaping of rhizosphere microbial community structure. J. Hazard. Mater. 2022, 422, 126936. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.; Bano, A. Modulation of Phytoremediation and Plant Growth by the Treatment with PGPR, Ag nanoparticle and untreated municipal wastewater. Int. J. Phytoremed. 2016, 18, 1258–1269. [Google Scholar] [CrossRef]
- Rizvi, A.; Khan, M.S. Biotoxic impact of heavy metals on growth, oxidative stress and morphological changes in root structure of wheat (Triticum aestivum L.) and stress alleviation by Pseudomonas aeruginosa strain CPSB1. Chemosphere 2017, 185, 942–952. [Google Scholar] [CrossRef]
- Kaur, P.; Singh, S.; Kumar, V.; Singh, N.; Singh, J. Effect of rhizobacteria on arsenic uptake by macrophyte Eichhornia crassipes (Mart.) solms. Int. J. Phytoremed. 2018, 20, 114–120. [Google Scholar] [CrossRef]
- Chen, B.; Luo, S.; Wu, Y.; Ye, J.; Wang, Q.; Xu, X. The effects of the endophytic bacterium Pseudomonas fluorescens Sasm05 and IAA on the plant growth and cadmium uptake of Sedum alfredii Hance. Front. Microbiol. 2017, 8, 2538. [Google Scholar] [CrossRef]
- Shabaan, M.; Asghar, H.N.; Akhtar, M.J.; Ali, Q.; Ejaz, M. Role of plant growth promoting rhizobacteria in the alleviation of lead toxicity to Pisum sativum L. Int. J. Phytoremed. 2021, 23, 837–845. [Google Scholar] [CrossRef]
- Ma, Y.; Rajkumar, M.; Oliveira, R.S.; Zhang, C.; Freitas, H. Potential of Plant Beneficial Bacteria and Arbuscular Mycorrhizal Fungi in Phytoremediation of Metal-Contaminated Saline Soils. J. Hazard. Mater. 2019, 379, 120813. [Google Scholar] [CrossRef]
- Tirry, N.; Kouchou, A.; El Omari, B.; Ferioun, M.; El Ghachtouli, N. Improved chromium tolerance of Medicago sativa by plant growth-promoting rhizobacteria (PGPR). J. Genet. Eng. Biotechnol. 2021, 19, 149. [Google Scholar] [CrossRef]
- Montalbán, B.; Thijs, S.; Lobo, M.C.; Weyens, N.; Ameloot, M.; Vangronsveld, J.; Pérez-Sanz, A. Cultivar and metal-specific effects of endophytic bacteria in Helianthus tuberosus exposed to Cd and Zn. Int. J. Mol. Sci. 2017, 18, 2026. [Google Scholar] [CrossRef] [PubMed]
- Karimi, A.; Khodaverdiloo, H.; Rasouli-Sadaghiani, M.H. Microbial-enhanced phytoremediation of lead contaminated calcareous soil by Centaurea cyanus L. CLEAN—Soil Air Water 2018, 46, 1700665. [Google Scholar] [CrossRef]
- Mishra, V.; Gupta, A.; Kaur, P.; Singh, S.; Singh, N.; Gehlot, P.; Singh, J. Synergistic effects of arbuscular mycorrhizal fungi and plant growth promoting rhizobacteria in bioremediation of iron contaminated soils. Int. J. Phytoremed. 2016, 18, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Anand, G.; Singh, N.; Kapoor, R. Arbuscular mycorrhiza augments arsenic tolerance in wheat (Triticum aestivum L.) by strengthening antioxidant defense system and thiol metabolism. Front. Plant Sci. 2017, 8, 906. [Google Scholar] [CrossRef]
- Singh, G.; Pankaj, U.; Chand, S.; Verma, R.K. Arbuscular mycorrhizal fungi-assisted phytoextraction of toxic metals by Zea mays L. from tannery sludge. Soil Sediment Contam. 2019, 28, 729–746. [Google Scholar] [CrossRef]
- Zhang, X.F.; Hu, Z.H.; Yan, T.X.; Lu, R.R.; Peng, C.L.; Li, S.S.; Jing, Y.X. Arbuscular mycorrhizal fungi alleviate cd phytotoxicity by altering Cd subcellular distribution and chemical forms in Zea mays. Ecotoxicol. Environ. Saf. 2019, 171, 352–360. [Google Scholar] [CrossRef]
- Peng, W.; Li, X.; Song, J.; Jiang, W.; Liu, Y.; Fan, W. Bioremediation of cadmium- and zinc-contaminated soil using Rhodobacter sphaeroides. Chemosphere 2018, 197, 33–41. [Google Scholar] [CrossRef]
- Carlos, M.H.J.; Stefani, P.V.Y.; Janette, A.M.; Melani, M.S.S.; Gabriela, P.O. Assessing the effects of heavy metals in ACC deaminase and IAA production on plant growth-promoting bacteria. Microbiol. Res. 2016, 188–189, 53–61. [Google Scholar] [CrossRef]
- Jin, Z.; Deng, S.; Wen, Y.; Jin, Y.; Pan, L.; Zhang, Y.; Black, T.; Jones, K.C.; Zhang, H.; Zhang, D. Application of Simplicillium chinense for Cd and Pb biosorption and enhancing heavy metal phytoremediation of soils. Sci. Total Environ. 2019, 697, 134148. [Google Scholar] [CrossRef]
- Sepehri, M.; Khatabi, B. Combination of siderophore-producing bacteria and Piriformospora indica provides an efficient approach to improve cadmium tolerance in alfalfa. Microb. Ecol. 2021, 81, 717–730. [Google Scholar] [CrossRef]
- Seifikalhor, M.; Hassani, S.B.; Aliniaeifard, S. Seed priming by cyanobacteria (Spirulina platensis) and salep gum enhances tolerance of maize plant against cadmium toxicity. J. Plant Growth Regul. 2020, 39, 1009–1021. [Google Scholar] [CrossRef]
- Cao, S.; Wang, W.; Zhao, Y.; Yang, S.; Wang, F.; Zhang, J.; Sun, Y. Enhancement of lead phytoremediation by perennial ryegrass (Lolium perenne L.) using agent of Streptomyces pactum Act12. J. Pet. Environ. Biotechnol. 2016, 7, 269–275. [Google Scholar] [CrossRef]
- Jeyasundar, P.G.S.A.; Ali, A.; Azeem, M.; Li, Y.; Guo, D.; Sikdar, A.; Abdelrahman, H.; Kwon, E.; Antoniadis, V.; Mani, V.M.; et al. Green remediation of toxic metals contaminated mining soil using bacterial consortium and Brassica juncea. Environ. Pollut. 2021, 277, 116789. [Google Scholar] [CrossRef] [PubMed]
- El-Shahir, A.A.; El-Tayeh, N.A.; Ali, O.M.; Abdel Latef, A.A.H.; Loutfy, N. The Effect of endophytic Talaromyces pinophilus on growth, absorption and accumulation of heavy metals of Triticum aestivum grown on sandy soil amended by sewage sludge. Plants 2021, 10, 2659. [Google Scholar] [CrossRef] [PubMed]
- Malik, A.; Butt, T.A.; Naqvi, S.T.A.; Yousaf, S.; Qureshi, M.K.; Zafar, M.I.; Farooq, G.; Nawaz, I.; Iqbal, M. Lead tolerant endophyte Trametes hirsuta Improved the growth and lead accumulation in the vegetative parts of Triticum aestivum L. Heliyon 2020, 6, e04188. [Google Scholar] [CrossRef]
- Li, X.; Zhang, X.; Wang, X.; Yang, X.; Cui, Z. Bioaugmentation-assisted phytoremediation of lead and salinity co-contaminated soil by Suaeda salsa and Trichoderma asperellum. Chemosphere 2019, 224, 716–725. [Google Scholar] [CrossRef]
- Belimov, A.A.; Shaposhnikov, A.I.; Azarova, T.S.; Makarova, N.M.; Safronova, V.I.; Litvinskiy, V.A.; Nosikov, V.V.; Zavalin, A.A.; Tikhonovich, I.A. Microbial consortium of pgpr, rhizobia and arbuscular mycorrhizal fungus makes pea mutant SGECdt comparable with indian mustard in cadmium tolerance and accumulation. Plants 2020, 9, 975. [Google Scholar] [CrossRef]
- Berg, G.; Rybakova, D.; Grube, M.; Köberl, M. The plant microbiome explored: Implications for experimental botany. J. Exp. Bot. 2016, 67, 995–1002. [Google Scholar] [CrossRef]
- Thijs, S.; Sillen, W.; Rineau, F.; Weyens, N.; Vangronsveld, J. Towards an enhanced understanding of plant-microbiome interactions to improve phytoremediation: Engineering the metaorganism. Front. Microbiol. 2016, 7, 341. [Google Scholar] [CrossRef]
- Vryzas, Z. The plant as metaorganism and research on next-generation systemic pesticides—Prospects and challenges. Front. Microbiol. 2016, 7, 1968. [Google Scholar] [CrossRef]
- Ahkami, A.H.; Allen White, R.; Handakumbura, P.P.; Jansson, C. Rhizosphere engineering: Enhancing sustainable plant ecosystem productivity. Rhizosphere 2017, 3, 233–243. [Google Scholar] [CrossRef]
- Ahmadi, K.; Zarebanadkouki, M.; Ahmed, M.A.; Ferrarini, A.; Kuzyakov, Y.; Kostka, S.J.; Carminati, A. Rhizosphere engineering: Innovative Improvement of root environment. Rhizosphere 2017, 3, 176–184. [Google Scholar] [CrossRef]
- Bell, T.H.; Cloutier-Hurteau, B.; Al-Otaibi, F.; Turmel, M.C.; Yergeau, E.; Courchesne, F.; St-Arnaud, M. Early rhizosphere microbiome composition is related to the growth and Zn uptake of willows introduced to a former landfill. Environ. Microbiol. 2015, 17, 3025–3038. [Google Scholar] [CrossRef] [PubMed]
- Johnson, N.C.; Wilson, G.W.T.; Bowker, M.A.; Wilson, J.A.; Miller, R.M. Resource limitation is a driver of local adaptation in mycorrhizal symbioses. Proc. Natl. Acad. Sci. USA 2010, 107, 2093–2098. [Google Scholar] [CrossRef] [PubMed]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; Van Der Putten, W.H. Going Back to the Roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef]
- Sasse, J.; Martinoia, E.; Northen, T. Feed your friends: Do plant exudates shape the root microbiome? Trends Plant Sci. 2018, 23, 25–41. [Google Scholar] [CrossRef]
- Tian, T.; Reverdy, A.; She, Q.; Sun, B.; Chai, Y. The role of rhizodeposits in shaping rhizomicrobiome. Environ. Microbiol. Rep. 2020, 12, 160–172. [Google Scholar] [CrossRef]
- Abdu, N.; Abdullahi, A.A.; Abdulkadir, A. Heavy metals and soil microbes. Environ. Chem. Lett. 2017, 15, 65–84. [Google Scholar] [CrossRef]
- Berg, J.; Brandt, K.K.; Al-Soud, W.A.; Holm, P.E.; Hansen, L.H.; Sørensen, S.J.; Nybroe, O. Selection for Cu-tolerant bacterial communities with altered composition, but unaltered richness, via long-term cu exposure. Appl. Environ. Microbiol. 2012, 78, 7438–7446. [Google Scholar] [CrossRef]
- Gołebiewski, M.; Deja-Sikora, E.; Cichosz, M.; Tretyn, A.; Wróbel, B. 16S rDNA pyrosequencing analysis of bacterial community in heavy metals polluted soils. Microb. Ecol. 2014, 67, 635–647. [Google Scholar] [CrossRef]
- Sobolev, D.; Begonia, M.F.T. Effects of heavy metal contamination upon soil microbes: Lead-induced changes in general and denitrifying microbial communities as evidenced by molecular markers. Int. J. Environ. Res. Public Health 2008, 5, 450–456. [Google Scholar] [CrossRef] [PubMed]
- Yergeau, E.; Sanschagrin, S.; Maynard, C.; St-Arnaud, M.; Greer, C.W. Microbial expression profiles in the rhizosphere of willows depend on soil contamination. ISME J. 2014, 8, 344–358. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, J.; Zhang, H.; Shi, W.; Liu, Y. Bacterial heavy-metal and antibiotic resistance genes in a copper tailing dam area in northern China. Front. Microbiol. 2019, 10, 1916. [Google Scholar] [CrossRef] [PubMed]
- Bell, T.H.; Joly, S.; Pitre, F.E.; Yergeau, E. Increasing phytoremediation efficiency and reliability using novel omics approaches. Trends Biotechnol. 2014, 32, 271–280. [Google Scholar] [CrossRef]
- Schenk, P.M.; Carvalhais, L.C.; Kazan, K. Unraveling plant-microbe interactions: Can multi-species transcriptomics help? Trends Biotechnol. 2012, 30, 177–184. [Google Scholar] [CrossRef]
- Jagtap, U.B.; Bapat, V.A. Genetic engineering of plants for heavy metal removal from soil. In Heavy Metal Contamination of Soils; Springer: Cham, Switzerland, 2015; pp. 433–470. ISBN 9783319145266. [Google Scholar]
- Lee, J.H. An overview of phytoremediation as a potentially promising technology for environmental pollution control. Biotechnol. Bioprocess Eng. 2013, 18, 431–439. [Google Scholar] [CrossRef]
- Faucon, M.P.; Houben, D.; Lambers, H. Plant Functional Traits: Soil and Ecosystem Services. Trends Plant Sci. 2017, 22, 385–394. [Google Scholar] [CrossRef]
- Faucon, M.P. Plant–Soil Interactions as Drivers of the Structure and Functions of Plant Communities. Diversity 2020, 12, 452. [Google Scholar] [CrossRef]
- Kapahi, M.; Sachdeva, S. Bioremediation Options for Heavy Metal Pollution. J. Health Pollut. 2019, 9, 191203. [Google Scholar] [CrossRef]
- Sun, W.; Cheng, K.; Sun, K.Y.; Ma, X. Microbially mediated remediation of contaminated sediments by heavy metals: A critical review. Curr. Pollut. Rep. 2021, 7, 201–212. [Google Scholar] [CrossRef]
- Njoku, K.L.; Akinyede, O.R.; Obidi, O.F. Microbial remediation of heavy metals contaminated media by Bacillus megaterium and Rhizopus stolonifer. Sci. Afr. 2020, 10, e00545. [Google Scholar] [CrossRef]
- Achal, V.; Pan, X.; Fu, Q.; Zhang, D. Biomineralization based remediation of As(III) contaminated soil by Sporosarcina ginsengisoli. J. Hazard. Mater. 2012, 201–202, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Imam, S.S.A.; Rajpoot, I.K.; Gajjar, B.; Sachdeva, A. Comparative study of heavy metal bioremediation in soil by Bacillus subtilis and Saccharomyces cerevisiae. Indian J. Sci. Technol. 2016, 9, 1–7. [Google Scholar] [CrossRef]
- Kang, C.H.; Kwon, Y.J.; So, J.S. Bioremediation of heavy metals by using bacterial mixtures. Ecol. Eng. 2016, 89, 64–69. [Google Scholar] [CrossRef]
- Chibuike, G.U.; Obiora, S.C. Heavy metal polluted soils: Effect on plants and bioremediation methods. Appl. Environ. Soil Sci. 2014, 2014, 752708. [Google Scholar] [CrossRef]
- Gupta, A.; Joia, J. Microbes as potential tool for remediation of heavy metals: A review. J. Microb. Biochem. Technol. 2016, 8, 364–372. [Google Scholar] [CrossRef]
- Hussain, N.; Abbasi, T.; Abbasi, S.A. Vermiremediation of an invasive and pernicious weed salvinia (Salvinia molesta). Ecol. Eng. 2016, 91, 432–440. [Google Scholar] [CrossRef]
- Mawang, C.I.; Azman, A.S.; Fuad, A.S.M.; Ahamad, M. Actinobacteria: An eco-friendly and promising technology for the bioaugmentation of contaminants. Biotechnol. Rep. 2021, 32, e00679. [Google Scholar] [CrossRef]
- Fauziah, S.H.; Agamuthu, P.; Hashim, R.; Izyani, A.K.; Emenike, C.U. Assessing the bioaugmentation potentials of individual isolates from landfill on metal-polluted soil. Environ. Earth Sci. 2017, 76, 401. [Google Scholar] [CrossRef]
- Singh, J.S.; Abhilash, P.C.; Singh, H.B.; Singh, R.P.; Singh, D.P. Genetically engineered bacteria: An emerging tool for environmental remediation and future research perspectives. Gene 2011, 480, 1–9. [Google Scholar] [CrossRef]
- Pant, G.; Garlapati, D.; Agrawal, U.; Prasuna, R.G.; Mathimani, T.; Pugazhendhi, A. Biological approaches practised using genetically engineered microbes for a sustainable environment: A review. J. Hazard. Mater. 2021, 405, 124631. [Google Scholar] [CrossRef]
- Liu, L.; Bilal, M.; Duan, X.; Iqbal, H.M.N. Mitigation of environmental pollution by genetically engineered bacteria—Current challenges and future perspectives. Sci. Total Environ. 2019, 667, 444–454. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.; Kuila, A. Bioremediation of heavy metals by microbial process. Environ. Technol. Innov. 2019, 14, 100369. [Google Scholar] [CrossRef]
- Azad, M.A.K.; Amin, L.; Sidik, N.M. Genetically engineered organisms for bioremediation of pollutants in contaminated sites. Chin. Sci. Bull. 2014, 59, 703–714. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, B.; Yu, Q. Genetic engineering-facilitated coassembly of synthetic bacterial cells and magnetic nanoparticles for efficient heavy metal removal. ACS Appl. Mater. Interfaces 2020, 12, 22948–22957. [Google Scholar] [CrossRef]
- Liu, J.; Zhu, N.; Zhang, Y.; Ren, T.; Shao, C.; Shi, R.; Li, X.; Ju, M.; Ma, T.; Yu, Q. Transcription profiling-guided remodeling of sulfur metabolism in synthetic bacteria for efficiently capturing heavy metals. J. Hazard. Mater. 2021, 403, 123638. [Google Scholar] [CrossRef]
- Wang, D.; Zheng, Y.; Xu, L.; Fan, X.; Wei, N.; Jin, N.; Huang, S.; Xiao, Q.; Wu, Z. Engineered cells for selective detection and remediation of Hg2+ based on transcription factor MerR regulated cell surface displayed systems. Biochem. Eng. J. 2019, 150, 107289. [Google Scholar] [CrossRef]
- Igiri, B.E.; Okoduwa, S.I.R.; Idoko, G.O.; Akabuogu, E.P.; Adeyi, A.O.; Ejiogu, I.K. Toxicity and bioremediation of heavy metals contaminated ecosystem from tannery wastewater: A review. J. Toxicol. 2018, 2018, 2568038. [Google Scholar] [CrossRef]
- Bazzi, W.; Abou Fayad, A.G.; Nasser, A.; Haraoui, L.P.; Dewachi, O.; Abou-Sitta, G.; Nguyen, V.K.; Abara, A.; Karah, N.; Landecker, H.; et al. Heavy metal toxicity in armed conflicts potentiates amr in A. baumannii by selecting for antibiotic and heavy metal co-resistance mechanisms. Front. Microbiol. 2020, 11, 68. [Google Scholar] [CrossRef]
- Mishra, J.; Singh, R.; Arora, N.K. Alleviation of heavy metal stress in plants and remediation of soil by rhizosphere microorganisms. Front. Microbiol. 2017, 8, 1706. [Google Scholar] [CrossRef]
- Yue, Z.B.; Li, Q.; Li, C.C.; Chen, T.H.; Wang, J. Component analysis and heavy metal adsorption ability of extracellular polymeric substances (EPS) from sulfate reducing bacteria. Bioresour. Technol. 2015, 194, 399–402. [Google Scholar] [CrossRef] [PubMed]
- Machuca, Á. Metal-chelating agents from ectomycorrhizal fungi and their biotechnological potential. In Diversity and Biotechnology of Ectomycorrhizae; Rai, M., Varma, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 347–369. ISBN 978-3-642-15196-5. [Google Scholar]
- Verma, S.; Bhatt, P.; Verma, A.; Mudila, H.; Prasher, P.; Rene, E.R. Microbial technologies for heavy metal remediation: Effect of process conditions and current practices. Clean Technol. Environ. Policy 2021, 1–23. [Google Scholar] [CrossRef]
- González-Chávez, M.C.; Carrillo-González, R.; Wright, S.F.; Nichols, K.A. The role of glomalin, a protein produced by arbuscular mycorrhizal fungi, in sequestering potentially toxic elements. Environ. Pollut. 2004, 130, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Lin, Z.; Pang, S.; Zhang, Y.; Bhatt, P.; Chen, S. Biosurfactant Is a powerful tool for the bioremediation of heavy metals from contaminated soils. J. Hazard. Mater. 2021, 418, 126253. [Google Scholar] [CrossRef] [PubMed]
- Lal, S.; Ratna, S.; Said, O.B.; Kumar, R. Biosurfactant and exopolysaccharide-assisted rhizobacterial technique for the remediation of heavy metal contaminated soil: An advancement in metal phytoremediation technology. Environ. Technol. Innov. 2018, 10, 243–263. [Google Scholar] [CrossRef]
- Ravindran, A.; Sajayan, A.; Priyadharshini, G.B.; Selvin, J.; Kiran, G.S. Revealing the efficacy of thermostable biosurfactant in heavy metal bioremediation and surface treatment in vegetables. Front. Microbiol. 2020, 11, 222. [Google Scholar] [CrossRef]
- Ayangbenro, A.S.; Babalola, O.O. Genomic analysis of Bacillus cereus NWUAB01 and its heavy metal removal from polluted soil. Sci. Rep. 2020, 10, 19660. [Google Scholar] [CrossRef]
- Ahuekwe, E.F.; Okoli, B.E.; Stanley, H.O.; Kinigoma, B. Evaluation of hydrocarbon emulsification and heavy metal detoxification potentials of sophorolipid biosurfactants produced from waste substrates using yeast and mushroom. In Proceedings of the SPE African Health, Safety, Security, Environment, and Social Responsibility Conference and Exhibition, Accra, Ghana, 4–6 October 2016; pp. 81–96. [Google Scholar] [CrossRef]
- Gomaa, E.Z.; El-Meihy, R.M. Bacterial biosurfactant from Citrobacter freundii MG812314.1 as a bioremoval tool of heavy metals from wastewater. Bull. Natl. Res. Cent. 2019, 43, 69. [Google Scholar] [CrossRef]
- Sun, G.L.; Reynolds, E.E.; Belcher, A.M. Using yeast to sustainably remediate and extract heavy metals from waste waters. Nat. Sustain. 2020, 3, 303–311. [Google Scholar] [CrossRef]
- Xu, Y.N.; Chen, Y. Advances in heavy metal removal by sulfate-reducing bacteria. Water Sci. Technol. 2020, 81, 1797–1827. [Google Scholar] [CrossRef]
- Choi, J.H.; Kim, I.H.; Kim, Y.K.; Oh, B.K. Effective bioremediation of cadmium (II), nickel (II), and chromium (VI) in a marine environment by using Desulfovibrio desulfuricans. Biotechnol. Bioprocess Eng. 2015, 20, 937–941. [Google Scholar] [CrossRef]
- Rana, K.; Rana, N.; Singh, B. Applications of sulfur oxidizing bacteria. In Physiological and Biotechnological Aspects of Extremophiles; Elsevier Inc.: Amsterdam, The Netherlands, 2020; pp. 131–136. ISBN 9780128183229. [Google Scholar]
- Wu, X.; Huang, P.; Dong, C.; Deng, X. Nickel bioaccumulation by a marine bacterium Brevibacterium sp. (X6) isolated from shenzhen bay, China. Mar. Pollut. Bull. 2021, 170, 112656. [Google Scholar] [CrossRef] [PubMed]
- Oves, M.; Khan, M.S.; Qari, H.A. Ensifer adhaerens for heavy metal bioaccumulation, biosorption, and phosphate solubilization under metal stress condition. J. Taiwan Inst. Chem. Eng. 2017, 80, 540–552. [Google Scholar] [CrossRef]
- Hansda, A.; Kumar, V.; Anshumali. A comparative review towards potential of microbial cells for heavy metal removal with emphasis on biosorption and bioaccumulation. World J. Microbiol. Biotechnol. 2016, 32, 170. [Google Scholar] [CrossRef]
- Danouche, M.; El Ghachtouli, N.; El Arroussi, H. Phycoremediation mechanisms of heavy metals using living green microalgae: Physicochemical and molecular approaches for enhancing selectivity and removal capacity. Heliyon 2021, 7, e07609. [Google Scholar] [CrossRef]
- Shahpiri, A.; Mohammadzadeh, A. Mercury Removal by Engineered Escherichia coli cells expressing different rice metallothionein isoforms. Ann. Microbiol. 2018, 68, 145–152. [Google Scholar] [CrossRef]
- Yu, Y.; Shi, K.; Li, X.; Luo, X.; Wang, M.; Li, L.; Wang, G.; Li, M. Reducing cadmium in rice using metallothionein surface-engineered bacteria WH16-1-MT. Environ. Res. 2022, 203, 111801. [Google Scholar] [CrossRef]
- Rehman, A.; Anjum, M.S. Multiple metal tolerance and biosorption of cadmium by Candida tropicalis isolated from industrial effluents: Glutathione as detoxifying agent. Environ. Monit. Assess. 2011, 174, 585–595. [Google Scholar] [CrossRef]
- Khullar, S.; Reddy, M.S. Arsenic toxicity and its mitigation in ectomycorrhizal fungus Hebeloma cylindrosporum through glutathione biosynthesis. Chemosphere 2020, 240, 124914. [Google Scholar] [CrossRef]
- Yong, X.; Chen, Y.; Liu, W.; Xu, L.; Zhou, J.; Wang, S.; Chen, P.; Ouyang, P.; Zheng, T. Enhanced cadmium resistance and accumulation in Pseudomonas putida KT2440 expressing the phytochelatin synthase gene of Schizosaccharomyces pombe. Lett. Appl. Microbiol. 2014, 58, 255–261. [Google Scholar] [CrossRef]
- Ahemad, M. Remediation of metalliferous soils through the heavy metal resistant plant growth promoting bacteria: Paradigms and prospects. Arab. J. Chem. 2019, 12, 1365–1377. [Google Scholar] [CrossRef]
- Sher, S.; Rehman, A. Use of heavy metals resistant bacteria—A strategy for arsenic bioremediation. Appl. Microbiol. Biotechnol. 2019, 103, 6007–6021. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Datta, S.; Chattyopadhyay, D.; Sarkar, P. Arsenic accumulating and transforming bacteria isolated from contaminated soil for potential use in bioremediation. J. Environ. Sci. Health—Part A Toxic/Hazard. Subst. Environ. Eng. 2011, 46, 1736–1747. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Adholeya, A. Detoxification and accumulation of chromium from tannery effluent and spent chrome effluent by Paecilomyces lilacinus Fungi. Int. Biodeterior. Biodegrad. 2011, 65, 309–317. [Google Scholar] [CrossRef]
- Narayani, M.; Shetty, K.V. Chromium-resistant bacteria and their environmental condition for hexavalent chromium removal: A review. Crit. Rev. Environ. Sci. Technol. 2013, 43, 955–1009. [Google Scholar] [CrossRef]
- Batool, R.; Yrjälä, K.; Hasnain, S. Hexavalent chromium reduction by bacteria from tannery effluent. J. Microbiol. Biotechnol. 2012, 22, 547–554. [Google Scholar] [CrossRef]
- Wiatrowski, H.A.; Ward, P.M.; Barkay, T. Novel reduction of mercury(II) by mercury-sensitive dissimilatory metal reducing bacteria. Environ. Sci. Technol. 2006, 40, 6690–6696. [Google Scholar] [CrossRef]
- Bolan, N.; Kunhikrishnan, A.; Thangarajan, R.; Kumpiene, J.; Park, J.; Makino, T.; Beth, M.; Scheckel, K. Remediation of heavy metal (loid)s contaminated soils—To mobilize or to immobilize? J. Hazard. Mater. 2014, 266, 141–166. [Google Scholar] [CrossRef]
- Di, X.; Beesley, L.; Zhang, Z.; Zhi, S.; Jia, Y.; Ding, Y. Microbial arsenic methylation in soil and uptake and metabolism of methylated arsenic in plants: A review. Int. J. Environ. Res. Public Health 2019, 16, 5012. [Google Scholar] [CrossRef]
- Verma, S.; Verma, P.K.; Chakrabarty, D. Arsenic bio-volatilization by engineered yeast promotes rice growth and reduces arsenic accumulation in grains. Int. J. Environ. Res. 2019, 13, 475–485. [Google Scholar] [CrossRef]
- Hu, H.; Lin, H.; Zheng, W.; Tomanicek, S.J.; Johs, A.; Feng, X.; Elias, D.A.; Liang, L.; Gu, B. Oxidation and methylation of dissolved elemental mercury by anaerobic bacteria. Nat. Geosci. 2013, 6, 751–754. [Google Scholar] [CrossRef]
- Soares Guimarães, L.H.; Segura, F.R.; Tonani, L.; Von-Zeska-Kress, M.R.; Rodrigues, J.L.; Calixto, L.A.; Silva, F.F.; Batista, B.L. Arsenic volatilization by Aspergillus sp. and Penicillium sp. isolated from rice rhizosphere as a promising eco-safe tool for arsenic mitigation. J. Environ. Manag. 2019, 237, 170–179. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, A.; Gupta, A.; Kaur, A.; Malik, D. Efficacy of Acinetobacter sp. B9 for simultaneous removal of phenol and hexavalent chromium from co-contaminated system. Appl. Microbiol. Biotechnol. 2014, 98, 9829–9841. [Google Scholar] [CrossRef] [PubMed]
- De, J.; Ramaiah, N.; Vardanyan, L. Detoxification of toxic heavy metals by marine bacteria highly resistant to mercury. Mar. Biotechnol. 2008, 10, 471–477. [Google Scholar] [CrossRef] [PubMed]
- Sharma, B.; Shukla, P. Lead bioaccumulation mediated by Bacillus cereus BPS-9 from an industrial waste contaminated site encoding heavy metal resistant genes and their transporters. J. Hazard. Mater. 2021, 401, 123285. [Google Scholar] [CrossRef]
- Chaturvedi, M.K. Studies on chromate removal by chromium-resistant Bacillus sp. isolated from tannery effluent. J. Environ. Prot. 2011, 2, 76–82. [Google Scholar] [CrossRef]
- Salehizadeh, H.; Shojaosadati, S.A. Removal of metal ions from aqueous solution by polysaccharide produced from Bacillus firmus. Water Res. 2003, 37, 4231–4235. [Google Scholar] [CrossRef]
- Muneer, B.; Iqbal, M.J.; Shakoori, F.R.; Shakoori, A.R. Tolerance and biosorption of mercury by microbial consortia: Potential use in bioremediation of wastewater. Pak. J. Zool. 2013, 45, 247–254. [Google Scholar]
- Fathima Benazir, J.; Suganthi, R.; Rajvel, D.; Padmini Pooja, M.; Mathithumilan, B. Bioremediation of Chromium in Tannery Effluent by Microbial Consortia. Afr. J. Biotechnol. 2010, 9, 3140–3143. [Google Scholar] [CrossRef]
- Mohapatra, R.K.; Parhi, P.K.; Pandey, S.; Bindhani, B.K.; Thatoi, H.; Panda, C.R. Active and passive biosorption of Pb(II) using live and dead biomass of marine bacterium Bacillus xiamenensis PbRPSD202: Kinetics and isotherm studies. J. Environ. Manag. 2019, 247, 121–134. [Google Scholar] [CrossRef]
- Bharagava, R.N.; Mishra, S. Hexavalent chromium reduction potential of Cellulosimicrobium sp. isolated from common effluent treatment plant of tannery industries. Ecotoxicol. Environ. Saf. 2018, 147, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.H.; Choi, J.H.; Joo, J.O.; Kim, Y.K.; Choi, J.W.; Oh, B.K. Development of a microbe-zeolite carrier for the effective elimination of heavy metals from seawater. J. Microbiol. Biotechnol. 2015, 25, 1542–1546. [Google Scholar] [CrossRef] [PubMed]
- Congeevaram, S.; Dhanarani, S.; Park, J.; Dexilin, M.; Thamaraiselvi, K. Biosorption of chromium and nickel by heavy metal resistant fungal and bacterial isolates. J. Hazard. Mater. 2007, 146, 270–277. [Google Scholar] [CrossRef]
- Juwarkar, A.A.; Nair, A.; Dubey, K.V.; Singh, S.K.; Devotta, S. Biosurfactant technology for remediation of cadmium and lead contaminated soils. Chemosphere 2007, 68, 1996–2002. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Wang, B.; Cai, Q.T.; Li, X.X.; Liu, M.; Hu, D.; Guo, D.B.; Wang, J.; Fan, C. Bioremediation of hexavalent chromium pollution by Sporosarcina saromensis M52 isolated from offshore sediments in Xiamen, China. Biomed. Environ. Sci. 2016, 29, 127–136. [Google Scholar] [CrossRef]
- Sedlakova-Kadukova, J.; Kopcakova, A.; Gresakova, L.; Godany, A.; Pristas, P. Bioaccumulation and biosorption of zinc by a novel Streptomyces K11 strain isolated from highly alkaline aluminium brown mud disposal site. Ecotoxicol. Environ. Saf. 2019, 167, 204–211. [Google Scholar] [CrossRef]
- Luna, J.M.; Rufino, R.D.; Sarubbo, L.A. Biosurfactant from Candida sphaerica UCP0995 exhibiting heavy metal remediation properties. Process Saf. Environ. Prot. 2016, 102, 558–566. [Google Scholar] [CrossRef]
- Basak, G.; Das, N. Characterization of sophorolipid biosurfactant produced by Cryptococcus sp. VITGBN2 and its application on Zn(II) removal from electroplating wastewater. J. Environ. Biol. 2014, 35, 1087–1094. [Google Scholar]
- Chatterjee, S.; Chatterjee, N.C.; Dutta, S. Bioreduction of chromium (VI) to chromium (III) by a novel yeast strain Rhodotorula mucilaginosa (MTCC 9315). Afr. J. Biotechnol. 2012, 11, 14920–14929. [Google Scholar] [CrossRef]
- Bano, A.; Hussain, J.; Akbar, A.; Mehmood, K.; Anwar, M.; Hasni, M.S.; Ullah, S.; Sajid, S.; Ali, I. Biosorption of heavy metals by obligate halophilic fungi. Chemosphere 2018, 199, 218–222. [Google Scholar] [CrossRef]
- Amini, M.; Younesi, H.; Bahramifar, N. Biosorption of nickel(II) from aqueous solution by Aspergillus niger: Response surface methodology and isotherm study. Chemosphere 2009, 75, 1483–1491. [Google Scholar] [CrossRef] [PubMed]
- Taştan, B.E.; Ertuǧrul, S.; Dönmez, G. Effective bioremoval of reactive dye and heavy metals by Aspergillus versicolor. Bioresour. Technol. 2010, 101, 870–876. [Google Scholar] [CrossRef] [PubMed]
- Noormohamadi, H.R.; Fat’hi, M.R.; Ghaedi, M.; Ghezelbash, G.R. Potentiality of white-rot fungi in biosorption of nickel and cadmium: Modeling optimization and kinetics study. Chemosphere 2019, 216, 124–130. [Google Scholar] [CrossRef] [PubMed]
- Abd El-Hameed, M.M.; Abuarab, M.E.; Abdel Mottaleb, S.; El-Bahbohy, R.M.; Bakeer, G.A. Comparative studies on growth and Pb(II) removal from aqueous solution by Nostoc muscorum and Anabaena variabilis. Ecotoxicol. Environ. Saf. 2018, 165, 637–644. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Saky, S.A.; Yang, Z.; Ho, S.H.; Chen, C.; Qin, L.; Zhang, G.; Wang, Y.; Lu, Y. The critical utilization of active heterotrophic microalgae for bioremoval of Cr(VI) in organics co-contaminated wastewater. Chemosphere 2019, 228, 536–544. [Google Scholar] [CrossRef]
- Chandrashekharaiah, P.; Debanjan, S.; Santanu, D.; Avishek, B. Cadmium biosorption and biomass production by two freshwater microalgae Scenedesmus acutus and Chlorella pyrenoidosa: An integrated approach. Chemosphere 2021, 269, 128755. [Google Scholar] [CrossRef]
- Pradhan, D.; Sukla, L.B.; Mishra, B.B.; Devi, N. Biosorption for removal of hexavalent chromium using microalgae Scenedesmus sp. J. Clean. Prod. 2019, 209, 617–629. [Google Scholar] [CrossRef]
- Mane, P.C.; Bhosle, A.B. Bioremoval of some metals by living algae Spirogyra sp. and Spirullina sp. from aqueous solution. Int. J. Environ. Res. 2012, 6, 571–576. [Google Scholar] [CrossRef]
- Cepoi, L.; Zinicovscaia, I.; Rudi, L.; Chiriac, T.; Miscu, V.; Djur, S.; Strelkova, L.; Grozdov, D. Spirulina platensis as renewable accumulator for heavy metals accumulation from multi-element synthetic effluents. Environ. Sci. Pollut. Res. 2020, 27, 31793–31811. [Google Scholar] [CrossRef]
- Xie, Y.; Fan, J.; Zhu, W.; Amombo, E.; Lou, Y.; Chen, L.; Fu, J. Effect of heavy metals pollution on soil microbial diversity and bermudagrass genetic variation. Front. Plant Sci. 2016, 7, 755. [Google Scholar] [CrossRef]
- Fajardo, C.; Costa, G.; Nande, M.; Botías, P.; García-Cantalejo, J.; Martín, M. Pb, Cd, and Zn soil contamination: Monitoring functional and structural impacts on the microbiome. Appl. Soil Ecol. 2019, 135, 56–64. [Google Scholar] [CrossRef]
- Li, X.; Meng, D.; Li, J.; Yin, H.; Liu, H.; Liu, X.; Cheng, C.; Xiao, Y.; Liu, Z.; Yan, M. Response of soil microbial communities and microbial interactions to long-term heavy metal contamination. Environ. Pollut. 2017, 231, 908–917. [Google Scholar] [CrossRef] [PubMed]
- Qiao, L.; Liu, X.; Zhang, S.; Zhang, L.; Li, X.; Hu, X.; Zhao, Q.; Wang, Q.; Yu, C. Distribution of the microbial community and antibiotic resistance genes in farmland surrounding gold tailings: A metagenomics approach. Sci. Total Environ. 2021, 779, 146502. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Shen, Q.; Wang, L.; Qiu, G.; Shi, J.; Xu, J.; Brookes, P.C.; Liu, X. Effects of Cd, Cu, Zn and their combined action on microbial biomass and bacterial community structure. Environ. Pollut. 2018, 243, 510–518. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Yang, G.; Hou, S.; Zhang, T.; Li, Z.; Liang, F. Distribution of ARGs and MGEs among glacial soil, permafrost, and sediment using metagenomic analysis. Environ. Pollut. 2018, 234, 339–346. [Google Scholar] [CrossRef]
- Feng, G.; Xie, T.; Wang, X.; Bai, J.; Tang, L.; Zhao, H.; Wei, W.; Wang, M.; Zhao, Y. Metagenomic Analysis of Microbial Community and Function Involved in Cd-Contaminated Soil. BMC Microbiol. 2018, 18, 11. [Google Scholar] [CrossRef]
- Jiang, X.; Liu, W.; Xu, H.; Cui, X.; Li, J.; Chen, J.; Zheng, B. Characterizations of heavy metal contamination, microbial community, and resistance genes in a tailing of the largest copper mine in China. Environ. Pollut. 2021, 280, 116947. [Google Scholar] [CrossRef]
- Hemmat-Jou, M.H.; Safari-Sinegani, A.A.; Mirzaie-Asl, A.; Tahmourespour, A. Analysis of microbial communities in heavy metals-contaminated soils using the metagenomic approach. Ecotoxicology 2018, 27, 1281–1291. [Google Scholar] [CrossRef]
- Misson, B.; Garnier, C.; Lauga, B.; Dang, D.H.; Ghiglione, J.F.; Mullot, J.U.; Duran, R.; Pringault, O. Chemical multi-contamination drives benthic prokaryotic diversity in the anthropized Toulon Bay. Sci. Total Environ. 2016, 556, 319–329. [Google Scholar] [CrossRef]
- Li, J.; Hu, H.W.; Ma, Y.B.; Wang, J.T.; Liu, Y.R.; He, J.Z. Long-term nickel exposure altered the bacterial community composition but not diversity in two contrasting agricultural soils. Environ. Sci. Pollut. Res. 2015, 22, 10496–10505. [Google Scholar] [CrossRef]
- Huang, C.C.; Liang, C.M.; Yang, T.I.; Chen, J.L.; Wang, W.K. Shift of bacterial communities in heavy metal-contaminated agricultural land during a remediation process. PLoS ONE 2021, 16, e0255137. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Yu, F.; Sun, X.; Wu, S.; Li, X.; Liu, T.; Li, Y. Long term effects of Lespedeza bicolor revegetation on soil bacterial communities in Dexing copper mine tailings in Jiangxi Province, China. Appl. Soil Ecol. 2018, 125, 192–201. [Google Scholar] [CrossRef]
- Li, Y.; Chen, L.; Wen, H. Changes in the composition and diversity of bacterial communities 13 years after soil reclamation of abandoned mine land in eastern china. Ecol. Res. 2015, 30, 357–366. [Google Scholar] [CrossRef]
- Moynahan, O.S.; Zabinski, C.A.; Gannon, J.E. Microbial community structure and carbon-utilization diversity in a mine tailings revegetation study. Restor. Ecol. 2002, 10, 77–87. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, W.; Liu, S.; Zhang, P.; Ye, C.; Liang, H. Revegetation of a barren rare earth mine using native plant species in reciprocal plantation: Effect of phytoremediation on soil microbiological communities. Environ. Sci. Pollut. Res. 2020, 27, 2107–2119. [Google Scholar] [CrossRef]
- Chen, Y.X.; Wang, Y.P.; Wu, W.X.; Lin, Q.; Xue, S.G. Impacts of chelate-assisted phytoremediation on microbial community composition in the rhizosphere of a copper accumulator and non-accumulator. Sci. Total Environ. 2006, 356, 247–255. [Google Scholar] [CrossRef]
- Liu, S.; Liu, W.; Yang, M.; Zhou, L.; Liang, H. The genetic diversity of soil bacteria affected by phytoremediation in a typical barren rare earth mined site of south china. Springer Plus 2016, 5, 1131. [Google Scholar] [CrossRef][Green Version]
- Wei, Z.; Hao, Z.; Li, X.; Guan, Z.; Cai, Y.; Liao, X. The effects of phytoremediation on soil bacterial communities in an abandoned mine site of rare earth elements. Sci. Total Environ. 2019, 670, 950–960. [Google Scholar] [CrossRef]
- Gupta, G.S.; Yadav, G.; Tiwari, S. Restoration of Wetland Ecosystem: A Trajectory Towards a Sustainable Environment; Upadhyay, A.K., Singh, R., Singh, D.P., Eds.; Springer: Singapore, 2020; pp. 195–226. ISBN 9789811376658. [Google Scholar]
- Sayqal, A.; Ahmed, O.B. Advances in Heavy Metal Bioremediation: An Overview. Appl. Bionics Biomech. 2021, 2021, 1609149. [Google Scholar] [CrossRef]
- Abatenh, E.; Gizaw, B.; Tsegaye, Z.; Wassie, M. Application of microorganisms in bioremediation-review. J. Environ. Microbiol. 2017, 1, 2–9. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, F.; Chen, J.; Sun, G. Arsenic removal from contaminated soil via biovolatilization by genetically engineered bacteria under laboratory conditions. J. Environ. Sci. 2011, 23, 1544–1550. [Google Scholar] [CrossRef]
- Bruneel, O.; Mghazli, N.; Sbabou, L.; Héry, M.; Casiot, C.; Filali-Maltouf, A. Role of microorganisms in rehabilitation of mining sites, focus on sub-Saharan African countries. J. Geochem. Explor. 2019, 205, 106327. [Google Scholar] [CrossRef]


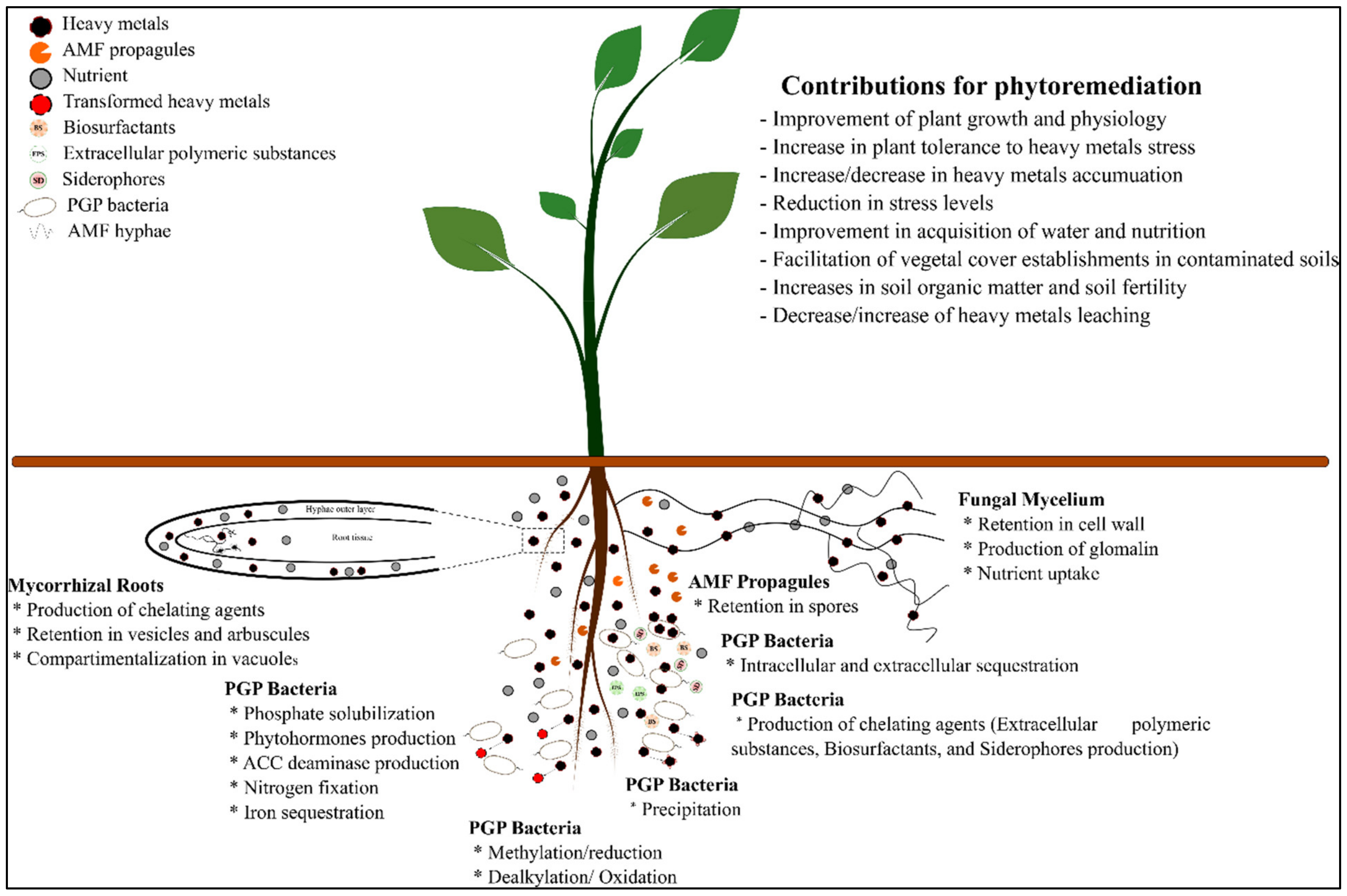
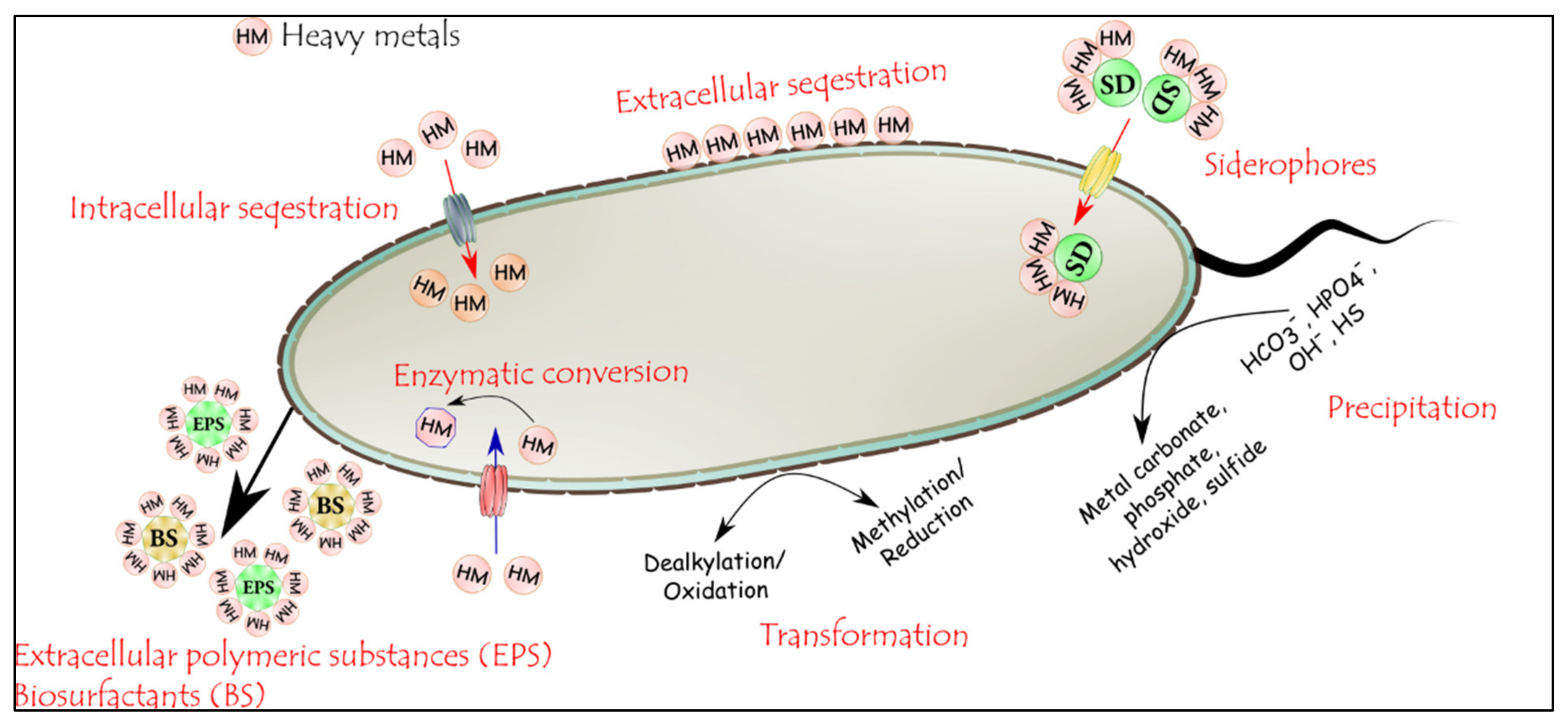
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raklami, A.; Meddich, A.; Oufdou, K.; Baslam, M. Plants—Microorganisms-Based Bioremediation for Heavy Metal Cleanup: Recent Developments, Phytoremediation Techniques, Regulation Mechanisms, and Molecular Responses. Int. J. Mol. Sci. 2022, 23, 5031. https://doi.org/10.3390/ijms23095031
Raklami A, Meddich A, Oufdou K, Baslam M. Plants—Microorganisms-Based Bioremediation for Heavy Metal Cleanup: Recent Developments, Phytoremediation Techniques, Regulation Mechanisms, and Molecular Responses. International Journal of Molecular Sciences. 2022; 23(9):5031. https://doi.org/10.3390/ijms23095031
Chicago/Turabian StyleRaklami, Anas, Abdelilah Meddich, Khalid Oufdou, and Marouane Baslam. 2022. "Plants—Microorganisms-Based Bioremediation for Heavy Metal Cleanup: Recent Developments, Phytoremediation Techniques, Regulation Mechanisms, and Molecular Responses" International Journal of Molecular Sciences 23, no. 9: 5031. https://doi.org/10.3390/ijms23095031
APA StyleRaklami, A., Meddich, A., Oufdou, K., & Baslam, M. (2022). Plants—Microorganisms-Based Bioremediation for Heavy Metal Cleanup: Recent Developments, Phytoremediation Techniques, Regulation Mechanisms, and Molecular Responses. International Journal of Molecular Sciences, 23(9), 5031. https://doi.org/10.3390/ijms23095031







